Abstract
Type I sulfatases require an unusual co- or post-translational modification for their activity in hydrolyzing sulfate esters. In eukaryotic sulfatases, an active site cysteine residue is oxidized to the aldehyde-containing Cα-formylglycine residue by the formylglycine-generating enzyme (FGE). The machinery responsible for sulfatase activation is poorly understood in prokaryotes. Here we describe the identification of a prokaryotic FGE from Mycobacterium tuberculosis. In addition, we solved the crystal structure of the Streptomyces coelicolor FGE homolog to 2.1Å resolution. The prokaryotic homolog exhibits remarkable structural similarity to human FGE, including the position of catalytic cysteine residues. Both biochemical and structural data indicate the presence of an oxidized cysteine modification in the active site that may be relevant to catalysis. In addition, we generated a mutant M. tuberculosis strain lacking FGE. Although global sulfatase activity was reduced in the mutant, a significant amount of residual sulfatase activity suggests the presence of FGE-independent sulfatases in this organism.
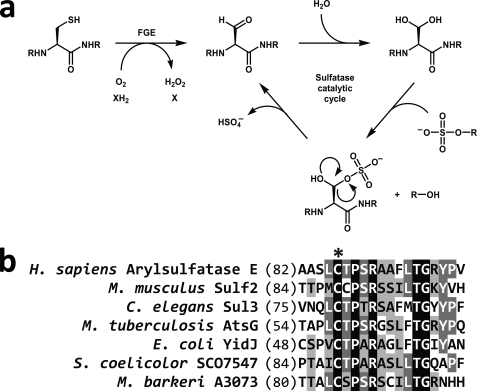
Type I sulfatases are members of an expanding family of enzymes that employ novel co- or post-translationally derived cofactors to facilitate catalysis (1, 2). The formylglycine (Fgly)4 residue positioned within the active site of type I sulfatases is thought to undergo hydration to a gem-diol, after which one of the hydroxyl groups acts as a catalytic nucleophile to initiate sulfate ester cleavage (Fig. 1a) (3). The FGly residue is located within a ∼13-residue consensus sequence termed the sulfatase motif (4) that defines this family of enzymes and is highly conserved throughout all domains of life (Fig. 1b). Although FGly is formed from cysteine residues in eukaryotic sulfatases, either cysteine (within the core motif CX[P/A]XR) or serine (SXPXR) can be oxidized to FGly in prokaryotic type I sulfatases. The co- or post-translational machineries necessary for these respective modifications appear to be different; FGE is able to activate CXPXR-type sulfatases (5–7) and anaerobic sulfatase-maturating enzyme is responsible for modifying SXPXR-type sulfatases and CXAXR-type sulfatases (8, 9). Some prokaryotes, such as Mycobacterium tuberculosis, have only CXPXR-type sulfatases (10), whereas other species have only SXPXR-type sulfatases or a combination of type I sulfatases (11). Some prokaryotes also contain FGly-independent sulfatases. These sulfatases function by distinct enzymatic mechanisms and are divided into two categories, Fe(II) α-ketoglutarate-dependent dioxygenase sulfatases (type II sulfatases) and metallo-β-lactamase sulfatases (type III sulfatases) (12–14). Unlike type I sulfatases, which share a high degree of sequence similarity, type II and III sulfatases have highly divergent sequences, complicating the discovery of these proteins by genomic search algorithms.
FIGURE 1.
Activation of sulfatases by formylglycine-generating enzyme (FGE) and proposed sulfatase mechanism. a, FGE activates sulfatases by oxidizing an active site cysteine to a 2-formylglycyl residue (FGly). Previously determined sulfatase crystal structures indicate that the active site FGly is hydrated, suggesting that sulfate ester cleavage is mediated by a transesterification elimination mechanism (3). b, the sulfatase motif is located toward the N terminus of sulfatases and targets the appropriate cysteine (*) for modification by FGE. Black indicates an exact residue match; dark gray indicates conserved residues; light gray indicates similar residues.
In higher eukaryotes, sulfatases are involved in a variety of essential tasks, including extracellular matrix remodeling and steroid titer regulation (1). In humans, FGE deficiency abolishes the activity of all 17 human sulfatases, leading to the fatal disorder multiple sulfatase deficiency (6, 15). Prokaryotic sulfatases have not been thoroughly investigated but are generally assumed to be used for sulfate scavenging (16). The acquisition and allocation of sulfur-containing compounds, including inorganic sulfate, are critical for many prokaryotes; the ability of M. tuberculosis to persist within human macrophages, in particular, is thought to be dependent on several sulfur-containing molecules such as mycothiol and sulfolipid-1 (17, 18). BLAST analysis (19) indicates that the M. tuberculosis genome contains six type I sulfatases, a number that is higher than most other bacterial genomes.
Several elegant biochemical and crystallographic studies on human FGE have analyzed the novel thiol-to-aldehyde oxidation catalyzed by this enzyme (6, 20). This recent work has identified several intriguing properties of the eukaryotic variant of this enzyme, including an active site that discriminates only the conserved residues within the core sulfatase motif, the presence and relative reactivities of 2 catalytically essential cysteine residues, the absence of cofactors, and the requirement of molecular oxygen for substrate turnover (6, 20). In this study, we have functionally identified the first prokaryotic FGE from M. tuberculosis and created an FGE-deficient strain in order to probe the function of type I sulfatases in this human pathogen. Additionally, we present the first structural and biochemical data of a prokaryotic FGE to further our understanding of the unique mechanism of this enzyme.
