Abstract
RecQ helicases are essential for the maintenance of genome stability. Five members of the RecQ family have been found in humans, including RECQ1, RECQ5, BLM, WRN, and RECQ4; the last three are associated with human diseases. At this time, only BLM and WRN helicases have been extensively characterized, and the information on the other RecQ helicases has only started to emerge. Our current paper is focused on the biochemical properties of human RECQ1 helicase. Recent cellular studies have shown that RECQ1 may participate in DNA repair and homologous recombination, but the exact mechanisms of how RECQ1 performs its cellular functions remain largely unknown. Whereas RECQ1 possesses poor helicase activity, we found here that the enzyme efficiently promotes DNA branch migration. Further analysis revealed that RECQ1 catalyzes unidirectional three-stranded branch migration with a 3′ → 5′ polarity. We show that this RECQ1 activity is instrumental in specific disruption of joint molecules (D-loops) formed by a 5′ single-stranded DNA invading strand, which may represent dead end intermediates of homologous recombination in vivo. The newly found enzymatic properties of the RECQ1 helicase may have important implications for the function of RECQ1 in maintenance of genomic stability.
RecQ helicases play an essential role in the maintenance of genome stability in all organisms (1–4). In humans, three of five known RecQ homologs (RECQ1, BLM, WRN, RECQ4, and RECQ5) have been implicated in distinct heritable diseases (5, 6). BLM is mutated in Bloom syndrome, which is characterized by growth retardation, sunlight sensitivity, immunodeficiency, genomic instability, and high incidence of cancer (7, 8). WRN is mutated in Werner syndrome, which is characterized by premature aging, genomic instability, and high incidence of cancer (9, 10). RECQ4 is mutated in Rothmund-Thomson syndrome, which is characterized by skin rash, small stature, skeletal dysplasias, chromosomal instability, premature aging, and high incidence of cancer (11–13). Thus, a common and foremost feature of these syndromes is genome instability and predisposition to cancer (6).
It is thought that RecQ helicases are implicated in regulation of recombination events, primarily homologous recombination (HR)2 (4, 14, 15). In Saccharomyces cerevisiae, deficiency in the sole RecQ homolog, Sgs1, leads to elevated levels of mitotic recombination, illegitimate recombination, sister chromatid exchange, and sensitivity to DNA damage by UV irradiation and other agents (4, 16). In Schizosaccharomyces pombe, mutants of the Sgs1 homolog Rqh1 show sensitivity to hydroxyurea and fail to recover from hydroxyurea arrest during S phase because of a high frequency of illegitimate recombination events (17). In humans, deficiency in BLM helicase leads to an increased frequency of sister chromatid exchanges and interhomolog exchanges (2, 7, 18).
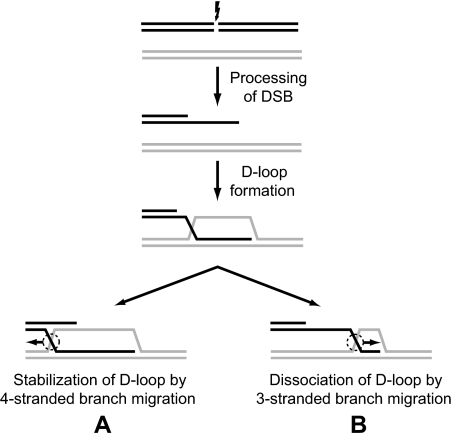
HR is responsible for the repair of DNA double-stranded breaks (DSB) and stalled replication forks (19–21). During DSB repair by HR, the DNA ends are processed by exonucleases to generate DNA with protruding ssDNA tails (22, 23). RAD51 recombinase then promotes invasion of these DNA molecules into homologous duplex DNA to generate joint molecules (D-loops) (Fig. 1). The invading 3′-ssDNA ends can then be used by DNA polymerase to prime DNA repair synthesis (22, 23). Next, depending on the pathway of HR, D-loops can be either extended by the four-stranded branch migration (Fig. 1, direction A) or disrupted (Fig. 1, direction B) either by three-stranded branch migration (24, 25) or alternatively through DNA unwinding by structure-specific DNA helicases.
FIGURE 1.
