Abstract
Transforming growth factor (TGF)-β1 plays an important role in the development of pulmonary fibrosis. In this study we examined the relationship between TGF-β1 stimulation and the expression of heparan sulfate (HS) 6-O-endosulfatase 1 (Sulf1) in cultured normal human lung fibroblasts (NHLFs) and in murine lungs in vivo. By removing 6-O-sulfates from specific HS intrachain sites on the cell surface, Sulf1 has been shown to modulate the activities of many HS binding growth factors and morphogens including fibroblast growth factor (FGF)-2. Real time reverse transcription-PCR analysis revealed that TGF-β1 increased Sulf1 expression in NHLFs in a dose- and time-dependent manner which was accompanied by a decrease in 6-O-sulfated disaccharides as revealed by high performance liquid chromatography analysis. Decreased ERK activation after FGF-2 stimulation was observed in TGF-β1-treated NHLFs compared with control cells without changes in HS-dependent FGF-2 binding or FGF-2·FR1c complex formation. To study the function of Sulf1, negative control or Sulf1-specific small interference RNA (siRNA)-transfected NHLFs were stimulated with TGF-β1. Enhanced Smad2/3 phosphorylation and elevated total Smad2 protein level were observed in Sulf1 siRNA-transfected cells and were accompanied by enhanced expression of α-smooth muscle actin and fibronectin. In addition, Sulf1 siRNA transfection enhanced the anti-proliferative effect of TGF-β1. Finally Sulf1 expression was up-regulated in the lungs of mice treated with adenovirus encoding active TGF-β1. Taken together, our data indicate that Sulf1 is a TGF-β1-responsive gene both in vitro and in vivo and may function as a negative regulator of TGF-β1-induced fibrogenesis.
Pulmonary fibrosis (PF)3 is characterized by increased number of myofibroblasts and subsequent tissue remodeling with extensive accumulation of collagen and other extracellular matrix (ECM) molecules in the lung resulting in loss of pulmonary function (1). Data from both patients and animal models support a pivotal role for transforming growth factor (TGF)-β1 in the development and progression of PF. TGF-β1 is present at sites of ECM gene expression in fibrotic human lungs (2, 3). The most studied animal disease model for PF, the bleomycin model, is largely driven by TGF-β1 (4, 5), and studies in mice administered with TGF-β1 adenoviral vectors in the lung show that overexpression of this cytokine alone is sufficient to induce PF (6).
In PF, TGF-β1 initiates and sustains fibroblast activation and transdifferentiation into myofibroblasts, which is exemplified by the de novo expression of α-smooth muscle actin (α-SMA) (7). The expression of α-SMA confers the myofibroblast contractile property, which is responsible for the distortion of normal lung architecture (8). Studies in both patients and animal models have also shown that myofibroblasts are the primary source of ECM gene expression in the active fibrotic sites (9, 10). In addition, compared with fibroblasts, myofibroblasts have reduced motility and proliferative capacity (11), and TGF-β1 provides myofibroblasts protection against apoptosis (12).
Heparan sulfate (HS) is the glycosaminoglycan moiety of the HS proteoglycans, the ubiquitous macromolecules associated with the cell surface, basement membrane, and the ECM (13, 14). HS polysaccharide chains are synthesized in the Golgi apparatus and contain repeating disaccharide units of uronic acid (iduronic or glucuronic acid) linked to N-acetylglucosamine. During HS biosynthesis, these disaccharides are selectively sulfated at the N, 6-O, and 3-O positions of the glucosamine and the 2-O position of the uronic acid residues by actions of the HS sulfotransferases (15). HS proteoglycans are involved in a wide range of biological processes through their HS chains, which bind to and modify the activities of a diverse repertoire of ligands including growth factors, morphogens, cytokines, chemokines, matrix proteins, and cell adhesion molecules (13-15). It is now well established that HS-protein interactions critically depend on the amount and the positions of the O-sulfate groups, in particular, the 6-O-sulfates, which form binding sites for proteins (14, 16). Thus, not surprisingly, HS 6-O-sulfation is dynamically regulated during embryonic development (17-20), aging (21), and carcinogenesis (22, 23).
HS 6-O-sulfation state can be regulated in two ways; that is, regulated expression of the HS 6-O-sulfotransferases (24) that function in the Golgi and further modification at the cell surface or the ECM by the newly identified HS 6-O-endosulfatases, Sulf1 and Sulf2 (25). The first Sulf, QSulf1, was discovered in a molecular cloning screen for Sonic hedgehog (Shh)-responsive genes in quail embryos and was shown to regulate cellular differentiation through modulation of Wnt signaling (26). Subsequently, Sulf1 and the closely related Sulf2 were identified in mammalian species (27, 28) and found to be misregulated in a variety of tumors (25). The influence of the Sulfs on the activity of HS-binding proteins active in tumorigenesis, such as vascular endothelial growth factor and fibroblast growth factor (FGF), suggests that modulation of Sulf activity could serve as a target for therapeutic intervention (29, 30).
TGF-β1 interacts strongly with heparin and highly sulfated HS (31-33). These interactions protect TGF-β1 from proteolytic degradation in vitro and potentiate the activity of TGF-β1 in supporting the anchorage-independent growth of rat kidney fibroblasts (31, 32). Importantly, selective loss of N-, 2-O-, or 6-O-sulfates all lead to reduced TGF-β1 activity (32). In lesion diffuse systemic sclerosis (dSSc) fibroblasts, syndecan-2 and syndecan-4 (members of the transmembrane HS proteoglycan, syndecan family) are markedly elevated compared with nonlesion dSSc or normal dermal fibroblasts, and HS side chains in these cells are required for TGF-β-induced contractile phenotype (34). The role of specific sulfation (N-, 2-O-, and/or 6-O-sulfation), however, was not addressed in this study. The above data indicate that HS plays an important role in TGF-β1-induced cellular responses, and changes in HS sulfation may modulate TGF-β1 function.
In this study we examined the relationship between TGF-β1 stimulation and the expression of Sulf1 and Sulf2 in normal human lung fibroblasts (NHLFs) and in murine lungs in vivo. Our data reveal that TGF-β1 strongly induces Sulf1 expression both in vitro and in vivo and that Sulf1 may function as a negative regulator of TGF-β1-induced fibrogenesis.
EXPERIMENTAL PROCEDURES
Cell Culture—Primary NHLFs and the culture reagent fibroblast growth medium-2 Bullet Kit were obtained from Cambrex Bioproducts (Walkersville, MD). The NHLFs were received at passage 3 and cultured in fibroblast growth medium-2 complete medium. Subconfluent NHLFs were rendered quiescent in fibroblast basal medium (FBM, Cambrex Bioproducts) containing 0.2% bovine serum albumin (BSA, Sigma) overnight before treatment with TGF-β1 and/or FGF-2 (R&D systems Inc., Minneapolis, MN) in the presence or absence of heparin (porcine intestinal mucosal heparin, Sigma). All experiments were performed in FBM containing 0.2% BSA and with cells at passages 5-6.
Real-time Quantitative Reverse Transcription-PCR—Messenger RNA (mRNA) of the gene of interest was quantified by quantitative real-time reverse transcription (RT)-PCR. Total RNA was isolated using the RNA mini plus kit (Qiagen Inc., Valencia, CA). Reverse transcription of 0.5-1 μg of total RNA was performed in a total volume of 20 μl using the iScript cDNA synthesis kit (Bio-Rad). One microliter of cDNA was PCR-amplified in 20-μl reactions containing primers at 200 or 400 nm in iQ SYBR Green Supermix (Bio-Rad). For 18 S rRNA amplification, 1:100 dilution of cDNA was used. PCR was performed for 35 cycles consisting of 95 °C for 15 s and 60 °C for 45 s using an iCycler iQ real time detection system (Bio-Rad). Dilution curves showed that PCR efficiency was 95-105% for all primer sets used. Negative controls included cDNA reaction without reverse transcriptase or RNA and PCR mix lacking cDNA to detect possible contamination. After amplification, specificity of the amplification was confirmed by melt curve analysis. Relative quantification was determined using the 2-ΔΔCt method (35) with data normalized to 18 S rRNA or the mRNA level of the 36B4 housekeeping gene. The PCR primers employed were designed using Beacon Designer 3.0 (Premier Biosoft International, Palo Alto, CA) and are listed in Table 1.