EXPERIMENTAL PROCEDURES
Preparation of Protein Expression Vectors—Supplemental Table S1 lists the oligonucleotides used in this study. The gene encoding M. tuberculosis FGE (Rv0712, encoding residues 2–299) was amplified from M. tuberculosis H37Rv genomic DNA and cloned into pET14b (Novagen) using NdeI and XhoI restriction sites. The gene encoding Streptomyces coelicolor FGE (SCO7548, encoding residues 2–314) was amplified from S. coelicolor A3(2) genomic DNA and cloned into pET151/d-TOPO (Invitrogen). Open reading frames Rv2407 (encoding residues 2–273), Rv3406 (encoding residues 2–295), and Rv3762c (encoding residues 2–626) were amplified from M. tuberculosis H37Rv genomic DNA. Rv2407 was ligated into pMAL-C2X (New England Biolabs) using BamHI and PstI restriction sites, and both Rv3406 and Rv3762c were ligated into pET28b (Novagen) using NdeI and XhoI restriction sites. Rv0484c, Rv0576, Rv1730c, Rv1980c, Rv2837c, Rv3137, Rv3205c, and Rv3849 were all amplified from M. tuberculosis H37Rv genomic DNA and cloned into pET151/D-TOPO. DNA sequencing was performed to confirm the fidelity of each gene product. Protein-encoding plasmids were transformed into BL21(DE3) cells (Invitrogen).
Site-directed Mutagenesis—Site-specific mutations in M. tuberculosis FGE and S. coelicolor FGE were produced with QuikChange PCR mutagenesis kit (Stratagene) using pET14b M. tuberculosis FGE or pET151 S. coelicolor FGE plasmids and the appropriate oligonucleotides (supplemental Table S1) in the mutagenesis reactions. Mutations were confirmed by DNA sequencing, and plasmids were transformed into BL21(DE3) cells for protein expression as described below.
Protein Expression and Purification—Clonal populations of BL21(DE3) cells harboring a His6-tagged protein-encoding plasmid were incubated in Luria Bertani medium with ampicillin or kanamycin with shaking at 37 °C until A600 = 0.5, at which time the temperature was lowered to 18 °C and 250 μm isopropyl-1-thio-β-d-galactopyranoside was added. After 12–16 h, cells were harvested and resuspended in 20 ml of lysis buffer (50 mm Tris, 500 mm NaCl, 10% glycerol, 20 mm imidazole, 1 mm dithiothreitol, 1 mm Tris(2-carboxyethyl)phosphine, 1 mm methionine, pH 7.5) per liter of culture and lysed by sonication. Cell lysate was treated with DNase I (10 μgml–1), cleared by centrifugation, and applied to a 1-ml HisTrap column (GE Healthcare). The column was washed with lysis buffer containing 35 mm imidazole, and His6-tagged protein was eluted using lysis buffer containing 250 mm imidazole. The eluted protein was concentrated to <2 ml and further purified on a Sephadex 16/60 S300 column (GE Healthcare) using lysis buffer without imidazole. Purified recombinant protein was subsequently concentrated to ∼20 mg ml–1. The identity and purity of M. tuberculosis and S. coelicolor FGE were assessed by electrospray ionization mass spectrometry (Bruker/Agilent Esquire). Rv2407 was not soluble in His6-tagged form and was alternatively fused to maltose-binding protein (MBP). Growth and lysis conditions for MBP-Rv2407-producing cells were the same as above except with the absence of imidazole in the lysis buffer. Cleared lysate was applied to amylose resin (New England Biolabs) in lysis buffer, washed in additional lysis buffer, and MBP-Rv2407 was eluted in lysis buffer with 10 mm maltose and subsequently concentrated. Maltose-binding protein was cleaved and removed from Rv2407 using Factor Xa (New England Biolabs) and amylose resin.
S. coelicolor FGE Crystallization—Attempts to crystallize FGE homologs from M. tuberculosis, Mycobacterium smegmatis, and Mycobacterium avium were not successful because of protein instability. S. coelicolor FGE was dialyzed into 10 mm Tris, 150 mm NaCl, 1 mm Tris(2-carboxyethyl)phosphine, pH 7.5. Crystals of His6-tagged S. coelicolor FGE were obtained using vapor diffusion by mixing 1 μl of dialyzed protein with 1 μl of crystallization solution (100 mm Tris, 2.4 m ammonium formate, 0.3% β-octylglucoside, 3.2% 2-butanol, pH 8.0) at room temperature. Crystals grew over a period of 2 weeks and were subsequently transferred to cryoprotectant consisting of crystallization solution with 20% glycerol prior to flash-freezing in liquid nitrogen.
S. coelicolor FGE Structure Determination—Data were collected at beamline 8.2.2 at the Advanced Light Source using an ADSC Quantum-Q315 CCD detector. Crystal quality was extremely variable, and only one crystal of more than 100 screened diffracted to better than 3.0 Å resolution. Diffraction data from this crystal were reduced and scaled to 2.1 Å resolution using HKL2000 (21); because of technical difficulties encountered with collection, data could only be collected to 83% overall completeness (Table 1). Initial phases were determined by molecular replacement using the human FGE (Protein Data Bank entry 1Y1E) as a search model in PHASER (22). The asymmetric unit contained five S. coelicolor FGE monomers in space group P3121. Initial stages of model refinement included cycles of simulated annealing with torsion angle dynamics and restrained B-factor refinement using CNS (23), followed by manual model rebuilding using O (24). Model building was aided by using bias-reduced prime-and-switch phased maps (25) (supplemental Fig. S1). Because of small structural differences observed between monomers in the asymmetric unit, non-crystallographic symmetry restraints were not used. The final cycles of refinement were carried out with twin lattice symmetry (26) restraints as implemented in REFMAC5 (27) using five twin lattice symmetry groups (corresponding to each FGE monomer in the asymmetric unit). Water molecules were added with ARP/WARP (28). The final model contained residues 18–305 in monomer A, residues 19–306 in monomer B, residues 20–306 in monomer C, residues 19–305 in monomer D, and residues 19–307 in monomer E. Final Rwork and Rfree values were 19.5 and 23.3%, respectively. Data collection and processing statistics are summarized in Table 1. All figures were generated with PyMOL.
TABLE 1.
