Abstract
Previous studies from our laboratory had indicated that cytochrome c-independent processing and activation of caspase-9 by caspase-8 contributed to early amplification of the caspase cascade in tumor necrosis factor (TNF)-α-treated murine cells. Here we show that murine caspase-9 is phosphorylated by casein kinase 2 (CK2) on a serine near the site of caspase-8 cleavage. CK2 has been shown to regulate cleavage of the pro-apoptotic Bid protein by phosphorylating serine residues near its caspase-8 cleavage site. Similarly, CK2 modification of Ser348 on caspase-9 appears to render the protease refractory to cleavage by active caspase-8. This phosphorylation did not affect the ability of caspase-9 to autoprocess. Substitution of Ser348 abolished phosphorylation but not cleavage, and a phospho-site mutant promoted apoptosis in TNF-α-treated caspase-9 knock-out mouse embryo fibroblasts. Furthermore, inhibition of CK2 activity and RNA interference-mediated knockdown of the kinase accelerated caspase-9 activation, whereas phosphatase inhibition delayed both caspase-9 activation and death in response to TNF receptor occupation. Taken together, these studies show that TNF receptor cross-linking promotes dephosphorylation of caspase-9, rendering it susceptible to processing by activated caspase-8 protein. Thus, our data suggest that modification of procaspase-9 to protect it from inappropriate cleavage and activation is yet another mechanism by which the oncogenic kinase CK2 promotes survival.
Activation of mammalian initiator caspase-9 in intrinsic apoptotic pathways occurs by “induced proximity” or dimerization and is dependent on mitochondrial cyt c2 release (1, 2). However, cyt c-independent mechanisms of activation of this caspase have also been demonstrated. Caspase-12 was able to process and activate caspase-9 in response to endoplasmic reticulum stress, whereas Sendai virus infection triggered a novel pathway of caspase-9 cleavage (3–5). Furthermore, early amplification of the caspase cascade in response to TNF receptor cross-linking on Bcl-xL-expressing murine pro B lymphoma cells, in which the mitochondrial amplification loop is essentially unavailable, occurred via cyt c-independent activation of caspase-9 (6). This study did not rule out dimerizaton of caspase-9 in this pathway but implicated active caspase-8 in its processing and activation. The involvement of caspase-8 in apoptosome-independent activation of caspase-9 in murine cells has recently been demonstrated in another study (7). The observation that caspase-8 could directly activate an initiator caspase of the intrinsic pathway without the involvement of mitochondria in death receptor-induced apoptosis suggested that some murine cells harbor both Type I and II characteristics (8) and implied that additional regulatory mechanisms could protect caspase-9 from inappropriate activation.
Both human and murine caspase-9 harbor a number of serines, threonines, and tyrosines, and a variety of kinases including ERK2, protein kinase A, protein kinase Cζ, and c-Abl tyrosine kinase have been implicated in regulating the activation of this protease (9–12). Phosphorylation of Ser196 on human caspase-9 by the oncogenic kinase Akt is believed to regulate its apoptotic activity, although the murine form lacks this serine (13, 14). Protein kinase CK2 (formerly casein kinase II), is a serine/threonine kinase that affects several signaling pathways (15, 16). Unlike the majority of protein kinases, CK2 is constitutively active and independent of second messengers or phosphorylation events. It phosphorylates a variety of substrates involved in cell cycle control, differentiation and proliferation (17, 18). It is a highly conserved, ubiquitous, and pleiotropic protein kinase comprising two catalytic subunits, α and/or α′, and two regulatory (β) subunits that form a heterotetrameric holoenzyme (19). The high constitutive activity of CK2 in cancer cells is believed to underlie its oncogenic potential (15, 16, 20). CK2 recognizes phospho-acceptor sites that are specified by clusters of acidic residues, with the one at position n+3 relative to the target amino acid playing a particularly crucial role (21). The frequency of aspartyl and glutamyl residues in its phosphoacceptor sites suggested that phosphorylation by CK2 could regulate caspase cleavage, which is known to occur at the carboxyl terminus of the acidic consensus E/DXD (17).
In the present study we show that CK2 modifies a serine in the vicinity of the primary processing site of murine caspase-9 in proliferating cells, protecting the protease from cleavage and activation by other caspases.
EXPERIMENTAL PROCEDURES
Cell Lines, Antibodies, and Plasmid Constructs—FL5.12 murine pro B lymphoma cells were cultured as described earlier (22). The generation of FL5.12-Neo and FL5.12-Bcl-xL cells has also been described previously (23). Electroporation of FL5.12 Bcl-xL cells with pGIPZ shRNAmir vectors was carried out under similar conditions, and transfectants were selected in the presence of 2 μg/ml puromycin. Caspase-9-/- MEFs (a gift from Richard Flavell, Yale University) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mm glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin sulfate, and 10 mm β–mercaptoethanol. Caspase-9 monoclonal antibodies were purchased from Stressgen Biotechnologies and NeoMarkers, Inc., anti-FLAG M2 antibody from Sigma, anti-actin monoclonal from EMD Biosciences, anti-CK2 from Upstate/Millipore, and anti-vinculin from Abcam, Inc. The caspase-8 antibody was a gift from Idun Pharmaceuticals, and polyclonal phosphoserine-specific antibodies were from Qiagen. Active recombinant caspase-3 and caspase-8 were purchased from Pharmingen. The pGIPZ shR-NAmir CK2 vectors and nonsilencing control were purchased from Open Biosystems (CK2-α clone identification code V2LMM_188343 and CK2-β clone identification code V2LMM_66668). Construction of full-length murine caspase-9 (mC-9) cDNA and the Myc-tagged version, mC-9myc has been previously described (6). Point mutants of mC-9, substituting Ser348 with alanine, glycine, or valine or Ser350 with alanine, were generated by a two-step PCR method. Inserts were ligated to BamHI and XhoI sites of the pcmyc vector (6). The FLAG-tagged murine caspase-9 construct was generated by inserting a HindIII/XhoI fragment containing full-length murine caspase-9 into HindIII/SalI sites of the pFLAG-CTC vector (Sigma).
In Vitro Caspase Cleavage Assay—Radiolabeled mC-9 and mutant proteins were synthesized in vitro using the TnT T7 transcription/translation system (Promega Biotech) and 15 μCi of [35S]methionine (Amersham Biosciences) per reaction. Translated proteins were cleaved at 37 °C for 60 min using 5 μl of in vitro translation mix as substrate in the presence of active human recombinant caspase-3 (15 milliunits/μl), or caspase-8 (150 milliunits/μl), in a 25 μl of total reaction volume. The reactions were attenuated with an equal volume of 2× Laemmli sample buffer containing reducing agent and resolved by SDS-PAGE, and the gels were fixed, dried, and autoradiographed.
Purification of Recombinant FLAG-tagged Murine Caspase-9 Protein—The recombinant plasmid, pFLAG-CTC/mC-9 was transformed into an expression host Escherichia coli BL21(DE3)pLysS (Novagen). The cell pellet from 250 ml of induced culture was lysed in 10 ml of lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100) containing protease inhibitor mixture, by a combination of freeze-thaw cycles and sonication, and the centrifuged supernatant was further clarified through a 0.22-μm filter (Millipore). The clarified supernatant was passed over an anti-FLAG M2 agarose affinity gel (Sigma) column several times, the column was then washed with 10–20 column volumes of Tris-buffered saline (50 mm Tris-HCl, pH 7.4, 150 mm NaCl), and bound fusion protein was eluted with six 1-ml aliquots of 0.1 m glycine, pH 3.5, into tubes containing 15–25 μl of 1 m Tris-HCl, pH 8.0. Eluted fractions were concentrated using Microcon YM-30 columns (Millipore).