TABLE 1.
Real-time PCR primer sequences
| Gene | Primer sequences (forward and reverse) | Product |
|---|---|---|
| bp | ||
| Human Sulf1 | 5′-TGCTCAAAGTGACGGGTTCTTGGT-3′ | 159 |
| 5′-GTTGGTCGGTTCAAATGCAGGGTT-3′ | ||
| Murine Sulf1 | 5′-GCGTCCTCTTGTCCACTCTG-3′ | 136 |
| 5′-TAGCCACTCCTTTGTATCACTCTG-3′ | ||
| Human Sulf2 | 5′-CTGTGGGAAGGCTGGGAAGG-3′ | 158 |
| 5′-TGAGAGTGCGTGCTTGCTTTC-3′ | ||
| Murine Sulf2 | 5′-CGAGGTGGACGGTGAGATATAC-3′ | 110 |
| 5′-CATCCTTGTCATCTTGGTCTTCAG-3′ | ||
| Human α-SMA | 5′-GAAGAAGAGGACAGCACTG-3′ | 144 |
| 5′-TCCCATTCCCACCATCAC-3′ | ||
| Human collagen I | 5′-CGGAGGAGAGTCAGGAAGG-3′ | 159 |
| 5′-CACAAGGAACAGAACAGAACAG-3′ | ||
| Human FN | 5′-GCTCTATTCCACCTTACAACAC-3′ | 154 |
| 5′-ACAACGATGCTTCCTGAGTC-3′ | ||
| Human/murine | 5′-CGACCTGGAAGTCCAACTAC-3′ | 109 (Ref. 36) |
| 36B4 | 5′-ATCTGCTGCATCTGCTTG-3′ | |
| Human/murine | 5′-GAGGGAGCCTGAGAAACGG-3′ | 68 |
| 18 S rRNA | 5′-GTCGGGAGTGGGTAATTTGC-3′ |
Analysis of Sulf1 mRNA Stability—For analysis of the rate of decay of Sulf1 mRNA, quiescent NHLFs were stimulated with 0.5 ng/ml TGF-β1 for 20 h. Transcription was then inhibited by the addition of the RNA polymerase II-specific inhibitor, 5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole (DRB; Sigma) at a final concentration of 50 μm. At various time points after the addition of DRB, total RNA was isolated, and the rate of mRNA degradation was subsequently determined using real-time RT-PCR as previously described (36).
Heparan Sulfate Disaccharide Analysis—Sulfated glycosaminoglycans from NHLFs cultured in 75-cm2 tissue culture flasks were metabolically labeled with 50 μCi/ml sodium [35S]sulfate (PerkinElmer Life Sciences) in FBM containing 0.2% BSA overnight at 37 °C. Media and cells were then collected separately and treated with Pronase (Sigma) at the concentration of 0.167 mg/ml in 40 mm sodium acetate, 0.32 m NaCl, pH 6.5, at 37 °C overnight. After filtration to remove insoluble materials, total glycosaminoglycans were affinity-purified by anion exchange chromatography with 0.5 ml of DEAE-Sepharose™ Fast Flow (GE Healthcare) packed in Poly-Prep columns (Bio-Rad). The columns were washed with 20 mm sodium acetate, 0.25 m NaCl, pH 6.0, and eluted with 20 mm sodium acetate, 1 m NaCl, pH 6.0. Glycosaminoglycan chains were recovered by ethanol precipitation and dissolved in water. The digestion of HS was carried out in 100 μl of 40 mm ammonium acetate, pH 7.0, containing 3.3 mm CaCl2 and 1 milliunit each of heparitinase I, II, and III (Seikagaku, Tokyo, Japan) at 37 °C overnight. The next morning samples were further boost-digested with 1 milliunit each of heparitinase I, II, and III at 37 °C for 1 h. Digestion was stopped by heating at 100 °C for 5 min. Samples were then filtered with Ultrafree-MC (5000 molecular weight limit, Millipore Corp. Bedford, MA), and the unsaturated disaccharides in the filtrates were resolved by ion-pairing reverse-phase HPLC (Protein & Peptide C18, Vydac, Deerfield, IL) with appropriate disaccharide standards (Seikagaku) as described (17).
Analysis of FGF-2 Binding and FGF-2·HS·FR1c Complex Formation—For analysis of HS-dependent FGF-2 binding and the assembly of the FGF-2·HS·FR1c signaling complex in situ, NHLFs grown in 8-well chamber slides were fixed for 20 min in 4% paraformaldehyde at room temperature. Before the binding assays, sections were incubated with phosphate-buffered saline (PBS) containing 2 m NaCl to remove any endogenous FGFs that may have been bound to the HS on the cell surface. Samples were then blocked in binding buffer (PBS containing 1 mm Ca2+, 0.5 mm Mg2+, and 0.5% BSA) for 1 h. All binding assays were performed at room temperature. To examine FGF-2 binding, cells in the chamber slides were incubated with 0.3 or 1.0 nm FGF-2 (R&D systems) in binding buffer for 1 h. For FGF-2·HS·FR1c complex formation, FGF-2 and FR1c-Fc (R&D systems) were added to the binding buffer together at 0.3 or 1.0 nm/each and applied to the chamber slides for 1 h. Slides were then washed three times in PBS containing Ca2+ and Mg2+. Bound FGF-2 and FGF-2·FR1c were detected with anti-FGF-2 or anti-Fc antibody (R&D systems) followed by an Alexa 488-conjugated donkey anti-goat secondary antibody (Invitrogen). Images were captured using a Nikon Eclipse 80i epifluorescent microscope equipped with a Cooke SensiCam CCD camera (Applied Scientific Instrumentation, Eugene, OR) and analyzed using IPLab v3.65a Scientific Image Processing Software (BD Biosciences). To confirm that the above binding assays were HS-dependent, selected wells were treated with a mixture of heparitinases I, II, and III (10 milliunits/ml of each in binding buffer) before the addition of FGF-2 or FGF-2·FR1c-Fc.
Small Interference RNA (siRNA) Transfection—siRNA against human Sulf1 (catalog #16708A and AM16704) and negative control siRNA (catalog #4611) were obtained from Ambion (Austin, TX). Transient transfections of siRNA were performed using Amaxa nucleofection technology (Amaxa, KoeIn, Germany). Briefly, 2 × 106 NHLFs were suspended in 100 μl of Nucleofector™ solution, 200 pmol of control or Sulf1 siRNA were then added, and the solution was mixed gently. Cells were transfected using the T-16 pulsing parameter and immediately transferred into 12-well plates containing pre-warmed culture medium. Cells were allowed to attach to the culture plate in fibroblast growth medium-2 for 4-5 h, washed twice with FBM and 0.2% BSA, and incubated in FBM and 0.2% BSA overnight before TGF-β1 treatment. Knockdown of Sulf1 was evaluated by real-time RT-PCR.
Immunoblot—Total cell extracts (10-20 μg) from NHLFs were separated on 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Invitrogen). Membranes were blocked in PBS containing 0.1% Tween 20 and 5% BSA and then probed with antibodies against phosphor-p44/42 (ERK) MAPK (Thr-202/Tyr-204), total p44/42 (ERK) MAPK, total and phosphorylated Smad2 and Smad3 (Cell Signaling, Beverly, MA), α-SMA, γ-tubulin (Sigma), and collagen I and fibronectin (Abcam Inc. Cambridge, MA). Membranes were subsequently incubated with appropriate horseradish peroxidase-conjugated secondary antibodies (Cell Signaling) before detection with enhanced chemiluminescence reagent (ECL plus, GE Healthcare). Quantification of the immunoblots was performed using NIH ImageJ software.