Data collection and refinement statistics
r.m.s., root mean square.
| Data collection | |
| Resolution (Å)a | 20-2.1 (2.1-2.17) |
| Wavelength (Å) | 1.00 |
| Space group | P3121 |
| Unit cell dimensions (a = b, c) (Å) | 142.4, 217.1 |
| Measured reflections | 123276 |
| Completeness (%) | 83.4 (80.9) |
| Redundancy | 2.6 (2.6) |
| Mosaicity (°) | 0.32 |
| I/σ | 15.8 (1.9) |
| Rsym (%)b | 5.7 (23.3) |
| Wilson B factor | 37.6 |
| Refinement | |
| Rwork (%)c | 19.5 |
| Rfree (%)c | 23.3 |
| Number of residues/waters | 1438/1017 |
| R.m.s. bonds (Å)/angles (°) | 0.008/1.062 |
| Ramachandran plot (%)d | 87.9/11.2/0.5/0.6e |
| Average B values | 41.5 |
Values in parentheses correspond to the highest resolution bin.
Rsym = 100*ΣhΣi|li(h) — 〈l(h)〉|/ΣhΣili(h), where li(h) is the ith measurement of reflection h and 〈l(h)〉 is the average value of the reflection intensity.
Rwork = 100*Σ||Fobs| — |Fcalc||/|Fobs|, where Fobs and Fcalc are the structure factor amplitudes from the data and the model, respectively. Rfree is Rwork with 5% of the reflections set aside throughout refinement.
Numbers correspond to the percentage of amino acid residues in the favored, allowed, generously allowed, and disallowed regions, respectively. Calculated using PROCHECK (42).
Seven residues were observed in stereochemically strained conformations either due to crystal packing contacts (Tyr-219 in monomers A and C) or hydrogen bonding interactions (Asn-232 in monomers A—E).
FGE Activity Assay—Wild type and mutant FGE from M. tuberculosis and S. coelicolor were purified as described above. The peptide substrate was synthesized by standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) solid phase synthesis methods and consisted of the 13-residue sequence LCSPSRGSLFTGR, a sulfatase consensus motif. The N terminus was acetylated, the C terminus was amidated, and the sequence was confirmed by mass spectrometry. Assay conditions were similar to those reported previously by Dierks et al. (6) in studies of human FGE. Anaerobic experiments were performed in the same manner except solutions were made anaerobic using an oxygen-scavenged gas manifold and reactions were started by mixing enzyme with substrate in an anaerobic glovebox. EDTA was added to the appropriate reactions at a concentration of 100 mm. Confirmation of FGly formation was performed by incubating 1 μl of desalted product with 1 μl of 5 mm biotin hydrazide (Sigma) for 30 min at room temperature. Samples were mixed 1:1 (v/v) with matrix solution (10 mg ml–1 α-cyano-4-hydroxy-cinnamic acid with 2 mm ammonium citrate) and analyzed by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (Applied Biosystems Voyager DE Pro).
Metal Detection—A multielement standard solution was prepared by appropriate dilution of inductively coupled plasma standards of calcium, copper, iron, manganese, magnesium, and zinc (Sigma). Metal content of M. tuberculosis and S. coelicolor FGE was analyzed by inductively coupled plasma-atomic emission spectroscopy (ICP-AES) using a PerkinElmer Optima 3000 DV. Absence of iron, copper, and zinc in S. coelicolor FGE was confirmed at beamline 8.3.1 at the Advanced Light Source. Absorption edges of these metals were examined using a double crystal monochromator and the x-ray fluorescence detector of the beamline.
M. tuberculosis FGE-deficient Strain Production—An unmarked, in-frame genetic deletion of the FGE-encoding open reading frame Rv0712 was created in M. tuberculosis H37Rv using allelic replacement (29). A 2-kb region upstream of Rv0712 was amplified and inserted into the mycobacterial delivery vector p2NILX between HindIII and XbaI restriction sites. p2NILX is derived from pNIL (29) and modified with the addition of an XbaI restriction site between KpnI and NotI restriction sites. A 2-kb region downstream of Rv0712 was amplified and inserted into p2NILX between XbaI and PacI restriction sites. Selection markers lacZ and sacB were digested from pGOAL17 and ligated into p2NILX using the PacI restriction site. The completed delivery vector was treated with UV light (120 mJ cm–2) and electroporated into electrocompetent M. tuberculosis H37Rv as described previously (30). Selection of the mutant was performed as described previously (29), and genotype was confirmed by Southern blot analysis (supplemental Fig. S2). The complemented strain was produced by transforming the Δfge strain with the integrating vector pMV306.kan containing the entire Rv0712 open reading frame under the control of the glutamine synthase promoter.
Sulfatase/phosphatase Assay—M. tuberculosis H37Rv strains were grown in 7H9 medium supplemented with albumin-dextrose-catalase (BD Biosciences) at 37 °C until A600 = 1.0. Cells were lysed by mechanical disruption using 0.1 mm zirconia beads (FastPrep; MP Biomedicals), and the crude lysate was cleared by centrifugation and filtered through a 0.22-μm membrane. Cleared lysate samples were normalized for total protein concentration (Bio-Rad AC/DC protein assay kit), and 50 μgof lysate protein was added to buffer (50 mm Tris, 500 mm NaCl, 100 μm MgCl2, 100 μm MnCl2, 100 μm CaCl2, pH 7.5), protease inhibitors (Protease Inhibitor mixture set III; EMD Bioscience), and 8 mm 4-methylumbelliferyl sulfate (4MUS; Sigma). Limpet sulfatase (Sigma) was used at a final concentration of 1 μgml–1 as a positive control. Escherichia coli lysate (strain TOP10; Invitrogen) was prepared in the same manner as M. tuberculosis lysate, and 50 μg of lysate protein was used per reaction. Reactions were incubated at 37 °C for 3 h and stopped by adding 4 volumes of 0.5 m Na2CO3/NaHCO3, pH 10.5. Sulfatase activity was measured using a fluorimeter (Gemini XL; Molecular Devices) using excitation and emission wavelengths of 360 and 460 nm, respectively. Lysate extracts exhibited steady state behavior. Sulfatase/phosphatase inhibitors were used according to the manufacturer's instructions and included microcystin, cantharidin, p-bromotetramisole, sodium vanadate, sodium molybdate, sodium tartrate, and imidazole (Phosphatase Inhibitor Mixtures 1 and 2; Sigma). Sulfatase activity of recombinant Rv2407, Rv3406, and Rv3762c was determined using the same conditions mentioned above, with the addition of 1 mm α-ketoglutarate, 200 μm ascorbate, and 100 μm FeCl2 to the buffer. Phosphatase activity was monitored as described above except with the substitution of 4-methylumbelliferyl phosphate for 4MUS.