The role of branch migration in processing of homologous recombination intermediates. The scheme demonstrates formation of joint molecules (D-loop) during DSB repair and their further processing by branch migration that may result in either D-loop stabilization (A) or dissociation (B).
Extensive biochemical studies have been performed to better understand the function of RecQ helicases, especially human RecQ homologs. RecQ helicases catalyze DNA unwinding with a3′ to 5′ polarity on a variety of DNA substrates, such as forked duplexes, triple helices, three- or four-way junctions, and G-quadruplex DNA (26–28). In addition to DNA unwinding, human RecQ helicases show a broad spectrum of activities consistent with their role in HR including disruption of the RAD51 presynaptic filament (BLM and RECQ5) (14, 15), branch migration of Holliday junctions (BLM and WRN) (29, 30), annealing of complementary ssDNA (all five helicases) (27, 31–34), dissociation of mobile D-loops, and dissolution of double Holliday junctions (BLM) (15, 35, 36).
Although no disease has been linked to RECQ1, this protein is the most abundant among the five human RecQ helicases (37), and cellular studies indicate that RECQ1 is biologically important for genome stability (38, 39). Embryonic fibroblasts from RECQ1-deficient mice displayed aneuploidy and chromosomal instability, accumulation of DNA strand breaks, and persistent Rad51 foci. Moreover, RECQ1 deficiency resulted in cellular sensitivity to ionizing radiation. Depletion of RECQ1 from human cells by RNA interference resulted in elevated sister chromatid exchange and DNA damage sensitivity. These findings are consistent with a role for RECQ1 in the processing of DNA repair and recombination intermediates.
Of the human RecQ helicases, RECQ1 is the smallest with a size of 649 residues (40, 41). RECQ1 possesses an ssDNA annealing activity, DNA-stimulated ATPase activity, and helicase activity. The helicase activity of RECQ1 is only capable of unwinding short duplex DNA substrates (42). RECQ1 forms oligomers to perform its helicase and strand annealing functions (43). RECQ1 physically interacts with RPA, a ubiquitous ssDNA-binding protein, which stimulates its helicase activity; the enzyme can unwind up to ∼500 bp in the presence of RPA (28). RECQ1 interacts physically and functionally with mismatch repair factors (MSH2/MSH6, EXO-1, and MLH1/PMS2) that regulate genetic recombination (44). RECQ1 is also reported to physically interact with topoisomerase IIIα (45), importin-α homologs (46), and RAD51 (39); however, the functional significance of these interactions remains to be elucidated.
In the current work, we found that RECQ1 possesses DNA branch migration activity. We demonstrate that RECQ1 promotes both the three-stranded and four-stranded branch migration but shows significantly greater efficiency in the three-stranded reaction. A specific feature of RECQ1-catalyzed branch migration is a strong preference toward the 3′ → 5′ direction in both the three-stranded and four-stranded reactions, which distinguishes RECQ1 from other known branch migration proteins such as BLM helicase and RAD54 that show no significant preference in branch migration directionality. We show that branch migration activity with the 3′ → 5′ polarity allows RECQ1 to specifically disrupt recombination intermediates (D-loops) formed by invasion of tailed DNA with the 5′-protruding ends. These D-loops, in contrast to the D-loops formed by invasion of tailed DNA with the 3′-protruding ends, cannot be readily extended by DNA polymerase and therefore may represent unproductive recombination intermediates during DSB repair. Therefore, RECQ1 branch migration may prevent accumulation of these unconventional and potentially toxic intermediates in vivo.
EXPERIMENTAL PROCEDURES
Proteins and DNA—Human RAD51, BLM, RAD54, and RECQ1 were purified as described (15, 27, 47). Supercoiled pUC19 dsDNA was purified as described (48). All of the oligonucleotides (IDT, Inc.) used in this study (supplemental Table S1) were purified, labeled, and stored as described previously (49). dsDNA substrates were prepared by annealing of equimolar amounts of oligonucleotides, as described (49).