Cell Proliferation Assay—Cell proliferation was examined using CellTiter 96 nonradioactive cell proliferation assay (Promega, Madison, WI) according to the manufacturer's protocol.
Treatment of Mice with Adenovirus Encoding Active TGF-β1—Eight-week-old male, pathogen-free C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Mice (5 per group) were anesthetized with isoflurane, and 1 × 108 plaque-forming units of the replication-deficient adenovirus encoding either GFP (control, AdGFP) or active TGF-β1 (AdTGF-β1223/225) (6, 37) in 50 μl of sterile PBS were introduced into the lungs via oropharyngeal aspiration as described (38). Mice were sacrificed 5 days later, and lung tissues were harvested, snap-frozen in liquid nitrogen, and stored at -80 °C. RNA was isolated from the right lungs and analyzed as described for NHLFs.
Statistical Analysis—Data were expressed as means ± S.E. of the mean. Statistical analyses were performed using unpaired Student's t test for two groups and analysis of variance followed by Bonferroni's or Dunnett's multiple comparison test when more than two groups were compared. Differences were considered statistically significant when p < 0.05. All experiments were repeated at least twice with similar results, and representatives are shown.
RESULTS
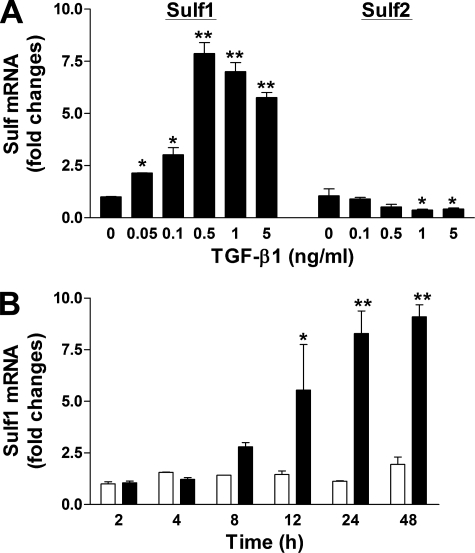
TGF-β1 Induces Sulf1 mRNA Expression in NHLFs—NHLFs express both Sulf1 and Sulf2 at relatively comparable basal levels (based on Ct values of real-time RT-PCR results, not shown). Upon TGF-β1 stimulation, Sulf1 expression was induced in a dose-dependent manner (Fig. 1A). Treatment of NHLFs with as little as 0.05 ng/ml TGF-β1 resulted in an increased level of Sulf1 mRNA, and 0.5 ng/ml TGF-β1 induced the maximum response (7-8-fold). In contrast, the expression of Sulf2 was reduced by TGF-β1, with about 50% remaining at 0.5 ng/ml (Fig. 1A). To further study the kinetics of TGF-β1 induction of Sulf1 mRNA, NHLFs were treated with 0.5 ng/ml TGF-β1 for different time periods, and Sulf1 mRNA levels were analyzed. This time-course study revealed a relatively slow induction of Sulf1 by TGF-β1 (Fig. 1B), with the earliest significant increase observed at 12 h after TGF-β1 exposure. Maximum induction was observed at 24 h, which was maintained at 48 h, the longest time point in this experiment.
FIGURE 1.
TGF-β1 induces Sulf1 expression in NHLFs in a dose- and time-dependent manner. A, NHLFs were rendered quiescent in FBM plus 0.2% BSA overnight before treatment with TGF-β1 at the indicated concentrations. Total RNA was isolated 24 h later and analyzed for Sulf1 and Sulf2 expression by real-time RT-PCR. -Fold change normalized to the 36B4 housekeeping gene is shown. Data are representative of two independent experiments in duplicate. p < 0.05 (*) and p < 0.01 (**) compared with control treatment with media alone. B, quiescent NHLFs were treated with TGF-β1 at 0.5 ng/ml. Total RNA from control (white bars) and TGF-β1 (black bars)-treated cells was isolated at different time points, and Sulf1 expression was analyzed as described in A. p < 0.05 (*) and p < 0.001 (**) compared with control at each time point.
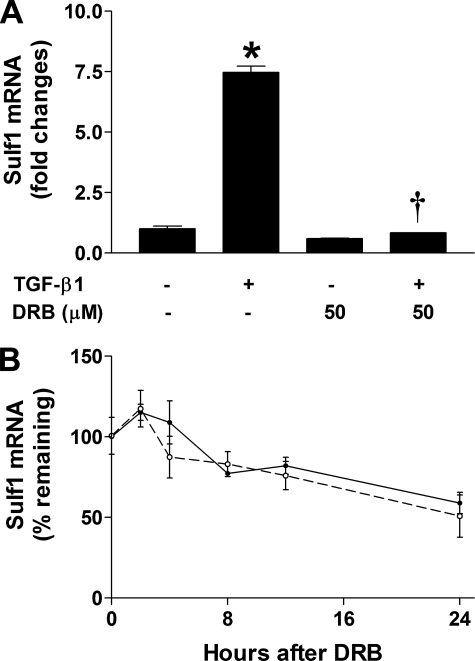
Increased Sulf1 mRNA levels may result from enhanced transcriptional activity, increased Sulf1 mRNA stability, or both. To examine the mechanisms of Sulf1 up-regulation by TGF-β1, we pretreated NHLFs with 50 μm DRB, an RNA polymerase II inhibitor, before the addition of TGF-β1 (in the presence of DRB) and analyzed Sulf1 mRNA expression 24 h later. As shown in Fig. 2A, inhibition of RNA polymerase II by DRB totally abolished TGF-β1-induced Sulf1 expression, which suggests that increased transcription is indeed responsible for TGF-β1-induced Sulf1 up-regulation. To examine whether TGF-β1 can stabilize Sulf1 mRNA, NHLFs were stimulated with TGF-β1 for 20 h before the addition of DRB to inhibit new mRNA synthesis, and the remaining Sulf1 mRNA was analyzed over time to determine the degradation rate of Sulf1. The results show (Fig. 2B) that Sulf1 has a relatively long half-life (∼24 h), and importantly, Sulf1 mRNA degrades at approximately the same rate in the presence or absence of TGF-β1. Taken together, the above results indicate that transcriptional activation rather than mRNA stabilization is responsible for increased Sulf1 mRNA levels by TGF-β1.
FIGURE 2.
TGF-β1 induces Sulf1 expression through increased transcriptional activity. A, inhibition of mRNA synthesis blocks TGF-β1-induced Sulf1 expression. Quiescent NHLFs were pretreated with the RNA polymerase II inhibitor, DRB (50 μm), for 1 h before the addition of TGF-β1 (0.5 ng/ml). Sulf1 expression was analyzed 24 h later as in Fig. 1A.*, p < 0.001 compared with control treatment with media alone; †, p < 0.001 compared with cells treated with TGF-β1 without DRB. B, Sulf1 mRNA has a similar half-life in the presence or absence of TGF-β1. Quiescent NHLFs were stimulated with (solid line) or without (dashed line) TGF-β1 (0.5 ng/ml) for 20 h before the addition of DRB (50 μm). Total RNA was extracted at the indicated time points, and the remaining Sulf1 mRNA was measured by real-time RT-PCR. Results were expressed as the percentage of mRNA remaining relative to the corresponding level before the addition of DRB.
The relative slow induction of Sulf1 by TGF-β1 (Fig. 1B) suggests that de novo protein synthesis might be required for TGF-β1-induced Sulf1 expression. To test this hypothesis, we stimulated NHLFs with TGF-β1 in the presence of protein translation inhibitors. Two protein translation inhibitors, cycloheximide and emetine, were used. Unexpectedly, the addition of either cycloheximide or emetine alone resulted in increased Sulf1 mRNA levels (2.8 ± 0.33- and 7.68 ± 0.45-fold increases over control at 10 μg/ml cycloheximide and 10 μg/ml emetine, respectively). The addition of both protein translation inhibitor and TGF-β1 resulted in a further increase (in the case of cycloheximide) or similar level (in the case of emetine) of Sulf1 mRNA. These results suggest that the transcription of Sulf1 mRNA may be under active suppression in unstimulated cells, and the addition of protein translation inhibitors blocks the synthesis of the protein factor(s) responsible for this suppression. TGF-β1 may induce Sulf1 expression through removal of this suppression as well, and the identity of the Sulf1 transcriptional suppressor(s) requires further investigation.