NBD Labeling—His6-tagged S. coelicolor FGE was treated with 1:50 (w/w) tobacco etch virus protease to remove the N-terminal His6 tag before 4-chloro-7-nitrobenz-2-oxa-1,3-diazole (NBD) labeling and mass spectrometric analyses. S. coelicolor FGE (45 μm) was incubated in buffer (25 mm potassium phosphate, 150 mm NaCl, pH 7.0) with 1 mm NBD (Invitrogen) for 30 min at room temperature (31). The sample was desalted by C18 reversed phase chromatography, and protein-NBD adducts were detected by mass spectrometry (Bruker/Agilent Esquire). Mapping of NBD adducts was performed by digesting NBD-reacted S. coelicolor FGE with 1:50 (w/w) trypsin, desalting by C18 reversed phase chromatography, and analyzing the resulting peptide fragments using electrospray ionization Fourier-transform ion cyclotron resonance mass spectrometry (Bruker 9.4T Apex III).
RESULTS AND DISCUSSION
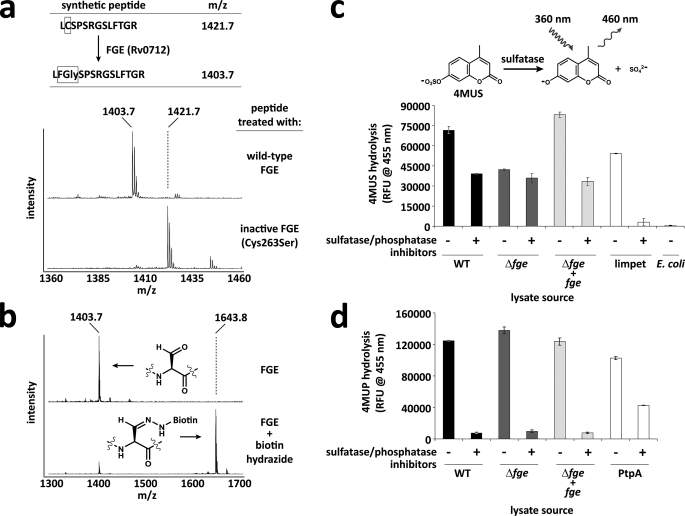
Using BLAST analysis, we identified M. tuberculosis H37Rv's open reading frame Rv0712 to be >30% identical to the human FGE SUMF1 (5, 6). Recombinant Rv0712 was able to modify a synthetic peptide containing the sulfatase motif as determined by mass spectrometry (Fig. 2a). The presence of FGly within the substrate was confirmed by treating the modified peptide with biotin hydrazide, which formed a covalent adduct with the peptide via hydrazone formation (Fig. 2b). Together these data implicate Rv0712 as the FGE of M. tuberculosis.
FIGURE 2.
Function of M. tuberculosis FGE (Rv0712) in vitro and in vivo. a, a synthetic peptide resembling a sulfatase motif was treated with recombinant M. tuberculosis FGE, and the resulting oxidation of cysteine to FGly was monitored by mass spectrometry. The C263S FGE mutant was inactive on the peptide substrate. Theionsatm/z 1427 and 1445 are sodium adducts of the modified and unmodified peptide, respectively. b, upon treatment with biotin hydrazide, the FGly-containing peptide forms a hydrazone adduct with biotin, resulting in a mass shift of +240 Da. c, lysates from wild-type (WT), Δfge, and complemented (Δfge + fge) strains of M. tuberculosis H37Rv were tested for sulfatase activity using the fluorogenic substrate 4-methylumbelliferyl sulfate (4MUS) with and without sulfatase/phosphatase inhibitors. Limpet sulfatase was used as a positive control. E. coli lysate was used as a negative control. d, lysates from WT, Δfge, and Δfge + fge strains of M. tuberculosis H37Rv were tested for phosphatase activity using the fluorogenic substrate 4-methylumbelliferyl phosphate with and without sulfatase/phosphatase inhibitors. The recombinant M. tuberculosis phosphatase PtpA was used as a positive control. Error bars represent S.D. ± mean between three independent experiments.
Similar to the human genome, the M. tuberculosis genome appears to encode only one functional copy of FGE. Therefore, we expected that the disruption of Rv0712 in M. tuberculosis would produce a sulfatase-deficient strain. Rv0712 was disrupted in M. tuberculosis H37Rv using homologous recombination and confirmed by Southern blot analysis (supplemental Fig. S2). Δfge M. tuberculosis was viable and demonstrated no obvious growth defects in vitro.
Sulfatase activity of the Δfge strain was compared with that of wild-type (WT) H37Rv and to the Δfge mutant into which FGE expression was restored by complementation. Crude lysates were generated from these three M. tuberculosis strains, and global sulfatase activity was determined using the general sulfatase substrate 4MUS. The Δfge strain exhibited a substantial, yet surprisingly incomplete, loss of sulfatase activity (Fig. 2c). We considered the possibility that the residual sulfatase activity resulted from phosphatases hydrolyzing 4MUS (32). However, when sulfatase activity was monitored in the presence of a mixture of broad spectrum sulfatase/phosphatase inhibitors, Δfge was not affected (Fig. 2c). Indeed, activity in lysates from WT and complemented Δfge was reduced by ∼40% in the presence of the inhibitor mixture, matching the sulfatase activity of Δfge in the absence of inhibitors. Given that the applied inhibitors are known to inhibit FGE-dependent sulfatases (33), these data suggest that M. tuberculosis possesses FGE-independent sulfatases.
To verify further that promiscuous phosphatases were not responsible for the residual sulfatase activity, we monitored phosphatase activity of crude lysates from each strain using 4-methylumbelliferyl phosphate. All three strains exhibited the same level of phosphatase activity in the absence of inhibitors, and activity was abolished in all strains in the presence of the inhibitors (Fig. 2d). These data further indicate that phosphatases are not accountable for the residual 4MUS hydrolysis activity observed in the Δfge strain and that FGE-dependent sulfatases are responsible for ∼40% of the total 4MUS hydrolysis activity in M. tuberculosis lysate.