In Vitro Kinase Assays—In vitro translated mC-9 protein or its point mutants were immunoprecipitated with either anti-Myc (Santa Cruz) or caspase-9 (NeoMarkers) antibodies and incubated at 30 °C for 1 h in kinase buffer (50 mm Tris/HCl, pH 7.5, 10 mm MgCl2, 1 mm dithiothreitol, 10 mm β-glycerophosphate, 10 mm ATP, and 5 μCi of [γ-32P]ATP) with or without 0.05 milliunit purified CK2, in the presence or absence of inhibitors, DRB (200 μm) heparin (28 μg/ml), and apigenin (400 μm). Kinase reactions were attenuated in Laemmli buffer and resolved by SDS-PAGE, and the gels were fixed, dried, and autoradiographed. For kinase assays with untagged human and mouse caspase-9, in vitro translated products were immunoprecipitated with caspase-9 antibody prior to the kinase reaction. FL5.12 cell lysates (5 μg of protein), prepared as described by Desagher et al. (25), were incubated with 5 μg of purified recombinant mC-9-FLAG for 20 min at 30 °C in 50 μl of kinase buffer containing 5 μCi of [γ-32P]ATP with or without inhibitors. The inhibitors used were as follows: 60 μm emodin, 400 μm apigenin, 1 μm wortmannin, 100 μm cold GTP, or 7 μg/ml heparin. The protein was then immunoprecipitated with an anti-FLAG antibody, resolved by SDS-PAGE, and visualized by autoradiography.
To determine caspase-9 autoprocessing in a cell-free system, 5 μl of a 25-μl cold kinase reaction were incubated alone, with caspase-9-/- cytosolic S100 extract (2) (50 μg) or with extract, plus 1 mm ATP and 0.2 μg of cyt c in Buffer A (20 mm Hepes-KOH, pH 7.5, 10 mm KCl, 1.5 mm MgCl2, 1 mm sodium EDTA, 1 mm sodium EGTA, 1 mm dithiothreitol, and 0.1 mm phenylmethylsulfonyl fluoride) in a total volume of 35 μl. The reactions were carried out for 30 min at 30 °C attenuated with Laemmli Buffer and Western blotted using anti-caspase-9 antibody (Stressgen). Phosphorylation of caspase-9 was confirmed in Western blots using anti-phosphoserine antibodies (Qiagen).
Cell Viability Assays—FL5.12 cells were harvested at regular intervals following the addition of TNF-α (5 ng/ml) and CHX (20 μg/ml) to the medium, and viability was determined by flow cytometric analysis of annexin V-fluorescein isothiocyanate and propidium iodide uptake. Inhibitors such as okadaic acid (1 μm), DRB (50 μm), or TBB (25 μm) were added 30 min to 3 h prior to the addition of TNF-α as indicated. Viability of shRNA transfectants was determined 24 h after withdrawal from IL-3-containing medium or exposure to 100 μm etoposide.
Western Blotting, Metabolic Labeling, and Immunoprecipitation—Cell pellets were washed in phosphate-buffered saline and lysed in radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with protease inhibitors. Lysates were centrifuged at 14,000 × g to remove cellular debris. For Western blotting experiments, equal amounts of protein were resolved by SDS-PAGE and transferred to nitrocellulose. Membrane blocking, washing, primary and secondary antibody incubations, and chemiluminescent detection were carried out according to the manufacturer's instructions. The blots were stripped for reuse by washing for 30 min to 2 h in TBS-T buffer (pH 2.5) at room temperature.
For the metabolic labeling experiments, FL5.12 cells were incubated with 0.5 mCi of [32P]orthophosphate (Amersham Biosciences) in phosphate-free medium in the presence or absence of DRB (50 μm) for 6 h at 37 °C. For apoptosis induction experiments, TNF-α/CHX or CHX alone were added to the culture medium 3 h following the addition of orthophosphate, and cells were incubated for an additional 3 h prior to harvesting. The cell pellets were washed in phosphate-buffered saline and lysed in NET-I buffer (100 mm NaCl, 1 mm EDTA, 20 mm Tris, pH 8.0, and 0.1% Igepal) supplemented with protease inhibitor mixture (Calbiochem). Immunoprecipitated complexes were washed in NET-I and eluted directly into gel loading buffer. Eluted proteins were resolved by SDS-PAGE, transferred to nitrocellulose, and first visualized by autoradiography. Total immunoprecipitated full-length and cleaved caspase-9 was detected by immunoblotting the membrane with the antibody against caspase-9 (Stressgen).
Caspase Activity Assays—The cells (5 × 106) were harvested following treatment with TNF-α (5 ng/ml) and CHX (20 μg/ml) for the time indicated, and the phosphate-buffered saline washed cell pellets were flash-frozen. Later, the pellets were thawed on ice for 10 min in 160 μl of lysis buffer with intermittent vortexing, centrifuged for 1 min at 10,600 × g, and tested for caspase activity in a colorimetric assay using LEHD-pNA (R & D Systems) as substrate. The reactions were allowed to develop for 2 h at 37 °C, and absorbance was measured at 405 nm.
Immunofluorescence Microscopy—Caspase-9 null mouse embryo fibroblasts were grown to 50% confluency in 8-chamber culture slides (BD Falcon). The cells were transfected with 0.5 μg of pCDNA 3.1, m-C-9 myc, or mutants LDAD (Ser348 to Ala) or SEPA (Asp353 to Ala), using 1 μl of Lipofectamine (Invitrogen) per well in serum-free medium. After ∼48 h the cells were fixed in 3.7% paraformaldehyde and permeabilized with ice cold methanol:acetone (1:1). After blocking in 1% bovine serum albumin, the permeabilized cells were incubated with anti-caspase-9 antibody for 1 h, then washed, and incubated with Alexa 488-tagged goat anti-mouse IgG (Invitrogen). The mounting medium (Vectastain) contained 4′,6′-diamino-2-phenylindole for visualizing nuclei. Fluorescence was visualized simultaneously with bright field optics using a FluoView 1000 Olympus multiphoton microscope.
RESULTS
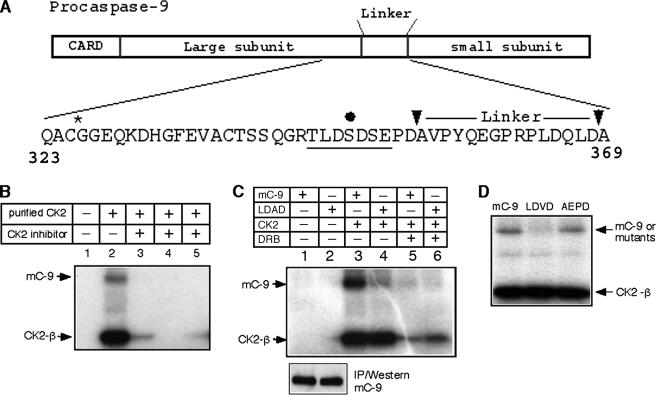
CK2 Phosphorylates Murine Caspase-9 on Ser348 in Vitro—Previous studies from our laboratory had determined that activated caspase-9 contributed to early amplification of the death receptor pathway in type II murine cells (6), independent of cyt c release. This likely involved cleavage at the auto-processing motif, SEPD (residues 350–353 in murine caspase-9; Fig. 1A), which serves as the primary substrate site for processing by active caspase-8 in vitro. We investigated the possibility that regulatory mechanisms were in place to prevent the inappropriate activation of this caspase in healthy proliferating cells.