Branch Migration Assay—32P-Labeled forked DNA intermediates (strands 169*/71 and 193*/117; here and below in this section 32P-labeled strands are marked by asterisks) and nonlabeled tailed DNAs (strands 170/171 and 194/195) were prepared by annealing. To prepare substrates for three-stranded branch migration that occurs in either the 3′ → 5′ or 5′ → 3′direction, DNA intermediates 169*/71 or 193*/117 (32 nm, molecules) were mixed with ssDNA oligonucleotide 201 or 379, respectively (48 nm, molecules), and incubated in buffer containing 25 mm Tris acetate, pH 7.5, 2 mm ATP, 5 mm magnesium acetate, 2 mm DTT, BSA (100 μg/ml), 15 mm phosphocreatine, and creatine phosphokinase (30 units/ml) for 15 min at 37 °C. To prepare partially movable three-stranded structure, forked DNA (strand 169*/71) was mixed with ssDNA oligonucleotide 248 according to the procedure described above.
To prepare DNA substrates for four-stranded branch migration, 169*/71 or 193*/117 DNA intermediates (32 nm, molecules) were mixed with strand 170/171 or 194/195, respectively (38 nm, molecules) and incubated in buffer containing 25 mm Tris acetate, pH 7.5, 1 mm ATP, 0.5 mm magnesium acetate, 2 mm DTT, BSA (100 μg/ml), 15 mm phosphocreatine, and creatine phosphokinase (30 units/ml) for 15 min at 37 °C.
To start the reaction, the indicated amounts of RECQ1 helicase were added for 30 min at 37 °C. The reactions were stopped by the addition of 1.5% SDS and proteinase K (800 μg/ml), mixed with a 0.10 volume of loading buffer (70% glycerol, 0.1% bromphenol blue), and analyzed by electrophoresis in 8% polyacrylamide gels in TBE buffer (89 mm Tris borate, pH 8.3, and 1 mm EDTA) at 135 V for 1.5 h. The gels were dried on DEAE-81 paper (Whatman) and quantified using a Storm 840 PhosphorImager (GE Healthcare).
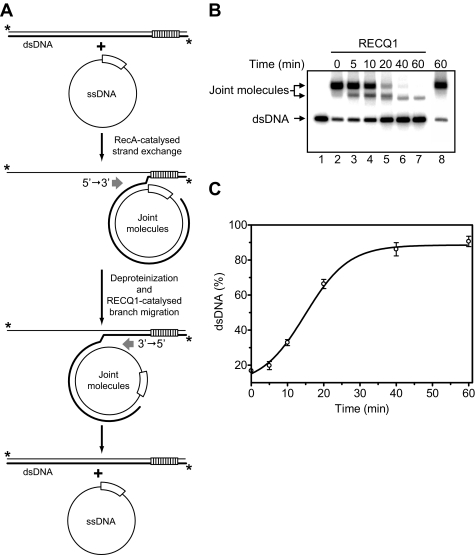
Branch Migration of Plasmid-based Joint Molecules—Joint molecules were prepared by carrying out a RecA-mediated three-stranded DNA exchange using pBSK(-) circular ssDNA and pBSK(+) linear dsDNA (see Fig. 5A) as follows. The RecA-ssDNA filaments were formed by incubating RecA protein (12 μm) with pBSK(-) circular ssDNA (20 μm, nucleotides) in buffer containing 25 mm Tris acetate, pH 7.5, 3 mm ATP, 15 mm MgCl2, 2 mm DTT, 100 μg/ml BSA, 15 mm phosphocreatine, and 30 units/ml creatine phosphokinase for 10 min at 37 °C. Then SSB (1.8 μm) was added, followed by a 5-min incubation. Three-stranded DNA exchange reaction was initiated by the addition of linear pBSK(+) dsDNA (15 μm, nucleotides) (super-coiled dsDNA cleaved with SpeI restriction endonuclease) to the RecA nucleoprotein filaments and continued for 20 min. Then the reactions were deproteinized by treatment with proteinase K (1.1 mg/ml) and 0.9% SDS for 15 min at 37 °C and purified by passing twice through S-400 Spin columns (GE Healthcare) equilibrated with 25 mm Tris acetate, pH 7.5, 50 mm NaCl, 5 mm MgCl2 at 23 °C. Branch migration of the purified joint molecules (0.4 nm, molecules) was carried out in buffer containing 25 mm Tris acetate, pH 7.5, 2 mm ATP, 3 mm magnesium acetate, 2 mm DTT, 100 μg/ml BSA, 15 mm phosphocreatine, and 30 units/ml creatine phosphokinase at 37 °C. RECQ1 protein (100 nm) was added to the reaction mixtures, followed by incubation for the indicated periods of time. The products of branch migration were deproteinized by the addition of 1.5% SDS and proteinase K (800 μg/ml), mixed with a 0.10 volume of loading buffer (70% glycerol, 0.1% bromphenol blue), and analyzed by electrophoresis in 1.3% agarose gels, visualized, and quantified as described above.