We next searched the M. tuberculosis genome for potential sources of FGE-independent sulfatase activity. The majority of known or putative prokaryotic sulfatases are of the type I class. However, some prokaryotes also have FGly-independent sulfatases that operate via different enzymatic mechanisms; these enzymes may not be sensitive to broad spectrum sulfatase inhibitors. Based on sequence similarity with known FGly-independent sulfatases from other prokaryotes, M. tuberculosis has three putative FGly-independent sulfatases encoded by open reading frames, Rv2407, Rv3406, and Rv3762c. We expressed recombinant forms of these proteins in E. coli, but the purified proteins exhibited no activity in the 4MUS assay, indicating that these putative sulfatases are probably not responsible for the residual sulfatase activity in Δfge M. tuberculosis (supplemental Fig. S3). Considering the lack of sequence similarity among FGly-independent sulfatases, M. tuberculosis may have other sulfatases not detectible by BLAST analysis. The identification of novel sulfatases in M. tuberculosis is therefore an important goal for the future.
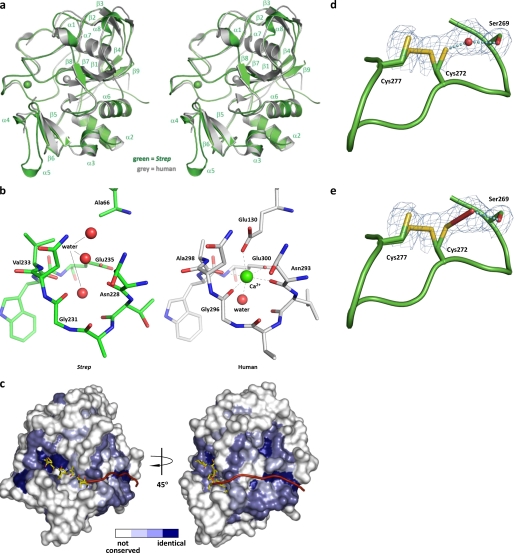
To understand better the unique enzymatic mechanism and substrate binding characteristics of prokaryotic FGEs, the structure of the M. tuberculosis FGE ortholog from S. coelicolor was determined to a resolution of 2.1 Å. The overall topology of the bacterial FGE is remarkably similar to the recently determined human FGE structure (20) (Fig. 3a). Similar to human FGE, S. coelicolor FGE has low secondary structure content, containing 16% α-helix and 12% β-sheet. Both share the novel “FGE fold,” but the S. coelicolor FGE variant contains only one Ca2+ ion as determined by coordination geometry and ICP-AES (supplemental Table S2). The human variant is stabilized by two Ca2+ ions; this difference may be partially due to a E66A substitution in S. coelicolor FGE that disrupts an appropriate coordination environment (Fig. 3b). However, the previously reported human paralog FGE structure contains a proline at the corresponding position yet still contains a Ca2+ ion at this site (34). Regardless, ICP-AES data indicate that enzymatically active M. tuberculosis FGE lacks both Ca2+ ions (supplemental Table S2), suggesting that the FGE fold may not require a divalent cation.
FIGURE 3.
Structure of S. coelicolor FGE. a, stereo superposition of S. coelicolor FGE (green) and human FGE (gray). S. coelicolor FGE secondary structure elements are indicated. Ca2+ ions are rendered as spheres. Overall root mean square deviation is 0.65 Å. Strep, S. coelicolor. b, comparison of the residues surrounding the potential Ca2+ binding site of S. coelicolor FGE and the second Ca2+ binding site of human FGE. Proper coordination geometry is impaired with a E66A mutation in S. coelicolor FGE. Water molecules are rendered as red spheres, and Ca2+ is rendered as a green sphere. c, surface representation of the putative exosite of S. coelicolor FGE. The surface is colored according to residue conservation between all known and putative FGEs (based on amino acid sequence alignment); white represents non-conserved residues, light blue represents weakly conserved residues, medium blue represents conserved residues, and dark blue represents identical residues. The 6-residue peptide substrate is modeled from the human FGE-peptide complex structure (37) (Protein Data Bank entry 2AIK). A hypothetical extended peptide substrate is represented as a red ribbon. d and e, active site cysteines 272 and 277 appear to exist in a partial disulfide bond. Cys-272 is shown in two alternate conformations. Electron density between Cys-272 and Ser-269 has been modeled as a water molecule (d) but also is consistent with formation of a hydroperoxide at partial occupancy (e). Monomer E is shown. Prime-and-switch map is contoured at 1.5 σ.
The active sites of the prokaryotic and human FGE are remarkably similar. Both are ∼20 Å in length, 12 Å in width, and 10 Å in depth and can accommodate only 6 of the 13 amino acids that define the sulfatase motif. Considering that the sulfatase motif extends toward the C terminus of the peptide substrate for another 8 residues beyond the core consensus sequence (CXPXR) (Fig. 1b), it is possible that FGE has evolved a secondary binding region to aid in substrate recognition. A similar, dual binding mechanism has been observed in other proteins such as thrombin and botulinum neurotoxin (35, 36). Indeed, a comparison of the conserved residues between a panel of FGEs, including S. coelicolor, M. tuberculosis, and human and other putative orthologs, reveals a region of high conservation that could accommodate binding of the C-terminal section of the sulfatase motif (Fig. 3c).