FIGURE 1.
Purified recombinant CK2 phosphorylates murine caspase-9 at Ser348in vitro. A, amino acid sequence in murine caspase-9 between the active site, QACGG (*), and the caspase-3 processing site. Arrows show the auto-processing and primary caspase-8 cleavage site, SEPD, and the caspase-3 site, DQLD. The filled black circle shows serine 348 predicted to be the target residue within a strong CK2 consensus motif (underlined). B–D, in vitro translated mC-9 protein was immunoprecipitated with anti-caspase-9 antibody and incubated in kinase buffer in the presence or absence of purified CK2. B, phosphorylation of mC-9 in the presence of CK2 inhibitors (lanes 3–5): DRB, heparin, and apigenin. C, top panel, CK2 phosphorylation of mC-9 and the LDAD mutant in the presence or absence of DRB in vitro. Lower panel, immunoprecipitation (IP)/Western of cold in vitro translated mC-9 and LDAD. D, CK2 phosphorylation of mC-9 and mutants AEPD and LDVD in vitro.
A scan of the sequence using NetPhos, a sequence- and structure-based prediction program of eukaryotic protein phosphorylation sites (24), revealed the first serine residue, Ser348, within the motif TLDSDSE in murine caspase-9, CK2 (Fig. 1A). CK2 is known to phosphorylate Bid on serine residues in the vicinity of the caspase-8 recognition site, rendering it resistant to cleavage by active caspase-8 in the absence of death receptor trimerization (25). In vitro kinase reactions using purified recombinant CK2 and in vitro translated murine caspase-9 as substrate (Fig. 1B) showed that caspase-9 was indeed a CK2 target. This phosphorylation was diminished in the presence of CK2 inhibitors. Auto-phosphorylation of the CK2 β-subunit (26) was also markedly reduced in presence of the inhibitors. Fig. 1C shows that phosphorylation was reduced at least 4-fold in the LDAD mutant (Ser348 to Ala), relative to wild type, whereas auto-phosphorylation of the CK2 β-subunit in these reactions was unchanged in the absence of the inhibitor, DRB. The immunoprecipitation/Western in the lower panel indicates that the reduced levels of phosphorylation of the LDAD mutant are not the result of decreased translation relative to caspase-9 or a failure of the immunoprecipitating caspase-9 antibody to recognize the mutant protein. The kinase assay in Fig. 1D, using the AEPD (Ser350 to Ala) mutant and an additional mutant LDVD (Ser348 to Val), demonstrated that, at least in vitro, Ser350 was not a CK2 target. We had previously shown that activated caspase-8 could process caspase-9 efficiently at the SEPD motif (Ref. 6; Fig. 1A, left arrowhead). We tested the sensitivity of some of the serine mutants in cleavage assays to determine whether Ser348 affects the ability of active caspase-8 to process caspase-9 at Asp353 in vitro. Supplemental Fig. S1 shows that S348G (LDGD), S348V (LDVD), S348A (or LDAD, not shown) mutants, as well as S350A (AEPD) are all cleaved efficiently by the active initiator caspase.
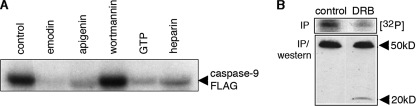
Murine Caspase-9 Is Phosphorylated by Endogenous CK2—Having established that Ser348 within the LDSD motif could be efficiently phosphorylated by purified recombinant CK2 enzyme in vitro, we determined whether endogenous CK2 phosphorylated the caspase-9 protein. Lysates prepared from FL5.12 pro B lymphoma cells were incubated with purified recombinant mC-9-FLAG in the absence or presence of inhibitors and immunoprecipitated with an anti-FLAG antibody. Caspase-9 phosphorylation was unaffected by the phosphatidylinositol 3-kinase inhibitor, wortmannin, whereas specific inhibitors of CK2, such as emodin, apigenin, and GTP (27), as well as heparin, which exhibits a broader specificity, all inhibited transfer of phosphate from [γ-32P]ATP to the purified recombinant protein, suggesting that CK2 was the endogenous kinase. To determine whether endogenous procaspase-9 could be phosphorylated by CK2, FL5.12 pro B lymphoma cells were incubated in the presence or absence of the CK2 inhibitor DRB (50 μm) and labeled with [32P]orthophosphate. Significant reduction in caspase-9 phosphorylation observed in the presence of DRB indicates that the protein is also a substrate for CK2 in vivo. The presence of the 20-kDa fragment in the immunoprecipitation/Western of DRB-treated lysates may suggest an increased susceptibility of the hypophosphorylated caspase to processing.
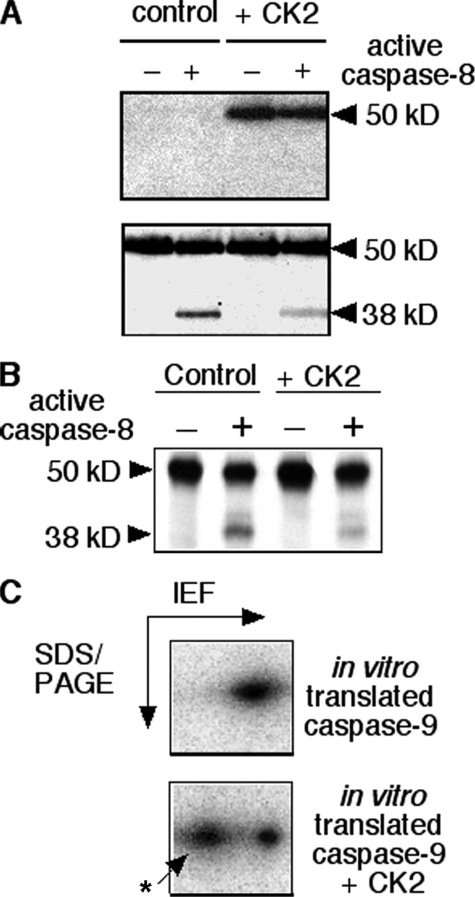
CK2-mediated Phosphorylation of Ser348 on Caspase-9 Interferes with Its Cleavage by Caspase-8—Caspase-9 is a tightly regulated initiator caspase in mammalian cells. It is auto-activated by induced proximity or dimerization following the release of mitochondrial cyt c and the assembly of the apoptosome in the cytosol (1, 2, 18, 28). However, a number of groups have determined that cyt c-independent mechanisms could also contribute to activation of this caspase (3, 5, 6). Previous studies in our laboratory had suggested that initiator caspase-8 cleaves and activates murine caspase-9 early during TNF receptor-induced apoptosis in murine cells. It was possible that the presence of a modified Ser348 on caspase-9 proximal to its cleavage site (see Fig. 1) inhibited the cleavage in a manner similar to that observed with pro-apoptotic protein, Bid (25). To test this possibility, purified recombinant mC-9-FLAG was incubated in kinase buffer containing [γ-32P]ATP in the presence or absence of recombinant CK2 and then subjected to a caspase-8 cleavage assay as described under “Experimental Procedures” and in the legends to Figs. 1 and 2. A representative experiment is shown in Fig. 3A. The top panel is an autoradiograph showing caspase-9 that had been phosphorylated by CK2 in vitro, and the lower panel is the same membrane showing caspase-9 and its cleavage products detected by chemiluminescence. Phosphorylation clearly resulted in a reduction in the levels of the 38-kDa cleavage product.