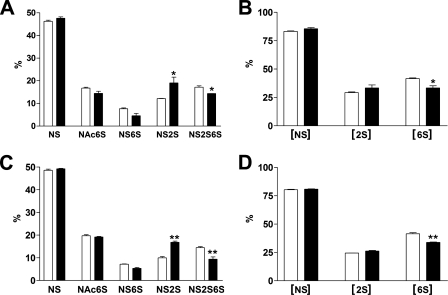
TGF-β1 Induces HS 6-O-Desulfation in NHLFs—To determine whether TGF-β1-induced Sulf1 mRNA expression leads to increased Sulf1 activity and thereby reduces 6-O-sulfation levels, sulfated disaccharide composition was analyzed by metabolic labeling of HS with sodium [35S]sulfate followed by HPLC analysis. As shown in Fig. 3, A and C, a significant reduction of trisulfated ΔUA2S-GlcNS6S (NS2S6S) was observed in TGF-β1-treated cells at both 24 h (A) and 48 h (C), with concurrent increase in ΔUA2S-GlcNS (NS2S). This is consistent with previous reports showing that Sulf1 preferentially removes 6-O-sulfates from trisulfated IdoA2S-GlcNS6S-containing sequences found within the S-domains of HS (39, 40). Fig. 3, B and D, show the percentage of disaccharides containing N-, 2-O-, or 6-O-sulfates, and the results indicate that only the level of the 6-O-sulfation was reduced by Sulf1 with the N- and 2-O-sulfation levels essentially unchanged. Disaccharide analysis of HS in the media from TGF-β1 treated cells showed similar results (data not shown). These structural data indicate that TGF-β1-mediated induction of Sulf1 indeed resulted in 6-O-desulfation of HS from NHLFs.
FIGURE 3.
TGF-β1 induces HS 6-O-desulfation in NHLFs. HS from control or TGF-β1-treated NHLFs were metabolically labeled overnight with 50 μCi/ml sodium [35S]sulfate in FBM plus 0.2% BSA. HS from both the media and cells were then isolated and digested to disaccharides as described under “Experimental Procedures.” The unsaturated disaccharides were resolved by ion-pairing reverse-phase HPLC. Data shown are sulfated disaccharide compositions (% of total) of cell-associated HS from control (white bars) or TGF-β1 (black bars)-treated NHLFs at 24 h (A and B) and 48 h (C and D). HS from the media gave similar results (not shown). S, sulfate; ΔUA, unsaturated uronic acid; GlcN, glucosamine; NS, ΔUA-GlcNS; Ac, acetyl; NAc6S, ΔUA-GlcNAc6S; NS6S, ΔUA-GlcNS6S; NS2S, ΔUA2S-GlcNS; NS2S6S, ΔUA2S-GlcNS6S; [NS], NS + NS6S + NS2S + NS2S6S; [2S], NS2S + NS2S6S; [6S], NAc6S + NS6S + NS2S6S. *, p < 0.05; **, p < 0.001.
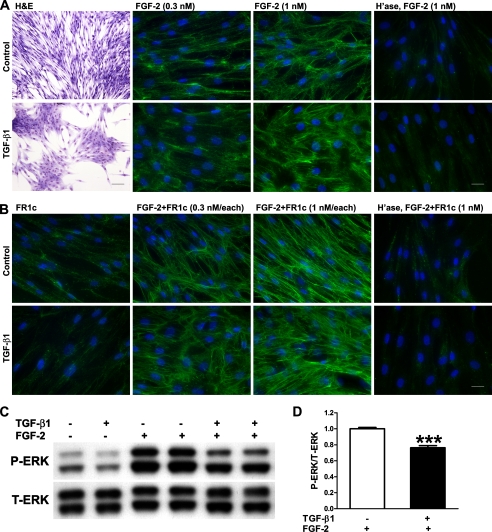
Dynamic changes in the HS sulfation state on the cell surface are directly linked with modulations in FGF signal transduction. Both biochemical and crystal structure studies have indicated the requirement of 6-O-sulfation for FGF-2-induced dimerization and activation of FGF receptor 1 (41, 42). Increased signaling and mitogenesis upon FGF stimulation have been reported in embryonic fibroblasts derived from the Sulf knock-out mice (43). Conversely, reduced FGF signaling was observed in Sulf-overexpressing cells as evidenced by reduced ERK activation upon FGF stimulation (40). To study the effect of TGF-β1-induced Sulf1 expression on FGF-2 signaling in NHLFs, we examined HS-dependent FGF-2 binding, FGF-2·FR1c complex formation, and FGF-2-induced ERK activation in these cells (Fig. 4). As expected, the expression of Sulf1 did not affect FGF-2 binding to cell surface HS (Fig. 4A) as 6-O-sulfation is not required in this interaction (40, 41). Different from the previous study (40), no differences were seen in HS-dependent FGF-2·FR1c complex formation between control and Sulf1 expressing (TGF-β1-treated) NHLFs (Fig. 4B). This is unexpected as 6-O-sulfation is required for FGF-2·HS·FR1c ternary complex formation. However, consistent with the previous study (40), the expression of Sulf1 did lead to a reduction (consistent, although modest) in FGF-2-induced ERK activation (Fig. 4, C and D).
FIGURE 4.
FGF-2-induced ERK activation is reduced in TGF-β1-treated NHLFs without changes in HS-dependent FGF-2 binding and FGF-2·FR1c complex formation. A, cell morphology (hematoxylin and eosin (H&E) staining; scale bar, 100 μm) and FGF-2 binding (scale bar, 25 μm) in control and TGF-β1-treated (0.5 ng/ml, 48 h) NHLFs. NHLFs adopted a flattened and contractile phenotype upon TGF-β1 treatment. No significant changes were observed in HS-dependent FGF-2 binding at the concentrations tested. Prior treatment with heparitinases (H'ase) abolished the FGF-2 binding, confirming that the observed binding is HS-dependent. B, FR1c binding and FGF-2·HS·FR1c complex formation (scale bar, 25 μm) in control and TGF-β1-treated (0.5 ng/ml, 48 h) NHLFs. FR1c alone did not bind strongly to cell surface HS with or without TGF-β1 treatment, and no significant differences were observed in FGF-2·HS·FR1c complex formation at the concentrations tested. Prior heparitinase treatment abolished the FGF-2·FR1c complex. C, FGF-2-induced ERK activation is reduced in TGF-β1-treated NHLFs. Control or TGF-β1-treated (0.5 ng/ml, 48 h) NHLFs were stimulated with FGF-2 (10 ng/ml) for 3 min. Total (T) and phosphorylated (P) ERK were assessed by Western blotting. D, quantification of ERK activation (ratio of phosphorylated ERK/total ERK). Phosphorylated ERK/total ERK ratio in control NHLFs treated with FGF-2 was set at 1.0. Combined results from three independent experiments in duplicate are shown. ***, p < 0.001.