FIGURE 5.
RECQ1 catalyzes three-stranded branch migration on long DNA substrates. A, the scheme illustrates disruption by RECQ1 DNA joint molecules that were prepared by RecA-promoted DNA strand exchange between pBSK(-) ssDNA and pBSK(+) linearized dsDNA. DNA strand exchange terminates at the border of the f1 origins (marked by empty and striped boxes), which are present in pBSK(+) and pBSK(-) plasmids in inverted orientations. The asterisks indicate 32P label at the DNA 5′-end. B, analysis of D-loop dissociation by RECQ1 (lanes 3–7) by electrophoresis in a 1% agarose gel. In controls (lanes 2 and 8), RECQ1 was substituted by storage buffer. Lane 1 shows migration of dsDNA. C, the data in B shown as a graph. The error bars indicate S.E.
DNA Helicase Assay—DNA helicase assay was used to exclude a possibility that formation of a DNA product in the three-stranded branch migration assay occurs through the mechanism that involves DNA melting and reannealing, as opposed to the branch migration mechanism. The reactions were carried out in buffer containing 25 mm Tris acetate, pH 7.5, 2 mm ATP, 5 mm magnesium acetate, 2 mm DTT, BSA (100 μg/ml), 15 mm phosphocreatine, creatine phosphokinase (30 units/ml), and 32P-labeled forked DNA 169*/71 (32 nm, molecules). The reaction mixture also contained ssDNA oligonucleotide 196 (48 nm, molecules) whose 5′-end is complementary to the 3′-end of oligonucleotide 71 that would be produced as a result of unwinding of the forked DNA. At the same time annealing of 169*/71 and 196 DNA intermediates, which would produce a substrate for branch migration, was precluded by incorporation of a heterologous sequence in the 3′-end of oligonucleotide 196 (Fig. 2B).
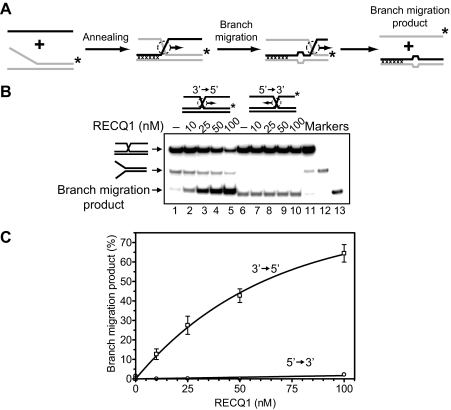
FIGURE 2.
RECQ1 helicase promotes the three-stranded DNA branch migration in the 3′ → 5′ direction. A, scheme depicts the three-stranded branch migration. Mutually heterologous terminal regions are marked by crosses. Unpaired ssDNA regions (four bases) that arise during branch migration are shown by widening in DNA duplex. These nonhomologous regions were introduced to block spontaneous branch migration. The asterisks indicate 32P label at the 5′-end DNA. B, the DNA products of the branch migration reactions were visualized by electrophoresis in a 8% polyacrylamide gel. Branch migration was initiated by addition of indicated amount of RECQ1 to appropriate DNA substrates followed by incubation for 30 min at 37 °C. In controls, storage buffer was added instead of RECQ1. Lanes 11–13 show migration of DNA markers. C, the data from B presented as a graph. The error bars indicate S.E.
The reactions were started by adding the indicated amounts of RECQ1 helicase and carried out for 30 min at 37 °C. The DNA products were deproteinized and analyzed by electrophoresis in 8% polyacrylamide gels, visualized, and quantified using a Storm 840 PhosphorImager (GE Healthcare).