FIGURE 2.
Phosphorylation of recombinant and endogenous caspase-9 is sensitive to inhibition by CK2-specific inhibitors. A, lysates from exponentially growing FL5.12 cells were incubated with purified, recombinant caspase-9-FLAG in kinase buffer containing [γ-32P]ATP with or without inhibitors, emodin, apigenin, wortmannin, cold GTP, or heparin. Caspase-9-FLAG was immunoprecipitated with an anti-FLAG antibody, resolved by SDS-PAGE, and visualized by autoradiography. B, FL5.12 Bcl-xL cells were cultured in the presence or absence of 50 μm DRB and labeled with [32P]orthophosphate at 37 °C for 4 h prior to harvesting. Caspase-9 was immunoprecipitated from harvested cells and visualized, first by autoradiography (top panel) and then by Western blotting (bottom panel).
FIGURE 3.
Phosphorylation of murine caspase-9 CK2 decreases its susceptibility to cleavage by active caspase-8. A, purified caspase-9-FLAG was incubated with [γ-32P]ATP in kinase buffer in the presence or absence of purified recombinant CK2. Aliquots of the kinase reactions above were used as the source of substrate in cleavage assays using active, recombinant caspase-8 protein. Attenuated reactions were SDS-PAGE-resolved, transferred to nitrocellulose, and autoradiographed to visualize phosphorylated caspase-9 (top panel). The same membrane was then immunoblotted with an antibody against caspase-9 (Stressgen) to visualize total full-length and cleaved protein and determine relative cleavage by caspase-8 of unphosphorylated and phosphorylated substrates. B, in vitro translated [35S]methionine-labeled mC-9 was used as substrate for phosphorylation by recombinant CK2. Kinase reactions were performed in the presence of cold ATP with or without CK2, following which caspase-8 cleavage reactions were carried out on aliquots of the kinase reactions. The results were visualized by autoradiography following resolution by SDS-PAGE. C, alternatively the kinase reactions were diluted in rehydration buffer and resolved by isoelectric focusing (IEF, pH range 5–8) in the first dimension, followed by electrophoresis in the second dimension using an 8–16% gradient.
Next, in vitro translated 35S-labeled caspase-9 was phosphorylated by CK2 in vitro in the presence of ATP and then subjected to an active caspase-8 cleavage assay. The attenuated cleavage reactions were resolved by SDS-PAGE and visualized by autoradiography (shown in Fig. 3B). Again, the level of the 38-kDa product is reduced in reactions in which caspase-9 was phosphorylated by CK2 prior to cleavage. Because phosphorylation of one to two residues on mC-9FLAG is predicted to increase its molecular mass only by 0.1–0.2 kDa, the modification did not cause a detectable shift in the mobility of caspase-9 in either 10 or 14% polyacrylamide gels. However, two-dimensional resolution of in vitro translated 35S-labeled, unphosphorylated, and phosphorylated caspase-9 (Fig. 3C) showed two major spots corresponding to isoelectric points of 5.8 and 5.9 (bottom panel). Densitometric analyses of a number of kinase assays determined that 30–60% of the recombinant caspase-9 was phosphorylated in the presence of purified recombinant CK2 in vitro (although the levels were much lower in whole cell lysates), and activated caspase-8 was able to cleave 30–35% of the unphosphorylated translated or purified recombinant mC-9 (not shown) within 30 min in the cleavage assays.
The observation that caspase-9 phosphorylated on Ser348 was refractory to processing at the SEPD motif strongly suggested that dephosphorylation of endogenous caspase-9 precedes its activation by caspase-8 in the death receptor pathway. To determine whether phosphorylation of murine caspase-9 affected its ability to autoprocess in the intrinsic apoptotic pathway following its recruitment to the apoptosome, CK2 phosphorylated caspase-9 or a control was incubated with extracts from caspase-9-/- MEFs in the presence or absence of ATP and cyt c (2). The results (supplemental Fig. S2) indicate that the autoprocessing ability of the unphosphorylated caspase is similar to that of CK2 phosphorylated form, suggesting that phosphorylation at Ser348 does not affect its activation through the mitochondrial pathway.
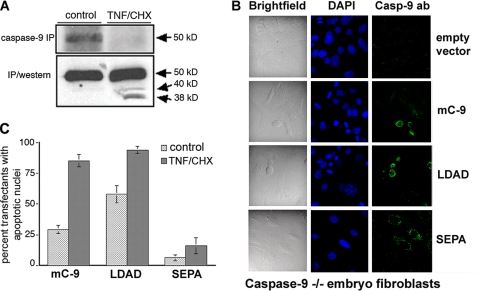
Caspase-9 Dephosphorylation and Cleavage Are Both Necessary and Sufficient for an Apoptotic Response to TNF-α in Murine pro B Cells and Embryo Fibroblasts—If endogenous caspase-9 could be processed and activated by a cyt c-independent mechanism early in the death receptor pathway (6), then TNF receptor trimerization could be expected to promote its dephosphorylation prior to cleavage. To test this, phosphate-starved FL5.12-Bcl-xL cells were labeled with [32P]orthophosphate (0.5 mCi) for 6 h. The overexpressed Bcl-xL in these cells prevents mitochondrial cyt c release (29, 30), allowing detection of the alternative pathway. Three hours following the addition of radiolabel, the culture medium was supplemented with TNF-αCHX or CHX alone. Fig. 4A shows that a decrease in levels of the phosphorylated form following TNF-α/CHX exposure (upper panel) was accompanied by processing of the dephosphorylated form (lower panel).
FIGURE 4.
Caspase-9 dephosphorylation and cleavage are both necessary and sufficient for an apoptotic response to TNF-α. A, Bcl-xL FL5.12 cells were labeled with [32P]orthophosphate (0.5 μCi) for 6 h. Three hours following the addition of radiolabel, the culture medium was supplemented with TNF-αCHX or CHX alone. Endogenous caspase-9 was immunoprecipitated, SDS-PAGE-resolved, transferred to nitrocellulose, and visualized, first by autoradiography (top panel), and then by Western blotting (lower panel) with an antibody against caspase-9 (AAM-139, Stressgen) that recognizes both full-length and processed protein. B, caspase-9-/- fibroblasts were transiently transfected with empty vector, wild type caspase-9, LDAD, or SEPA constructs in chamber slides and either incubated with TNF-αCHX (or CHX alone, not shown) 36 h post-transfection. The cells were fixed and stained 4 h later and observed using an Olympus Fluoview 1000 multiphoton confocal microscope. C, quantification of data from the transfection experiments. The y axis indicates percent caspase-9 expressing green fluorescent cells with condensed (apoptotic) nuclei (mean and standard error, n = 3).