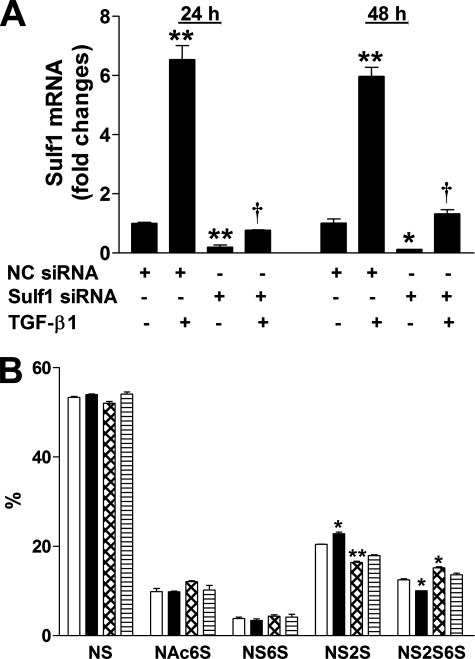
Sulf1 siRNA Blocks TGF-β1-induced Sulf1 Expression in NHLFs—To investigate the role of Sulf1 in TGF-β1-induced fibrotic responses in NHLFs, we used siRNA to block the overexpression of Sulf1. Initially, we tested three siRNA sequences against Sulf1 (Ambion, catalog# 16708A), and all three sequences reduced Sulf1 expression to variable extents (not shown). The sequence that resulted in the highest and most consistent knockdown (siRNA ID #121032) was used in all subsequent experiments. Fig. 5A shows Sulf1 mRNA levels in NHLFs transfected with the negative control (NC, sequence that does not target any known mRNA) siRNA or siRNA specific against Sulf1. The results show that Sulf1 siRNA significantly reduced base-line Sulf1 mRNA levels at both 24 and 48 h (10∼20% remaining compared with NC siRNA-treated cells); induction of Sulf1 by TGF-β1 in NC cells was similar to NHLFs without any treatment, indicating that the transfection procedure did not alter NHLF response to TGF-β1, and most importantly, Sulf1 siRNA transfection reduced Sulf1 mRNA to control levels at both 24 and 48 h after TGF-β1 stimulation.
FIGURE 5.
Sulf1 siRNA blocks TGF-β1-induced Sulf1 expression and increases HS 6-O-sulfation. A, NC or Sulf1 siRNA-transfected NHLFs were stimulated with TGF-β1 (0.5 ng/ml) for 24 or 48 h, and Sulf1 expression was analyzed by real-time RT-PCR as described in Fig. 1A. p < 0.05 (*) and p < 0.01 (**) in comparison to NC siRNA-transfected cells without TGF-β1 treatment; †, p < 0.01 compared with NC siRNA-transfected cells treated with TGF-β1. B, sulfated disaccharide compositions (% of total) of cell associated HS from NC siRNA-transfected (white bars, control; black bars, TGF-β1 treated) or Sulf1 siRNA-transfected (cross-hatch bars, control; horizontal bars, TGF-β1 treated) NHLFs at 48 h. p < 0.01 (*) and p < 0.001 (**) in comparison to NC siRNA-transfected cells without TGF-β1 treatment. NS, ΔUA-GlcNS.
We also analyzed sulfated disaccharide composition in the siRNA-transfected NHLFs, and the results (Fig. 5B) are consistent with Sulf1 mRNA levels in these cells (Fig. 5A). In NC siRNA-transfected cells, a reduction of NS2S6S with concurrent increase in NS2S was observed upon TGF-β1 treatment, similar to untransfected NHLFs (Fig. 3). In Sulf1 siRNA-transfected cells without TGF-β1 treatment, an increase in NS2S6S with concurrent decrease in NS2S was seen, reflecting the reduced basal Sulf1 level (Fig. 5A). In Sulf1 siRNA-transfected cells treated with TGF-β1, the level of NS2S6S was similar to control level (NC siRNA-transfected cells without TGF-β1 treatment). It is important to note that under our experimental conditions, Sulf1 mRNA is still increased by TGF-β1 in Sulf1 siRNA-transfected cells, but the Sulf1 level is equivalent to NC siRNA-transfected cells without TGF-β1 treatment.
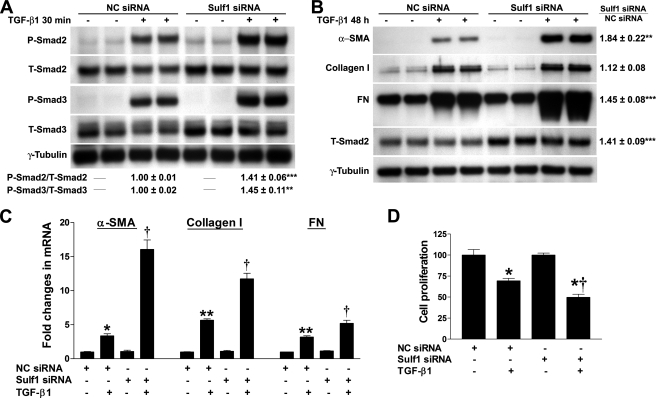
Sulf1 Functions as a Negative Regulator of TGF-β1-induced Fibrotic Responses in NHLFs—Smad signaling is central for the biological functions of TGF-β1 (44). Upon binding to its receptors, TGF-β1 initiates Smad2 and Smad3 phosphorylation (44). To study the role of Sulf1 in TGF-β1-induced fibrogenesis, we first examined Smad2 and Smad3 phosphorylation upon TGF-β1 stimulation in NC and Sulf1 siRNA-treated NHLFs. As shown in Fig. 6A, both Smad2 and Smad3 phosphorylation was enhanced in Sulf1 siRNA-treated cells compared with NC cells. Down-regulation of total Smad2 and Smad3 protein levels was observed in both NC and Sulf1 siRNA-transfected cells after TGF-β1 treatment, as expected after ligand stimulation. Interestingly, the basal level of total Smad2 level was elevated in Sulf1 siRNA-transfected cells. The increase in the total protein level of Smad3 was not consistent.
FIGURE 6.
Sulf1 silencing renders NHLFs more responsive to TGF-β1. A, NC or Sulf1 siRNA-transfected NHLFs were treated with TGF-β1 (0.5 ng/ml) for 30 min. Total protein extracts were prepared, and Smad2/3 protein levels and Smad2/3 activation (phosphorylation) were assessed by Western blotting.γ-Tubulin was used as the loading control. Levels of Smad activation (ratio of phosphorylated (P)-Smad/total (T)-Smad) were calculated based on data from three independent experiments in duplicate, and the phosphorylated Smad/total Smad ratio in NC siRNA-transfected cells treated with TGF-β1 was set at 1.0. **, p < 0.01; ***, p < 0.001. B, NC or Sulf1 siRNA-transfected NHLFs were treated with TGF-β1 (0.5 ng/ml) for 48 h. Total protein extracts were prepared, and the protein levels of α-SMA, type I collagen, FN, and total Smad2 were assessed by Western blotting. -Fold changes in TGF-β1-induced α-SMA, type I collagen, FN expression in Sulf1 siRNA-transfected versus NC siRNA-transfected cells normalized to γ-tubulin were shown. -Fold changes in total Smad2 expression (basal, without TGF-β1 treatment) normalized to γ-tubulin was also shown. Statistics were based on data from three independent experiments in duplicate. **, p < 0.01; ***, p < 0.001. C, the expression of α-SMA, type I collagen, and FN were also assessed at mRNA level by real-time RT-PCR as described in Fig. 1A. p < 0.05 (*) and p < 0.01 (**) in comparison to NC siRNA-transfected cells without TGF-β1 treatment; †, p < 0.05 compared with NC siRNA-transfected cells treated with TGF-β1. D, NC or Sulf1 siRNA-transfected NHLFs were treated with TGF-β1 (0.5 ng/ml) for 48 h, and cell proliferation was examined using CellTiter 96 nonradioactive cell proliferation assay and expressed as the percentage of the control without TGF-β1 treatment. *, p < 0.001 compared with NC siRNA or Sulf1 siRNA-transfected cells without TGF-β1 treatment; †, p < 0.001 compared with NC siRNA-transfected cells treated with TGF-β1.