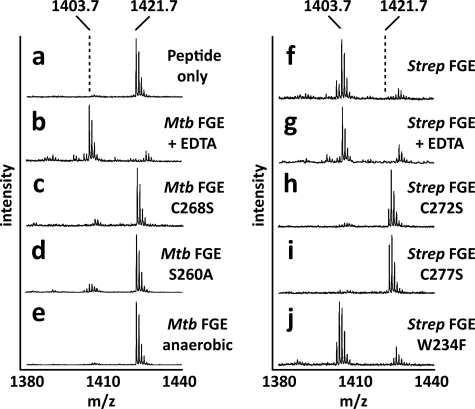
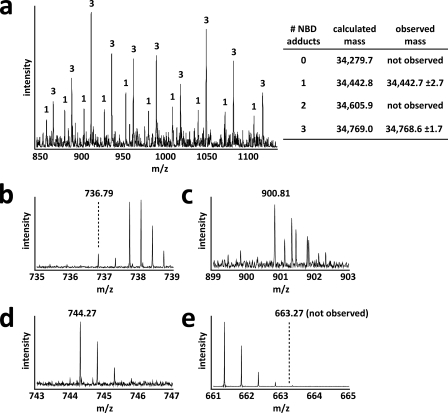
FGE is thought to catalyze the oxidation of a thiol to an aldehyde using 2 conserved cysteine residues within its active site (20, 37). These cysteines in M. tuberculosis (Cys-263 and Cys-268) and S. coelicolor FGE (Cys-272 and Cys-277) are required for substrate turnover as serine mutants were unable to generate FGly in vitro (Fig. 2a and Fig. 4, c, h, i). Interestingly, prime-and-switch phased maps showed Cys-272 and Cys-277 to be engaged in a partial disulfide bond (Fig. 3, d and e). Biochemical confirmation of these partial disulfides was provided by treating the three solvent-exposed cysteines of native S. coelicolor FGE with the thiol-labeling reagent NBD. Two distinct populations corresponding to S. coelicolor FGE with either one or three NBD adducts were detected by intact protein mass spectrometry (Fig. 5), confirming the partial disulfide status of these residues. A previous study also investigated the reactivity of the human FGE equivalents of Cys-272 and Cys-277 using the thiol-reactive reagent iodoacetamide (37).
FIGURE 4.
M. tuberculosis and S. coelicolor FGE activity is dependent upon molecular oxygen but independent of metal cofactors. a, a synthetic peptide resembling the sulfatase motif (mass, 1421.7 Da) was used as a substrate for FGE. Conversion of cysteine to FGly resulted in a loss of 18 Da that was detected by mass spectrometry. b and g, metal chelator EDTA had no effect on activity. c, h, i, loss of active site cysteines in M. tuberculosis and S. coelicolor FGE abolished activity. d, loss of active site Ser-260 in M. tuberculosis FGE significantly reduced activity. e, M. tuberculosis FGE was inactive in the absence of molecular oxygen. f, WT S. coelicolor FGE was able to oxidize the synthetic peptide. j, active site Trp-234 in S. coelicolor FGE is not essential for catalytic activity. The ion at m/z 1427 is the sodium adduct of the FGly-containing product peptide. Mtb, M. tuberculosis; Strep, S. coelicolor.
FIGURE 5.
Active site cysteines 272 and 277 in S. coelicolor FGE are engaged in a partial disulfide bond. a, S. coelicolor FGE was labeled with NBD, and adducts were detected by mass spectrometry. Two populations of NBD-modified S. coelicolor FGE were observed. One population had a single NBD adduct, which corresponds to disulfide connected Cys-272 and Cys-277 (see b). The other population had three NDB adducts corresponding to Cys-272 and Cys-277 thiols (see c). Multiple charge states are shown for both populations. Numbers above each ion peak indicate the number of NBD adducts attributed to each population. b–d, NBD adducts were mapped to each surface-exposed thiol using mass spectrometry after trypsinolysis. Shown are the +2 charge state ions corresponding to disulfide-connected Cys-272 and Cys-277 (b, calculated mass, 1,471.58; observed mass, 1,471.58), NBD adducts on both Cys-272 and Cys-277 (c, calculated mass, 1,799.58; observed mass, 1,799.61), and an NBD adduct on Cys-301 (d, calculated mass, 1486.52; observed mass, 1,486. 54). e, Cys-301 was not observed as a free thiol (expected position of +2 charge state ion shown). Cys-301 was an additional surface-exposed thiol that served as an internal control to assess labeling efficiency.
In addition to the two active site cysteines, S. coelicolor and M. tuberculosis FGE require molecular oxygen for catalysis as no FGly formation was observed in reactions performed in an anaerobic environment (Fig. 4e and data not shown). As a member of the oxygenase family, FGE might be expected to contain a transition metal, such as iron or copper, or an organic cofactor such as FADH for activation of molecular oxygen. However, analysis by ICP-AES and x-ray absorption edge scanning indicated that active M. tuberculosis and S. coelicolor FGE lack all redox active metals (supplemental Table S2 and data not shown). Additionally, these FGEs do not require addition of metals to function in assays in vitro and can function in the presence of EDTA (Fig. 4, b and g). Similarly, UV-visible absorption spectroscopy did not reveal the presence of chromophoric organic cofactors (data not shown). Together with electron density information for S. coelicolor FGE, these data indicate that prokaryotic FGEs, similar to human FGE, do not use exogenous cofactors for catalysis.
As an alternate means of activating molecular oxygen, FGE may function similarly to other cofactor-less oxygenases and make unique use of conventional residues (38). Conserved residues within reactive distance from the catalytic cysteine pair of S. coelicolor FGE include Trp-234 and Ser-269. Roeser et al. (37) hypothesized that Trp-234 may function to activate molecular oxygen, similar to the proposed mechanism of O2 reduction by catalytic antibodies (39). However, mutation of Trp-234 to Phe or Ala did not abolish activity (Fig. 4j and data not shown), indicating that molecular oxygen activation must be achieved by another route. Activity was severely reduced by the S269A equivalent mutation in M. tuberculosis FGE (S260A) (Fig. 4d), similar to the equivalent human FGE mutant (S333A). It has been speculated that this active site serine acts as a nucleophile during catalysis (20), but no experimental evidence has as yet confirmed this hypothesis.