We had previously identified SEPD, the autoprocessing site on mC-9, as the primary cleavage site for activated caspase-8 in vitro (6). These data and the result in Fig. 4A suggested that both dephosphorylation and processing at this site were critical for the TNF response. To determine this, we expressed caspase-9 and relevant point mutants in knock-out cells. Caspase-9 knock-out MEFs are resistant to low doses (5 ng/ml) of TNF-α, although they do mount an apoptotic response in 24 h to concentrations about 10 times higher (31). The MEFs were transiently transfected with empty vector or plasmids encoding mC-9, LDAD, or SEPA and 36 h post-transfection were incubated with TNF-α (5 ng/ml) and CHX for an additional 5 h. The cells were then fixed, stained, and observed under a confocal microscope. Condensed nuclei stained with 4′,6′-diamino-2-phenylindole were indicative of apoptotic cells. Fig. 4B shows that expression of mC-9 or the nonphosphorylatable (LDAD) mutant sensitized C-9-/- MEFs to low doses of ligand within 4 h of exposure to TNF-α, whereas cells expressing the noncleavable mutant (SEPA) or the empty vector continued to be unresponsive. Fig. 4C shows a quantification of the data, expressed as the percentage of transfected (green fluorescent) cells with apoptotic nuclei, from three separate experiments.
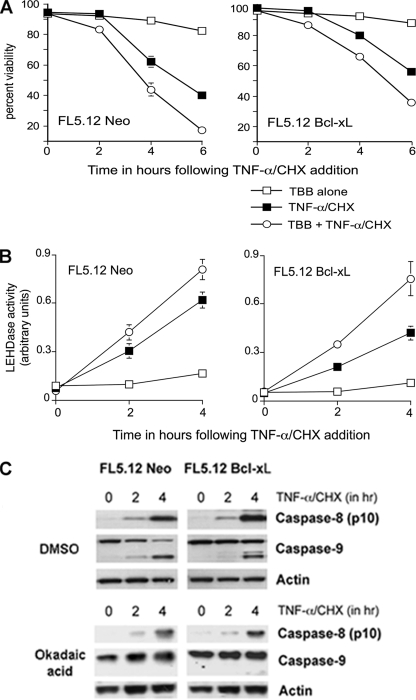
TNF-α-induced Caspase-9 Processing and Activation in FL5.12 Cells Is Accelerated by CK2-specific Inhibitors and Inhibited by the Serine Phosphatase Inhibitor Okadaic Acid—It has been reported that CK2 inhibits Fas ligand and TNF-α-induced apoptosis (25, 32, 33). We determined whether inhibition of endogenous CK2 would enhance the rate of cell death induced by TNF receptor cross-linking in FL5.12 cells. The results in Fig. 5A show that preincubation with the CK2-specific inhibitor TBB accelerated the onset of apoptosis in TNF-α-treated FL5.12 cells as assessed by flow cytometry. The acceleration in control cells could be attributed to enhanced cleavage of Bid and early release of mitochondrial cyt c. However, the mitochondrial route is largely blocked in Bcl-xL-overexpressing cells at early time points, suggesting that CK2 inhibition might also accelerate caspase-9 cleavage and activation. Caspase-9 activity was measured in cell pellets collected at 2-h intervals following the addition of ligand (Fig. 5B). The results indicate that the accelerated response to TNF-α in cells preincubated with CK2 inhibitor TBB is due in large part to the early activation of caspase-9, particularly in Bcl-xL-overexpressing cells.
FIGURE 5.
CK2-specific inhibitor accelerates caspase-9 activation, and phosphatase inhibitor delays caspase-9 processing in response to TNF-α/CHX. A, FL5.12 cells were exposed to TNF-α/CHX for 6 h following incubation in medium containing either the CK2-specific inhibitor TBB or vehicle (dimethyl sulfoxide, DMSO) for 3 h. The graph shows the percentage of viability of cells harvested at 0, 2, 4, and 6 h following the addition of ligand (mean and standard error, n = 3). B, graph showing caspase-9 activity in Neo and Bcl-xL cells following exposure to TNF-α/CHX in the presence or absence of TBB, as determined by colorimetric substrate (LEHD-pNA) cleavage assays. C, Western blots of lysates (50 μg of protein) of 0-, 2-, and 4-h time points from the above experiment, showing processed caspase-8, p10 fragment (top panel of each set), full-length and cleaved caspase-9 (center), and loading control, actin (bottom). The membranes were sequentially stripped and reprobed.
The studies described above strongly suggest that dephosphorylation of murine caspase-9 precedes its cleavage and activation in TNF receptor-induced cell death. The dephosphorylation of CK2 targets that occurs in response to a death stimulus is crucial for the normal control of cell signaling and is accomplished in part by the activation of protein phosphatases such as protein phosphatase 2A (34, 35). However, the naturally occurring protein phosphatase 2A inhibitor okadaic acid (OA) appears not to abrogate, and may even enhance, caspase-8 activation in response to Fas ligand (36). Okadaic acid delayed the onset of TNF-induced apoptosis in both controls and Bcl-xL-overexpressing cells (supplemental Fig. S3). Keeping in perspective the possibility that OA could affect a number of phosphatase dependent pathways, we asked whether the protection of TNF-treated FL5.12 by OA, at least in part, included caspase-8 or its downstream targets. Western blots of cell lysates were tested for the presence of cleaved caspases-8 and 9 (Fig. 5C) following TNF cross-linking in the presence or absence of the phosphatase inhibitor. The presence of the small (p10) cleavage product of caspase-8 in cells treated with TNF, both in the absence or in the presence of OA, suggested that the inhibitory effect was downstream of this initiator caspase. Processing of caspase-9, on the other hand, was detected only in the absence of phosphatase inhibition. The lack of caspase-9 cleavage fragments in lysates of OA-treated cultures further confirmed that dephosphorylation of caspase-9 contributed to its processing and activation and suggested that a protein phosphatase 2A-like phosphatase was likely to be involved.
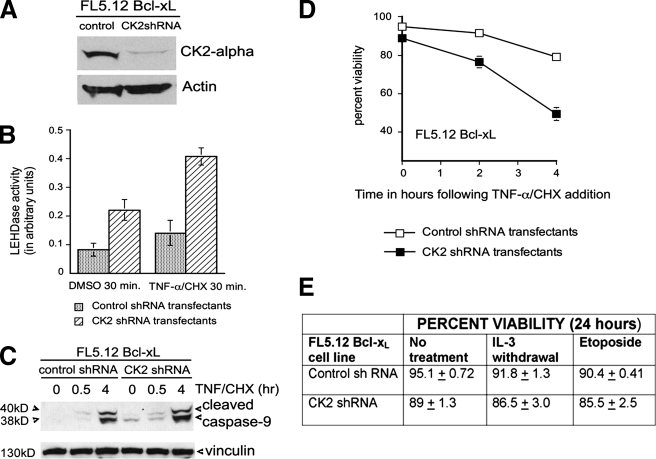
Loss of CK2 Promotes Early Activation of Caspase-9 in the TNF Pathway but Does Not Affect Bcl-xL Protection against Inducers of Intrinsic Apoptotic Pathways—To further confirm the loss of CK2 kinase activity on caspase-9 function, we generated knockdowns by transfecting FL5.12 cells with CK2-specific or nonsilencing pGIPZ shRNAmir vectors and selecting for puromycin resistance (Fig. 6A). The CK2 negative transfectants exhibited a low, but detectable, rate of cell death (Fig. 6D) and an increased basal level of caspase-9 activity compared with controls (Fig. 6, B and C), even in the absence of inducers. Following TNF treatment, CK2 negative cells showed increased caspase-9 cleavage and activation within 30 min (Fig. 6, B and C), as well as an accelerated loss of viability compared with controls (Fig. 6D). Bcl-xL has been shown to protect cells against apoptosis induced by growth factor deprivation and DNA-damaging compounds such as etoposide (22, 37). Fig. 6E shows that the loss of CK2 did not affect the ability of Bcl-xL to protect against these inducers of intrinsic apoptotic pathways for at least 24 h, although the cells did not survive beyond 72 h (not shown). This may be attributable to detrimental effects of CK2 loss on its targets in other signaling pathways. Taken together, these results support the hypothesis that phosphorylation of caspase-9 by CK2 protects it from cleavage and activation by an upstream initiator caspase but does not affect its ability to auto-activate in intrinsic apoptotic pathways.