We then examined the expression of downstream TGF-β1-responsive genes including α-SMA, type I collagen, and fibronectin (FN). Consistent with the observed enhancement of Smad2/3 signaling, protein levels of both α-SMA and FN were significantly elevated in Sulf1 siRNA-treated cells 48 h after TGF-β1 stimulation (Fig. 6B). In contrast, only a slight increase in type I collagen was observed in Sulf1 knockdown cells. Interestingly, total Smad2 protein level remained elevated in Sulf1 siRNA-treated cells at 48 h. Similar results were observed at 24 h post-TGF-β1 stimulation (data not shown). The protein levels of type I collagen and FN in the conditioned media were also examined, and the results were similar to those from the total cell extracts (not shown). We also examined mRNA levels of α-SMA, type I collagen, and FN by real-time RT-PCR, which showed enhanced expression of all three in Sulf1 siRNA-treated cells (Fig. 6C).
TGF-β1 exerts anti-proliferative effects on NHLFs (Fig. 6D). Consistent with enhanced Smad2/3 signaling, the anti-proliferative action of TGF-β1 was also enhanced by pretreatment of NHLFs with Sulf1 siRNA (50.3 ± 3.6% reduction in proliferation in Sulf1 siRNA-treated cells compared with 30.6 ± 2.9% reduction in NC siRNA-treated cells, p < 0.001).
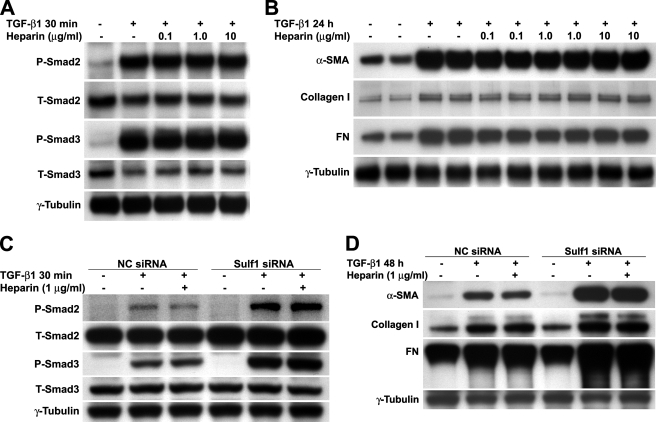
Heparin Does Not Enhance TGF-β1 Signaling in NHLFs—Lyon et al. (32) have shown in a previous study that heparin and highly sulfated HS potentiate the biological activity of TGF-β1 through antagonism of the binding and inactivation of TGF-β1 by α2-macroglobulin in the serum rather than by modulation of growth factor-receptor interactions, as is the case in FGF/FGF receptor signaling. To examine whether the 6-O-sulfation state directly influences NHLF response to TGF-β1, we treated these cells with TGF-β1 in the presence of heparin. In all the experiments presented in this study, serum-free media were used so that our results are not complicated by any component from the serum. Consistent with the previous study, heparin (0.1 to 10 μg/ml) did not alter the level of Smad2/3 phosphorylation (Fig. 7A) nor did it change the protein levels of α-SMA, type I collagen, or FN (Fig. 7B) induced by TGF-β1. Heparin alone had no effect (not shown).
FIGURE 7.
Heparin does not directly enhance TGF-β1 signaling in NHLFs. A, quiescent NHLFs were treated with TGF-β1 (0.5 ng/ml) for 30 min in the presence or absence of heparin at indicated concentrations. Total protein extracts were prepared, and Smad2/3 protein levels and Smad2/3 activation (phosphorylation) were assessed by Western blotting. P, phosphorylated; T, total. B, NHLFs were treated as in A. Total protein extracts were isolated 24 h later, and the expression levels of α-SMA, type I collagen, and FN were assessed by Western blotting. C, NC siRNA and Sulf1 siRNA-transfected NHLFs were treated with TGF-β1 (0.5 ng/ml) for 30 min in the presence or absence of 1 μg/ml heparin. Total protein extracts were prepared, and Smad2/3 protein levels and Smad2/3 activation (phosphorylation) were assessed by Western blotting. D, NC siRNA and Sulf1 siRNA-transfected NHLFs were treated as in C. Total protein extracts were isolated 48 h later, and the expression levels of α-SMA, type I collagen, and FN were assessed by Western blotting.
We also examined the TGF-β1 responses in siRNA-transfected cells in the presence or absence of heparin. Similar to untransfected cells (Fig. 7, A and B), heparin did not alter TGF-β1-induced Smad activation (Fig. 7C) or downstream gene expression (Fig. 7D) in either NC siRNA- or Sulf1 siRNA-transfected cells.
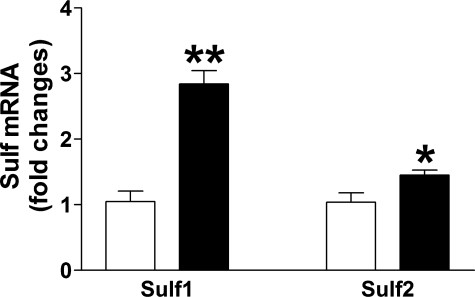
TGF-β1 Induces Sulf1 Expression in Vivo—Previous studies have shown that expression of active TGF-β1 in the lungs of mice or rats by means of adenoviral vectors results in inflammation and persistent fibrosis (6, 37). To test whether TGF-β1 induces Sulf1 expression in vivo, we adopted this model. Consistent with the in vitro findings, mice that received adenovirus encoding active TGF-β1 had a 3-fold increase in the expression of Sulf1 in the lungs compared with mice treated with the GFP control vector (Fig. 8). The expression of Sulf1 in lung fibroblasts was likely to be higher since whole lung RNA extracts were used in the analysis. A slight but significant up-regulation of Sulf2 (45% increase over control) was also observed in TGF-β1-treated mice. In addition, the expression of connective tissue growth factor and type I collagen were up-regulated 11.5- and 8.6-fold, respectively, in TGF-β1-treated mice (data not shown), confirming the in vivo fibrogenic activity of the TGF-β1 virus.
FIGURE 8.
TGF-β1 induces lung Sulf1 expression in vivo. C57BL6 mice were treated with adenovirus encoding GFP (control, white bars) or active TGF-β1 (black bars). Mice were sacrificed 5 days later and total RNA was isolated from the right lungs. Sulf1 and Sulf2 expression were assessed by real-time RT-PCR. Data shown are the mean ± S.E. of five mice from each group. -Fold changes of Sulf expression were normalized to 18S. *, p < 0.05; **, p < 0.001.
DISCUSSION
The HS 6-O-endosulfatases, Sulf1 and Sulf2, are unique in that they edit HS sulfation patterns on the cell surface and in the ECM, and thus, readily modulate HS-dependent cell signaling events and ECM remodeling. In this study we show for the first time that TGF-β1, a profibrotic cytokine, induces Sulf1 expression in NHLFs and in murine lungs in vivo and that Sulf1 may function as a negative regulator of TGF-β1-induced fibrogenesis.
The enhanced expression of Sulf1 in NHLFs was accompanied by decreased expression of Sulf2. Similar compensatory changes have been reported in mouse embryonic fibroblasts derived from Sulf knock-out animals (43), supporting the notion that Sulf1 and Sulf2 are co-regulated at the level of gene expression. Further kinetic studies of Sulf1 expression in NHLFs suggest that TGF-β1 induces Sulf1 expression at the level of gene transcription (Fig. 2), possibly through the removal of transcriptional suppression as suggested by our data using protein synthesis inhibitors. The increased Sulf1 expression was accompanied by HS 6-O-desulfation as revealed by disaccharide analysis. Although the extent of 6-O-desulfation is less dramatic (20∼30% reduction compared with the control) than what was previously reported in cells overexpressing Sulf1 or Sulf2 (39, 40), we believe that our results are more biologically relevant, as the expression of Sulf1 is induced by a cytokine rather than forced expression in previous studies. Although no quantitative data were shown in the previous studies (39, 40), it is very likely that a much higher expression of Sulf1 (compared with the ∼7-fold induction by TGF-β1 in NHLFs in our study) was achieved with the forced expression.