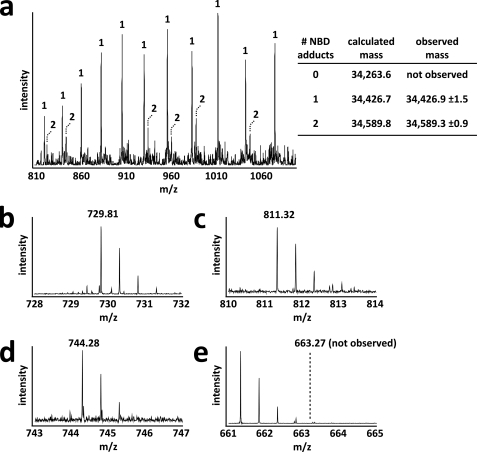
Interestingly, Cys-272 itself may be involved in activating molecular oxygen. All 5 modeled Cys-272 residues within the asymmetric unit of S. coelicolor FGE have extra electron density extending away from their alternate, non-disulfide-bound conformations. Prime-and-switch phased maps indicate that this extra density could be modeled as one water molecule or a hydroperoxide moiety with partial occupancy (Fig. 3, d and e). NBD-labeling experiments using the C277S S. coelicolor FGE mutant indicate that Cys-272 is not a reactive thiol in a subpopulation of enzyme molecules (Fig. 6), suggesting that the extra density corresponds to a moiety covalently bound to Cys-272, such as hydroperoxide. Previously published structures of the human FGE have also suggested that this extra density could be hydroperoxide or cysteine sulfenic acid combined with a bound water molecule (20). However, the latter fits the observed electron density in S. coelicolor FGE poorly. Furthermore, no sulfenic acid was detected when S. coelicolor FGE was treated with NBD (Figs. 5 and 6). The presence of hydroperoxide modeled with partial occupancy cannot be ruled out based on the observed electron density of S. coelicolor FGE. However, mass spectrometric analysis of intact S. coelicolor and M. tuberculosis FGE revealed no mass anomaly (supplemental Fig. S4 and data not shown), suggesting that if Cys-272 is modified, the modification is transient or acid-labile. In the current structure, we conservatively have modeled this density as water but note the possibility of hydroperoxide formation as being consistent with a role for Cys-272 and Ser-269 in activating O2.
FIGURE 6.
A subpopulation of S. coelicolor FGE C277S does not contain a reduced Cys-272 residue. a, S. coelicolor FGE C277S was labeled with NBD, and adducts were detected by mass spectrometry. Two populations of NBD-modified S. coelicolor FGE C277S were observed. One population corresponds to Cys-272 as a free thiol with a single NBD adduct on Cys-301 (see b and d). The other population had NDB adducts at both Cys-272 and Cys-301 (see c and d). Multiple charge states are shown for both populations. Numbers above each ion peak indicate the number of NBD adducts attributed to each population. b–d, NBD adducts were mapped to each surface-exposed thiol using mass spectrometry after trypsinolysis. Shown are the +2 charge state ions corresponding to Cys-272 as a free thiol (b, calculated mass, 1,457.61; observed mass, 1,457.60), Cys-272 with an NBD adduct (c, calculated mass, 1,620.62; observed mass, 1,620.60), and Cys-301 with an NBD adduct (d, calculated mass, 1,486.54; observed mass, 1486.54). e, Cys-301 was not observed as a free thiol (expected position of +2 charge state ion shown). Cys-301 was an additional surface-exposed thiol that served as an internal control to assess labeling efficiency.
CONCLUSION
In summary, we have functionally characterized prokaryotic FGE from M. tuberculosis and solved the structure of the ortholog from S. coelicolor. Our studies indicate that FGE-activated sulfatases account for approximately half the total 4MUS hydrolysis activity in M. tuberculosis lysate, suggesting that this organism possesses FGE-independent sulfatases that have yet to be identified. Defining the complete repertoire of sulfatases from M. tuberculosis (and other prokaryotes) is the subject of future studies and will provide a platform for defining the role of sulfatases in the life cycle and pathogenesis of bacteria.
In addition to defining a new niche of enzyme mechanisms in which thiol reactivity might be directly exploited in molecular oxygen activation, FGE may offer new avenues for protein engineering. The aldehyde group has unique electrophilic reactivity that has been exploited in other contexts for selective bioconjugation (40). The sulfatase motif can therefore serve as a genetically encoded chemical tag, as we have recently reported (41). Notably, the prokaryotic FGEs studied here are soluble proteins that are readily expressed in heterologous systems. By contrast, human FGE is an endoplasmic reticulum-associated protein that modifies its substrates as they enter the secretory pathway. The availability of a structure for prokaryotic FGEs, as presented here, may provide the basis for engineering new substrate specificities and creating a spectrum of tailored sulfatase motifs.
Supplementary Material
Author's Choice—Final version full access.
The atomic coordinates and structure factors (code 2Q17) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grants AI051662 and GM059907 (to C. R. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Tables S1 and S2.
Footnotes
The abbreviations used are: FGly, formylglycine; FGE, formylglycine generating enzyme; WT, wild type; 4MUS, 4-methylumbelliferyl sulfate; ICP-AES, inductively coupled plasma-atomic emission spectroscopy; NBD, 4-chloro-7-nitrobenz-2-oxa-1,3-diazole.