FIGURE 6.
Loss of CK2 promotes early activation of caspase-9 in the TNF pathway but does not affect the ability of Bcl-xL to protect against inducers of intrinsic apoptotic pathways. A, Western blot showing expression of CK2 α in FL5.12 Bcl-xL cells transfected with a pGIPZ nonsilencing shRNAmir vector or pGIPZ CK2 (α and β) shRNAmir vectors. The blots were stripped and reprobed with antibody against actin. B, graph showing caspase-9 activity in control and CK2 shRNA transfectants prior to and after 30 min of exposure to TNF-α/CHX. C, Western blots of lysates from the shRNA transfectants (50 μg of protein), following 0-, 0.5-, and 4-h TNF-α/CHX treatment, showing cleaved caspase-9 (top panel) and loading control, vinculin (bottom panel), after stripping. D, percentage of viability of shRNA transfected cells harvested at 0, 2, and 4 h following the addition of ligand (mean and standard error, n = 3). E, table showing percent viability of the above transfectants 24 h following IL-3 withdrawal or incubation with 100 μm etoposide (mean and standard error, n = 3).
DISCUSSION
Caspases, a family of cysteinyl aspartate-specific proteases, are the executioners of apoptotic cell death (38). Caspases are synthesized as zymogens with a prodomain of variable length followed by a large subunit and a small subunit and are activated through proteolysis at the carboxyl-terminal aspartic acid residues in specific tetrapeptide processing motifs (39). Although initiator caspases (caspases-8 and -9) undergo auto-activation, effector caspases (caspases-3, -6, and -7) are activated following processing at specific cleavage sites by other activated caspases. In the intrinsic apoptotic pathway auto-activation of initiator caspase-9 by induced proximity or homodimerization is dependent on the release of mitochondrial cyt c and occurs following its recruitment to the apoptosome (2, 40). However, growing evidence points to alternate mechanisms of activation for this caspase that involve neither cyt c release nor Apaf-1 activation (3, 5–7). Previous studies from our laboratory had suggested that murine caspase-9 could be directly cleaved and activated by initiator caspase-8 during death receptor-induced apoptosis, implying that caspase-9 was functioning not as an initiator but as an effector caspase early in this apoptotic pathway (Ref. 6 and Fig. 5). In the current study we have described a mechanism by which the sensitivity of caspase-9 to cleavage by caspase-8 is controlled by phosphorylation. We show that residue Ser348 on murine caspase-9 is specifically phosphorylated by CK2 and that the phosphorylation protects it from cleavage at Asp353 in the processing motif.
To our knowledge, this is the first example of phospho-regulation of caspase-9 activation in an extrinsic apoptotic pathway. A variety of kinases, including ERK2, protein kinase A, protein kinase Cζ and c-Abl tyrosine kinase, have been implicated in the regulation of mammalian caspase-9 function in the intrinsic pathway. For instance, ERK/mitogen-activated protein kinase (MAPK)-mediated phosphorylation of human caspase-9 at a conserved threonine is sufficient to block its processing (9), and protein kinase A inhibits cyt c-dependent recruitment of pro-caspase-9 to the apoptosome (11). Phosphorylation of Ser144 on caspase-9 by protein kinase Cζ during hyperosmotic stress also prevents the activation of the intrinsic apoptotic pathway (10), whereas c-Abl-mediated tyrosine phosphorylation of caspase-9 on residue 153 contributes to its DNA damage-induced autoprocessing and, subsequently, to apoptosis (12). It is interesting to note that human caspase-9 is not phosphorylated by CK2 in vitro (supplemental Fig. S4A). Early indications that human and murine caspase-9 may be regulated differently came from a study showing that the Akt phosphorylation site (at Ser196), presumed to be involved in the inactivation of human caspase-9, is absent in the murine form (13, 14). Furthermore, the 46-kDa human caspase-9 protein lacks residues 99–131 of its murine counterpart, a 50-kDa protein. Although human protein harbors a serine residue at a similar position, flanking residues do not conform to the CK2 requirement for acidic residue clusters (21) and thus constitute a weak consensus for the kinase. Alignment of a few caspase-9 sequences in the vicinity of the CK2 motif shows that rat and dog caspase-9 also harbor strong CK2 consensi (supplemental Fig. S4B). The absence of a strong CK2 target consensus in the human protein is not surprising for another reason. Human cells have been classified as cells of type I or type II based on the nature of their response to death receptor activation by ligands such as Fas and TNF-α. Survival members of the Bcl-2 family, by virtue of their ability to prevent cyt c release, therefore, impart almost total protection against apoptosis induced via Fas receptor cross-linking to type II cells. The activation of caspase-9 in the presence of Bcl-xL and in the absence of cyt c release during TNF-induced apotosis precludes classification of some murine cells as Type II cells (6). In vitro cleavage assays had previously shown that human caspase-9 is cleaved less efficiently than the murine protein by active caspase-8 and that DQLD (Asp330), rather than PEPD (Asp315), was the preferred processing site (6). This difference in susceptibility between human and murine caspase-9 to both processing by active caspase-8 as well as to phosphomodification of a proximal serine may help explain the difference in response to death receptor activation between human cells of type II and murine cells that elude strict classification as Type II cells.