Sulfation at the 6-O-position on the HS chain has been shown to be important for a number of developmental processes as well as pathological conditions. The fact that it is the only known sulfate moiety post-synthetically edited underscores the importance of the Sulfs in the regulation of HS-dependent biological processes. Multiple studies have demonstrated that a gain or loss of 6-O-sulfation carried out by the Sulfs can alter the outcome of HS-ligand interactions. The morphogen Wnt shows high affinity binding to 6-O-sulfated HS that prevents its functional interactions with its cognate receptor Frizzled. By removing 6-O-sulfates, QSulf1 positively influences Wnt signaling by restoring active Wnt-Frizzled interaction (39), which in turn leads to the activation of MyoD, a Wnt-induced regulator of muscle specification (26). Sulf activity also has an activating effect on bone morphogenetic protein (BMP) signaling by releasing HS-associated BMP antagonist Noggin from the cell surface, thus allowing BMP to interact with its cognate receptor (45). In contrast, Sulf activity inhibits FGF-2 signaling by removing the 6-O-sulfate moiety necessary for high affinity FGF2-HS-FGF receptor ternary complex formation. This negative regulatory function results in inhibition of embryonic mesoderm formation and angiogenesis (46). Consistent with previous studies, the expression of Sulf1 induced by TGF-β1 results in reduced FGF-2 signaling as revealed by reduced ERK activation in NHLFs. Surprisingly, we did not observe changes in FGF-2·HS·FR1c complex formation in TGF-β1-treated cells. This could be due to the relative moderate 6-O-desulfation induced by TGF-β1 or the limitation of the binding assay (sensitivity).
To examine whether Sulf1 exerts a positive or a negative effect on TGF-β1-induced fibrogenesis, we blocked TGF-β1-induced Sulf1 expression by siRNA. Enhanced Smad2/3 phosphorylation as well as enhanced expression of α-SMA and FN in Sulf1 knockdown cells indicate that Sulf1 functions as a negative regulator of TGF-β1. Although the expression of type I collagen was enhanced at the mRNA level, only a slight increase in type I collagen protein was observed in Sulf1 knockdown cells, suggesting that additional factors might be involved at the level of protein translation. Indeed, the expression of type I collagen is posttranscriptionally regulated at multiple steps (47).
To further test whether 6-O-sulfates directly enhance TGF-β1 signaling, we treated NHLFs with TGF-β1 in the presence of heparin, a highly sulfated form (including 6-O-sulfates) of HS. Consistent with a previous report (32), heparin did not alter Smad2/3 phosphorylation or the expression of downstream TGF-β1-responsive genes. This indicates that 6-O-sulfation of HS does not directly influence the interaction between TGF-β1 and its receptors. Alternatively, cell surface localization of the 6-O-sulfates is important in modulating TGF-β1 function.
TGF-β1 is the second molecule so far identified to induce Sulf1 expression. The first molecule known to induce Sulf1 is Shh, as Sulf1 was first discovered as a Shh-responsive gene (26). Different from Shh whose signaling is not influenced by Sulf1, Sulf1 induced by TGF-β1 negatively regulates TGF-β1-mediated cellular responses as reported in this study. The mechanism of increased expression and activation of Smad2/3 in Sulf1 knockdown cells is unclear. Regulation at transcriptional level is possible, as a recent study showed that overexpression of Sulf1 in hepatocellular carcinoma cells increases histone H4 acetylation, and down-regulation of Sulf1 using shRNA constructs conversely decreased it, thus changing the balance between histone deacetylase and histone acetyltransferase activity in the nucleus (48). Alteration at cell surface TGF-β receptor level is also possible, as overexpression of syndecan-2, a transmembrane HS proteoglycan, in renal papillary fibroblasts results in increased expression of type I and type II TGF-β receptors (TβRI and TβRII) (49). Finally, Sulf1 expression level could also be linked to the balance between stimulatory and inhibitory Smad signaling. These possibilities are currently being investigated in our laboratory.
In our in vivo experiments, TGF-β1 treatment resulted in increases in both Sulf1 and Sulf2 expression. This seems to be contradictory to our in vitro data showing that TGF-β1 induces an increase in Sulf1 and a decrease in Sulf2 expression in NHLFs. In contrast to fibroblasts, which express both Sulf1 and Sulf2, A549 cells, adenocarcinoma cells originated from alveolar type II epithelial cells, mainly express Sulf2, and interestingly enough, TGF-β1 augments Sulf2 expression ∼5-fold in these cells.4 It is possible that TGF-β1 induces Sulf2 expression in alveolar type II cells in vivo, resulting in the observed increase in Sulf2 expression in these mice.
In summary, we report in the present study that TGF-β1 induces Sulf1 expression in both cultured lung fibroblasts and in murine lungs in vivo and that Sulf1 may function as a negative regulator of TGF-β1-induced fibrotic responses. It will be important to define whether Sulf1 is induced in patients suffering from PF. Future studies using Sulf knock-out animals are also justified to clarify the roles of the Sulfs in the pathobiology of PF.
Acknowledgments
We thank Dr. Patricia Sime for providing the adenoviral stocks encoding AdGFP and AdTGF-β1223/225.
This work was supported, in whole or in part, by National Institutes of Health Grant HL083480. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PF, pulmonary fibrosis; ΔUA, unsaturated uronic acid; α-SMA, α-smooth muscle actin; BSA, bovine serum albumin; DRB, 5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole; ECM, extracellular matrix; FBM, fibroblast basal medium; FGF, fibroblast growth factor; FN, fibronectin; HS, heparan sulfate; IdoA, iduronic acid; NC, negative control; NHLF, normal human lung fibroblast; NS, ΔUA-GlcNS; NS2S, ΔUA2S-GlcNS; NS2S6S, ΔUA2S-GlcNS6S; NS6S, ΔUA-GlcNS6S; PBS, phosphate-buffered saline; RT, reverse transcription; Shh, Sonic hedgehog; TGF-β1, transforming growth factor-β1; HPLC, high performance liquid chromatography; MAPK, mitogen-activated protein kinase; ERK, extracellular signal-regulated kinase; PBS, phosphate-buffered saline; siRNA, small interference RNA; GFP, green fluorescent protein.