References
- 1.Hanson, S. R., Best, M. D., and Wong, C. H. (2004) Angew. Chem. Int. Ed. 43 5736–5763 [DOI] [PubMed] [Google Scholar]
- 2.Okeley, N. M., and van der Donk, W. A. (2000) Chem. Biol. 7 R159–R171 [DOI] [PubMed] [Google Scholar]
- 3.Lukatela, G., Krauss, N., Theis, K., Selmer, T., Gieselmann, V., von Figura, K., and Saenger, W. (1998) Biochemistry 37 3654–3664 [DOI] [PubMed] [Google Scholar]
- 4.Dierks, T., Lecca, M. R., Schlotterhose, P., Schmidt, B., and von Figura, K. (1999) EMBO J. 18 2084–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosma, M. P., Pepe, S., Annunziata, I., Newbold, R. F., Grompe, M., Parenti, G., and Ballabio, A. (2003) Cell 113 445–456 [DOI] [PubMed] [Google Scholar]
- 6.Dierks, T., Schmidt, B., Borissenko, L. V., Peng, J., Preusser, A., Mariappan, M., and von Figura, K. (2003) Cell 113 435–444 [DOI] [PubMed] [Google Scholar]
- 7.Schmidt, B., Selmer, T., Ingendoh, A., and von Figura, K. (1995) Cell 82 271–278 [DOI] [PubMed] [Google Scholar]
- 8.Szameit, C., Miech, C., Balleininger, M., Schmidt, B., von Figura, K., and Dierks, T. (1999) J. Biol. Chem. 274 15375–15381 [DOI] [PubMed] [Google Scholar]
- 9.Berteau, O., Guillot, A., Benjdia, A., and Rabot, S. (2006) J. Biol. Chem. 281 22464–22470 [DOI] [PubMed] [Google Scholar]
- 10.Mougous, J. D., Green, R. E., Williams, S. J., Brenner, S. E., and Bertozzi, C. R. (2002) Chem. Biol. 9 767–776 [DOI] [PubMed] [Google Scholar]
- 11.Sardiello, M., Annunziata, I., Roma, G., and Ballabio, A. (2005) Hum. Mol. Genet. 14 3203–3217 [DOI] [PubMed] [Google Scholar]
- 12.Muller, I., Kahnert, A., Pape, T., Sheldrick, G. M., Meyer-Klaucke, W., Dierks, T., Kertesz, M., and Uson, I. (2004) Biochemistry 43 3075–3088 [DOI] [PubMed] [Google Scholar]
- 13.Kahnert, A., and Kertesz, M. A. (2000) J. Biol. Chem. 275 31661–31667 [DOI] [PubMed] [Google Scholar]
- 14.Hagelueken, G., Adams, T. M., Wiehlmann, L., Widow, U., Kolmar, H., Tummler, B., Heinz, D. W., and Schubert, W. D. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 7631–7636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diez-Roux, G., and Ballabio, A. (2005) Annu. Rev. Genomics Hum. Genet. 6 355–379 [DOI] [PubMed] [Google Scholar]
- 16.Kertesz, M. A. (2000) FEMS Microbiol. Rev. 24 135–175 [DOI] [PubMed] [Google Scholar]
- 17.Sareen, D., Newton, G. L., Fahey, R. C., and Buchmeier, N. A. (2003) J. Bacteriol. 185 6736–6740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, L., Goren, M. B., Holzer, T. J., and Andersen, B. R. (1988) Infect Immun. 56 2876–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997) Nucleic Acids Res. 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dierks, T., Dickmanns, A., Preusser-Kunze, A., Schmidt, B., Mariappan, M., von Figura, K., Ficner, R., and Rudolph, M. G. (2005) Cell 121 541–552 [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307–326 [DOI] [PubMed] [Google Scholar]
- 22.Storoni, L. C., McCoy, A. J., and Read, R. J. (2004) Acta Crystallogr. 60 432–438 [DOI] [PubMed] [Google Scholar]
- 23.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. 54 905–921 [DOI] [PubMed] [Google Scholar]
- 24.Jones, T. A., Zou, J. Y., Cowan, S. W., and Kjeldgaard, M. (1991) Acta Crystallogr. Sect. A 47 Pt. 2, 110–119 [DOI] [PubMed] [Google Scholar]
- 25.Terwilliger, T. C. (2004) Acta Crystallogr. 60 2144–2149 [DOI] [PubMed] [Google Scholar]
- 26.Winn, M. D., Isupov, M. N., and Murshudov, G. N. (2001) Acta Crystallogr. 57 122–133 [DOI] [PubMed] [Google Scholar]
- 27.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. 53 240–255 [DOI] [PubMed] [Google Scholar]
- 28.Lamzin, V. S., and Wilson, K. S. (1993) Acta Crystallogr. 49 129–147 [DOI] [PubMed] [Google Scholar]
- 29.Parish, T., and Stoker, N. G. (2000) Microbiology 146 Pt. 8, 1969–1975 [DOI] [PubMed] [Google Scholar]
- 30.Hatfull, G. F., and Jacobs, W. R., Jr. (eds) (2000) Molecular Genetics of Mycobacteria, pp. 313–320, ASM Press, Washington, DC
- 31.Ellis, H. R., and Poole, L. B. (1997) Biochemistry 36 15013–15018 [DOI] [PubMed] [Google Scholar]
- 32.O'Brien, P. J., and Herschlag, D. (1998) J. Am. Chem. Soc. 120 12369–12370 [Google Scholar]
- 33.Stankiewicz, P. J., and Gresser, M. J. (1988) Biochemistry 27 206–212 [DOI] [PubMed] [Google Scholar]
- 34.Dickmanns, A., Schmidt, B., Rudolph, M. G., Mariappan, M., Dierks, T., von Figura, K., and Ficner, R. (2005) J. Biol. Chem. 280 15180–15187 [DOI] [PubMed] [Google Scholar]
- 35.Hageman, T. C., and Scheraga, H. A. (1974) Arch. Biochem. Biophys. 164 707–715 [DOI] [PubMed] [Google Scholar]
- 36.Breidenbach, M. A., and Brunger, A. T. (2004) Nature 432 925–929 [DOI] [PubMed] [Google Scholar]
- 37.Roeser, D., Preusser-Kunze, A., Schmidt, B., Gasow, K., Wittmann, J. G., Dierks, T., von Figura, K., and Rudolph, M. G. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 81–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fetzner, S. (2002) Appl. Microbiol. Biotechnol. 60 243–257 [DOI] [PubMed] [Google Scholar]
- 39.Wentworth, P., Jr., Jones, L. H., Wentworth, A. D., Zhu, X., Larsen, N. A., Wilson, I. A., Xu, X., Goddard, W. A., III, Janda, K. D., Eschenmoser, A., and Lerner, R. A. (2001) Science 293 1806–1811 [DOI] [PubMed] [Google Scholar]
- 40.Lemieux, G. A., and Bertozzi, C. R. (1998) Trends Biotechnol. 16 506–513 [DOI] [PubMed] [Google Scholar]
- 41.Carrico, I. S., Carlson, B. L., and Bertozzi, C. R. (2007) Nat. Chem. Biol. 3 321–322 [DOI] [PubMed] [Google Scholar]
- 42.Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283–291 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.