CK2 is a ubiquitous and constitutively active kinase with oncogenic potential. In addition to modifying hundreds of known substrates with cell growth, differentiation, and cell cycle functions (17, 18), this kinase counters apoptosis promoted by DNA damaging agents as well as death receptors (33, 41). A number of proteins have been reported to become refractory to caspase cleavage upon CK2-mediated phosphorylation. The kinase (along with another serine/threonine kinase, CK1) phosphorylates Bid, a Bcl-2 family protein and crucial intermediate in death receptor pathways, at serine residues in the vicinity of the caspase-8 recognition site, rendering it resistant to cleavage by activated caspase-8 (25). In addition, Max, connexin 45.6, HS1, and presenilin-2 have been shown to lose susceptibility to cleavage by caspases upon CK2-mediated phosphorylation (42–45). ARC, a protein that inhibits caspase-8 activity when phosphorylated, has also been identified as a CK2 target (46). Recently procaspase-2 has emerged as a CK2 target and shown to be activated by dimerization independent of the PIDDosome (a complex comprising death domain-containing protein, PIDD, and the adaptor protein, RAIDD) following dephosphorylation (47). In the current study, we have identified yet another pro-apoptotic protein that is rendered resistant to caspase-8 by CK2-mediated modification. Thus, CK2 appears to exert its survival role by controlling the activity of pro-apoptotic caspase substrates in specific signaling pathways. We suggest, furthermore, that constitutive activation of this kinase ensures that the CK2 target site remains modified and protected unless specifically dephosphorylated in response to an apoptotic stimulus. The appearance of a 20-kDa (late) processing product of caspase-9 in cells incubated with the CK2 inhibitor, DRB (Fig. 2B), and the presence of the 38-kDa cleavage product in untreated CK2 negative cells (Fig. 6C) suggests that the dephosphorylated protein may be susceptible to low levels of active caspases in the cell. Studies have shown that caspase-8 is active at a low basal level in a number of cells (48). The absence of the 38- and 40-kDa intermediates in Fig. 2B is currently unclear, although it is tempting to speculate that in the absence of a death stimulus the 38-kDa cleavage product of caspase-8 activity may be the only intermediate generated and that even this is quickly processed down to the 20-kDa active subunit in FL5.12 cells.
Exposure of murine FL5.12 pro B lymphoma cells to TNF-α promotes dephosphorylation of caspase-9, rendering it susceptible to proteolytic processing. Inhibition of CK2 activity in the presence of the apoptotic trigger accelerates both caspase-9 activity and cell death. Although the dephosphorylation of caspase-9 during death receptor activation is likely to be the combined result of activated phosphatase(s) and CK2 down-regulation or inhibition, it is significant that in protecting FL5.12 cells, a naturally occurring inhibitor of serine/threonine protein phosphatases (okadaic acid) specifically inhibits caspase-9 cleavage without affecting auto-processing and activation of caspase-8, the initiator caspase (Fig. 5). This experiment also confirms that activated caspase-9 is a major contributor to death induced by TNF-α in these cells. The inability of caspase-9-negative MEF transfectants expressing the noncleavable SEPA mutant to activate apoptosis in response to TNF treatment (unlike wild type caspase and LDAD expressing MEFs) is further evidence that processing of murine caspase-9 is essential for the death response (Fig. 4B).
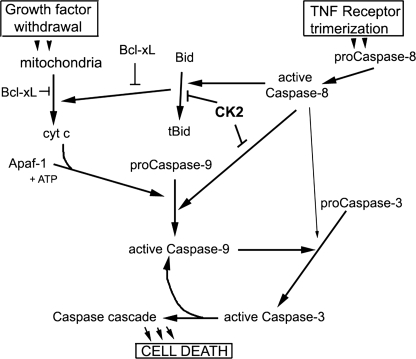
In conclusion, we have shown that a serine residue adjacent to the primary site of caspase-8 cleavage on murine caspase-9 is modified by the survival protein kinase, CK2, in vitro and in vivo. We have also shown that this modification lowers the susceptibility of caspase-9 to cleavage but does not affect caspase-9 auto-processing (model in Fig. 7). Phosphatase inhibitors delay TNF-induced apoptosis in control cells as well as in cells overexpressing the Bcl-xL protein. Inhibition of intracellular CK2 or loss of the kinase accelerates apoptosis and promotes early caspase-9 activation in response to TNF-α and cycloheximide but does not affect the ability of anti-apoptotic protein Bcl-xL to protect against inducers of mitochondrial apoptotic pathways. These studies strongly suggest that modification of murine procaspase-9 in the vicinity of its processing motif protects the protease from mistimed or inappropriate activation by other caspases and further emphasize the role of the ubiquitous kinase CK2 in promoting cell survival.
FIGURE 7.
Simplified model of mitochondrial and TNF receptor-activated apoptotic pathways in murine cells. The kinase CK2 controls activation of caspase-9 in receptor-activated apoptotic pathways where it is processed by initiator caspase-8 but not in mitochondrial pathways where it is activated by autoprocessing.
Supplementary Material
Acknowledgments
We thank Dr. Richard Flavell (Yale University) for the caspase-9 knock-out embryo fibroblasts.
This work was supported, in whole or in part, by National Institutes of Health NCI Grant CA-15062 (to K. A.). This work was also supported by National Science Foundation Award MCB-0350070 (to A. K.), by a National Science Foundation Research Experience for Undergraduates supplement (to T. N. P.), and by the Medical Research Fund of the Department of Veterans Affairs. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: cyt c, cytochrome c; CK2, casein kinase 2; DRB, 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole; CHX, cyclohexamide; TBB, 4,5,6,7-tetrabromobenzotriazole;TNF,tumornecrosisfactor;ERK,extracellular signal-regulated kinase; MEF, mouse embryonic fibroblast; OA, okadaic acid.
References
- 1.Stennicke, H. R., Deveraux, Q. L., Humke, E. W., Reed, J. C., Dixit, V. M., and Salvesen, G. S. (1999) J. Biol. Chem. 274 8359-8362 [DOI] [PubMed] [Google Scholar]
- 2.Zou, H., Li, Y., Liu, X., and Wang, X. (1999) J. Biol. Chem. 274 11549-11556 [DOI] [PubMed] [Google Scholar]
- 3.Bitzer, M., Armeanu, S., Prinz, F., Ungerechts, G., Wybranietz, W., Spiegel, M., Bernlohr, C., Cecconi, F., Gregor, M., Neubert, W. J., Schulze-Osthoff, K., and Lauer, U. M. (2002) J. Biol. Chem. 277 29817-29824 [DOI] [PubMed] [Google Scholar]