X. Yue and J. A. Lasky, unpublished data.
References
- 1.Lasky, J. A., and Brody, A. R. (2000) Environ. Health Perspect. 108 Suppl 4, 751-762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khalil, N., O'Connor, R. N., Flanders, K. C., and Unruh, H. (1996) Am. J. Respir. Cell Mol. Biol. 14 131-138 [DOI] [PubMed] [Google Scholar]
- 3.Broekelmann, T. J., Limper, A. H., Colby, T. V., and McDonald, J. A. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 6642-6646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao, J., Shi, W., Wang, Y.-L., Chen, H., Bringas, P., Jr., Datto, M. B., Frederick, J. P., Wang, X.-F., and Warburton, D. (2002) Am. J. Physiol. Lung Cell. Mol. Physiol. 282 585-593 [DOI] [PubMed] [Google Scholar]
- 5.Raghow, B., Irish, P., and Kang, A. H. (1989) J. Clin. Investig. 84 1836-1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sime, P. J., Xing, Z., Graham, F. L., Csaky, K. G., and Gauldie, J. (1997) J. Clin. Investig. 100 768-776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phan, S. H. (2002) Chest 122 Suppl. 6, 286-289 [Google Scholar]
- 8.Zhang, H. Y., Gharaee-Kermani, M., Zhang, K., Karmiol, S., and Phan, S. H. (1996) Am J. Pathol. 148 527-537 [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhn, C., and McDonald, J. A. (1991) Am J. Pathol. 138 1257-1265 [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang, K., Rekhter, M. D., Gordon, D., and Phan, S. H. (1994) Am J. Pathol. 145 114-125 [PMC free article] [PubMed] [Google Scholar]
- 11.Hetzel, M., Bachem, M., Anders, D., Trischler, G., and Faehling, M. (2005) Lung 183 225-237 [DOI] [PubMed] [Google Scholar]
- 12.Zhang, H. Y., and Phan, S. H. (1999) Am. J. Respir. Cell Mol. Biol. 21 658-665 [DOI] [PubMed] [Google Scholar]
- 13.Rapraeger, A. C. (2002) Methods Cell Biol. 69 83-109 [DOI] [PubMed] [Google Scholar]
- 14.Esko, J. D., and Lindahl, U. (2001) J. Clin. Investig. 108 169-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esko, J. D., and Selleck, S. B. (2002) Annu. Rev. Biochem. 71 435-471 [DOI] [PubMed] [Google Scholar]
- 16.Nakato, H., and Kimata, K. (2002) Biochim. Biophys. Acta 1573 312-318 [DOI] [PubMed] [Google Scholar]
- 17.Yue, X., Schultheiss, T. M., McKenzie, E. A., and Rosenberg, R. D. (2004) Glycobiology 14 745-755 [DOI] [PubMed] [Google Scholar]
- 18.Allen, B. L., and Rapraeger, A. C. (2003) J. Cell Biol. 163 637-648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walz, A., McFarlane, S., Brickman, Y. G., Nurcombe, V., Bartlett, P. F., and Holt, C. E. (1997) Development 124 2421-2430 [DOI] [PubMed] [Google Scholar]
- 20.Brickman, Y. G., Ford, M. D., Gallagher, J. T., Nurcombe, V., Bartlett, P. F., and Turnbull, J. E. (1998) J. Biol. Chem. 273 4350-4359 [DOI] [PubMed] [Google Scholar]
- 21.Feyzi, E., Saldeen, T., Larsson, E., Lindahl, U., and Salmivirta, M. (1998) J. Biol. Chem. 273 13395-13398 [DOI] [PubMed] [Google Scholar]
- 22.Jayson, G. C., Lyon, M., Paraskeva, C., Turnbull, J. E., Deakin, J. A., and Gallagher, J. T. (1998) J. Biol. Chem. 273 51-57 [DOI] [PubMed] [Google Scholar]
- 23.Safaiyan, F., Lindahl, U., and Salmivirta, M. (1998) Eur. J. Biochem. 252 576-582 [DOI] [PubMed] [Google Scholar]
- 24.Habuchi, H., Nagai, N., Sugaya, N., Atsumi, F., Stevens, R. L., and Kimata, K. (2007) J. Biol. Chem. 282 15578-15588 [DOI] [PubMed] [Google Scholar]
- 25.Lamanna, W. C., Kalus, I., Padva, M., Baldwin, R. J., Merry, C. L., and Dierks, T. (2007) J. Biotechnol. 129 290-307 [DOI] [PubMed] [Google Scholar]
- 26.Dhoot, G. K., Gustafsson, M. K., Ai, X., Sun, W., Standiford, D. M., and Emerson, C. P., Jr. (2001) Science 293 1663-1666 [DOI] [PubMed] [Google Scholar]
- 27.Morimoto-Tomita, M., Uchimura, K., Werb, Z., Hemmerich, S., and Rosen, S. D. (2002) J. Biol. Chem. 277 49175-49185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohto, T., Uchida, H., Yamazaki, H., Keino-Masu, K., Matsui, A., and Masu, M. (2002) Genes Cells 7 173-185 [DOI] [PubMed] [Google Scholar]
- 29.Uchimura, K., Morimoto-Tomita, M., Bistrup, A., Li, J., Lyon, M., Gallagher, J., Werb, Z., and Rosen, S. D. (2006) BMC Biochem. 7 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai, J. P., Chien, J. R., Moser, D. R., Staub, J. K., Aderca, I., Montoya, D. P., Matthews, T. A., Nagorney, D. M., Cunningham, J. M., Smith, D. I., Greene, E. L., Shridhar, V., and Roberts, L. R. (2004) Gastroenterology 126 231-248 [DOI] [PubMed] [Google Scholar]
- 31.McCaffrey, T. A., Falcone, D. J., Vicente, D., Du, B., Consigli, S., and Borth, W. (1994) J. Cell. Physiol. 159 51-59 [DOI] [PubMed] [Google Scholar]
- 32.Lyon, M., Rushton, G., and Gallagher, J. T. (1997) J. Biol. Chem. 272 18000-18006 [DOI] [PubMed] [Google Scholar]
- 33.McCaffrey, T. A., Falcone, D. J., and Du, B. (1992) J. Cell. Physiol. 152 430-440 [DOI] [PubMed] [Google Scholar]
- 34.Chen, Y., Shi-wen, X., van Beek, J., Kennedy, L., McLeod, M., Renzoni, E. A., Bou-Gharios, G., Wilcox-Adelman, S., Goetinck, P. F., Eastwood, M., Black, C. M., Abraham, D. J., and Leask, A. (2005) Am. J. Pathol. 167 1699-1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak, K. J., and Schmittgen, T. D. (2001) Methods 25 402-408 [DOI] [PubMed] [Google Scholar]
- 36.Sullivan, D. E., Ferris, M., Pociask, D., and Brody, A. R. (2005) Am. J. Respir. Cell Mol. Biol. 32 342-349 [DOI] [PubMed] [Google Scholar]
- 37.Warshamana, G. S., Pociask, D. A., Fisher, K. J., Liu, J. Y., Sime, P. J., and Brody, A. R. (2002) Int. J. Exp. Pathol. 83 183-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lakatos, H. F., Burgess, H. A., Thatcher, T. H., Redonnet, M. R., Hernady, E., Williams, J. P., and Sime, P. J. (2006) Exp. Lung Res. 32 181-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ai, X., Do, A. T., Lozynska, O., Kusche-Gullberg, M., Lindahl, U., and Emerson, C. P., Jr. (2003) J. Cell Biol. 162 341-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai, Y., Yang, Y., MacLeod, V., Yue, X., Rapraeger, A. C., Shriver, Z., Venkataraman, G., Sasisekharan, R., and Sanderson, R. D. (2005) J. Biol. Chem. 280 40066-40073 [DOI] [PubMed] [Google Scholar]
- 41.Guimond, S., Maccarana, M., Olwin, B. B., Lindahl, U., and Rapraeger, A. C. (1993) J. Biol. Chem. 268 23906-23914 [PubMed] [Google Scholar]
- 42.Schlessinger, J., Plotnikov, A. N., Ibrahimi, O. A., Eliseenkova, A. V., Yeh, B. K., Yayon, A., Linhardt, R. J., and Mohammadi, M. (2000) Mol. Cell 6 743-750 [DOI] [PubMed] [Google Scholar]
- 43.Lamanna, W. C., Baldwin, R. J., Padva, M., Kalus, I., Ten Dam, G., van Kuppevelt, T. H., Gallagher, J. T., von Figura, K., Dierks, T., and Merry, C. L. (2006) Biochem. J. 400 63-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Massague, J., and Gomis, R. R. (2006) FEBS Lett. 580 2811-2820 [DOI] [PubMed] [Google Scholar]
- 45.Viviano, B. L., Paine-Saunders, S., Gasiunas, N., Gallagher, J., and Saunders, S. (2004) J. Biol. Chem. 279 5604-5611 [DOI] [PubMed] [Google Scholar]
- 46.Wang, S., Ai, X., Freeman, S. D., Pownall, M. E., Lu, Q., Kessler, D. S., and Emerson, C. P., Jr. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 4833-4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindquist, J. N., Marzluff, W. F., and Stefanovic, B. (2000) Am. J. Physiol. Gastrointest. Liver Physiol. 279 471-476 [DOI] [PubMed] [Google Scholar]
- 48.Lai, J. P., Yu, C., Moser, C. D., Aderca, I., Han, T., Garvey, T. D., Murphy, L. M., Garrity-Park, M. M., Shridhar, V., Adjei, A. A., and Roberts, L. R. (2006) Gastroenterology 130 2130-2144 [DOI] [PubMed] [Google Scholar]
- 49.Chen, L., Klass, C., and Woods, A. (2004) J. Biol. Chem. 279 15715-15718 [DOI] [PubMed] [Google Scholar]