- 4.Meyer, K., Basu, A., Saito, K., Ray, R. B., and Ray, R. (2005) Virology 336 198-207 [DOI] [PubMed] [Google Scholar]
- 5.Morishima, N., Nakanishi, K., Takenouchi, H., Shibata, T., and Yasuhiko, Y. (2002) J. Biol. Chem. 277 34287-34294 [DOI] [PubMed] [Google Scholar]
- 6.McDonnell, M. A., Wang, D., Khan, S. M., Vander Heiden, M. G., and Kelekar, A. (2003) Cell Death Differ. 10 1005-1015 [DOI] [PubMed] [Google Scholar]
- 7.Gyrd-Hansen, M., Farkas, T., Fehrenbacher, N., Bastholm, L., Hoyer-Hansen, M., Elling, F., Wallach, D., Flavell, R., Kroemer, G., Nylandsted, J., and Jaattela, M. (2006) Mol. Cell Biol. 26 7880-7891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scaffidi, C., Schmitz, I., Zha, J., Korsmeyer, S. J., Krammer, P. H., and Peter, M. E. (1999) J. Biol. Chem. 274 22532-22538 [DOI] [PubMed] [Google Scholar]
- 9.Allan, L. A., Morrice, N., Brady, S., Magee, G., Pathak, S., and Clarke, P. R. (2003) Nat. Cell Biol. 5 647-654 [DOI] [PubMed] [Google Scholar]
- 10.Brady, S. C., Allan, L. A., and Clarke, P. R. (2005) Mol. Cell Biol. 25 10543-10555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin, M. C., Allan, L. A., Lickrish, M., Sampson, C., Morrice, N., and Clarke, P. R. (2005) J. Biol. Chem. 280 15449-15455 [DOI] [PubMed] [Google Scholar]
- 12.Raina, D., Pandey, P., Ahmad, R., Bharti, A., Ren, J., Kharbanda, S., Weichselbaum, R., and Kufe, D. (2005) J. Biol. Chem. 280 11147-11151 [DOI] [PubMed] [Google Scholar]
- 13.Cardone, M. H., Roy, N., Stennicke, H. R., Salvesen, G. S., Franke, T. F., Stanbridge, E., Frisch, S., and Reed, J. C. (1998) Science 282 1318-1321 [DOI] [PubMed] [Google Scholar]
- 14.Fujita, E., Jinbo, A., Matuzaki, H., Konishi, H., Kikkawa, U., and Momoi, T. (1999) Biochem. Biophys. Res. Commun. 264 550-555 [DOI] [PubMed] [Google Scholar]
- 15.Ahmed, K., Gerber, D. A., and Cochet, C. (2002) Trends Cell Biol. 12 226-230 [DOI] [PubMed] [Google Scholar]
- 16.Seldin, D. C., Landesman-Bollag, E., Farago, M., Currier, N., Lou, D., and Dominguez, I. (2005) Mol. Cell Biochem. 274 63-67 [DOI] [PubMed] [Google Scholar]
- 17.Meggio, F., and Pinna, L. A. (2003) FASEB J. 17 349-368 [DOI] [PubMed] [Google Scholar]
- 18.Canton, D. A., and Litchfield, D. W. (2006) Cell Signal. 18 267-275 [DOI] [PubMed] [Google Scholar]
- 19.Litchfield, D. W. (2003) Biochem. J. 369 1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tawfic, S., Yu, S., Wang, H., Faust, R., Davis, A., and Ahmed, K. (2001) Histol. Histopathol. 16 573-582 [DOI] [PubMed] [Google Scholar]
- 21.Pinna, L. A. (2002) J. Cell Sci. 115 3873-3878 [DOI] [PubMed] [Google Scholar]
- 22.Boise, L. H., Gonzalez-Garcia, M., Postema, C. E., Ding, L., Lindsten, T., Turka, L. A., Mao, X., Nunez, G., and Thompson, C. B. (1993) Cell 74 597-608 [DOI] [PubMed] [Google Scholar]
- 23.Kelekar, A., Chang, B. S., Harlan, J. E., Fesik, S. W., and Thompson, C. B. (1997) Mol. Cell. Biol. 17 7040-7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blom, N., Gammeltoft, S., and Brunak, S. (1999) J. Mol. Biol. 294 1351-1362 [DOI] [PubMed] [Google Scholar]
- 25.Desagher, S., Osen-Sand, A., Montessuit, S., Magnenat, E., Vilbois, F., Hochmann, A., Journot, L., Antonsson, B., and Martinou, J. C. (2001) Mol. Cell 8 601-611 [DOI] [PubMed] [Google Scholar]
- 26.Sarno, S., Marin, O., Boschetti, M., Pagano, M. A., Meggio, F., and Pinna, L. A. (2000) Biochemistry 39 12324-12329 [DOI] [PubMed] [Google Scholar]
- 27.Niefind, K., Putter, M., Guerra, B., Issinger, O. G., and Schomburg, D. (1999) Nat. Struct. Biol. 6 1100-1103 [DOI] [PubMed] [Google Scholar]
- 28.Adrain, C., Slee, E. A., Harte, M. T., and Martin, S. J. (1999) J. Biol. Chem. 274 20855-20860 [DOI] [PubMed] [Google Scholar]
- 29.Adams, J. M., and Cory, S. (1998) Science 281 1322-1326 [DOI] [PubMed] [Google Scholar]
- 30.Vander Heiden, M. G., Chandel, N. S., Williamson, E. K., Schumacker, P. T., and Thompson, C. B. (1997) Cell 91 627-637 [DOI] [PubMed] [Google Scholar]
- 31.Hakem, R., Hakem, A., Duncan, G. S., Henderson, J. T., Woo, M., Soengas, M. S., Elia, A., de la Pompa, J. L., Kagi, D., Khoo, W., Potter, J., Yoshida, R., Kaufman, S. A., Lowe, S. W., Penninger, J. M., and Mak, T. W. (1998) Cell 94 339-352 [DOI] [PubMed] [Google Scholar]
- 32.Olsen, B. B., Jessen, V., Hojrup, P., Issinger, O. G., and Boldyreff, B. (2003) FEBS Lett. 546 218-222 [DOI] [PubMed] [Google Scholar]
- 33.Wang, G., Ahmad, K. A., and Ahmed, K. (2005) Mol. Cell Biochem. 274 201-205 [DOI] [PubMed] [Google Scholar]
- 34.Goldberg, Y. (1999) Biochem. Pharmacol. 57 321-328 [DOI] [PubMed] [Google Scholar]
- 35.Lebrin, F., Bianchini, L., Rabilloud, T., Chambaz, E. M., and Goldberg, Y. (1999) Mol. Cell Biochem. 191 207-212 [PubMed] [Google Scholar]
- 36.Chatfield, K., and Eastman, A. (2004) Biochem. Biophys. Res. Commun. 323 1313-1320 [DOI] [PubMed] [Google Scholar]
- 37.Minn, A. J., Rudin, C. M., Boise, L. H., and Thompson, C. B. (1995) Blood 86 1903-1910 [PubMed] [Google Scholar]
- 38.Thornberry, N. A., and Lazebnik, Y. (1998) Science 281 1312-1316 [DOI] [PubMed] [Google Scholar]
- 39.Nicholson, D. W. (1999) Cell Death Differ. 6 1028-1042 [DOI] [PubMed] [Google Scholar]
- 40.Pop, C., Timmer, J., Sperandio, S., and Salvesen, G. S. (2006) Mol. Cell 22 269-275 [DOI] [PubMed] [Google Scholar]
- 41.Guo, C., Yu, S., Davis, A. T., Wang, H., Green, J. E., and Ahmed, K. (2001) J. Biol. Chem. 276 5992-5999 [DOI] [PubMed] [Google Scholar]
- 42.Krippner-Heidenreich, A., Talanian, R. V., Sekul, R., Kraft, R., Thole, H., Ottleben, H., and Luscher, B. (2001) Biochem. J. 358 705-715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruzzene, M., Brunati, A. M., Sarno, S., Donella-Deana, A., and Pinna, L. A. (1999) FEBS Lett. 461 32-36 [DOI] [PubMed] [Google Scholar]
- 44.Walter, J., Grunberg, J., Schindzielorz, A., and Haass, C. (1998) Biochemistry 37 5961-5967 [DOI] [PubMed] [Google Scholar]
- 45.Yin, X., Jedrzejewski, P. T., and Jiang, J. X. (2000) J. Biol. Chem. 275 6850-6856 [DOI] [PubMed] [Google Scholar]
- 46.Li, P. F., Li, J., Muller, E. C., Otto, A., Dietz, R., and von Harsdorf, R. (2002) Mol. Cell 10 247-258 [DOI] [PubMed] [Google Scholar]
- 47.Shin, S., Lee, Y., Kim, W., Ko, H., Choi, H., and Kim, K. (2005) EMBO J. 24 3532-3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, L., Alva, A., Su, H., Dutt, P., Freundt, E., Welsh, S., Baehrecke, E. H., and Lenardo, M. J. (2004) Science 304 1500-1502 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.