Dissociation of Synthetic Mobile and Nonmobile D-loops—32P-Labeled branched DNA intermediates (254/373* or 254/209*) were prepared by annealing. To prepare mobile and nonmobile D-loops branched DNA intermediates 254/373* (32 nm, molecules) were mixed with either oligonucleotide 293 or 255 (48 nm, molecules) and incubated in buffer containing 25 mm Tris acetate, pH 7.5, 2 mm ATP, 5 mm magnesium acetate, 2 mm DTT, BSA (100 μg/ml), 15 mm phosphocreatine, and creatine phosphokinase (30 units/ml) for 10 min at 37 °C and then for 10 min at 30 °C. To prepare mobile D-loop, which can migrate in the 5′ → 3′ direction, a branched DNA intermediate (254/209*) was mixed with ssDNA oligonucleotide 293 according to the procedure described above. The reactions were started by adding the indicated amounts of RECQ1 helicase and carried out for 30 min at 30 °C. When indicated, a nonradioactive oligonucleotide 209 (48 nm, molecules) was included in the reaction mixture with nonmobile D-loops to prevent their reannealing after disruption by RecQ1. The products of D-loop dissociation were deproteinized and analyzed by electrophoresis in 6% polyacrylamide gels, visualized, and quantified using a Storm 840 PhosphorImager (GE Healthcare).
Disruption of Deproteinized Plasmid D-loops by RECQ1—D-loops with 3′-or5′-invaded ssDNA were prepared in a RAD51-promoted reaction, as previously described (24) using 32P-labeled single-stranded 100-mer oligonucleotides (oligonucleotide 209 or 373), in which the 64 base regions either from the 3′-or5′-ends were homologous to the same region of pUC-19 supercoiled DNA. D-loops were deproteinized and purified as in Ref. 24.
Dissociation of the deproteinized D-loops (150 pm, molecules) by RECQ1 (7.5 nm, or indicated otherwise) was carried out in buffer containing 25 mm Tris acetate, pH 7.5, 1 mm ATP, 1 mm magnesium acetate, 2 mm DTT, BSA (100 μg/ml), 15 mm phosphocreatine, and creatine phosphokinase (30 units/ml). The reaction mixtures were incubated for the indicated periods of time at 37 °C. The products of dissociation were deproteinized and analyzed by electrophoresis in 1% agarose gels, visualized, and quantified using a Storm 840 PhosphorImager (GE Healthcare).
Disruption of “Native” D-loops—To form D-loops, 32P-labeled ssDNA (oligonucleotide 209 or 373; 30 nm, molecules) was preincubated with RAD51 or RAD51 K133R (1 μm) proteins in buffer containing 25 mm Tris acetate, pH 7.5, 1 mm ATP, 1 mm calcium chloride (1 mm magnesium acetate for RAD51 K133R), 2 mm DTT, and BSA (100 μg/ml) for 15 min at 37 °C. D-loop formation was initiated by the addition of pUC19 supercoiled dsDNA (50 μm, nucleotides) followed by a 15-min incubation. Then in the case when RAD51 was used, to remove Ca2+, the reaction mixture was passed through a Bio-Gel P-6 spin column (Bio-Rad) equilibrated with buffer containing 25 mm Tris acetate, pH 7.5, 1 mm ATP, 1 mm magnesium acetate, 2 mm DTT, BSA (100 μg/ml), 15 mm phosphocreatine, and creatine phosphokinase (30 units/ml). Dissociation of native D-loops (without removal of RAD51) was initiated by the addition of RECQ1 (in indicated concentrations) and carried out for 30 min at 37 °C. In the case when RAD51 K133R was used, the ATP regeneration system was incorporated to the reaction mixtures before the addition of RECQ1 helicase. The products of D-loop dissociation were deproteinized, analyzed by electrophoresis in a 1% agarose gel, visualized, and quantified using a Storm 840 PhosphorImager (GE Healthcare).
RESULTS
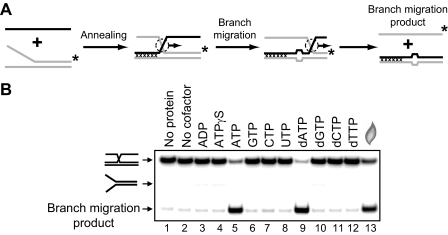
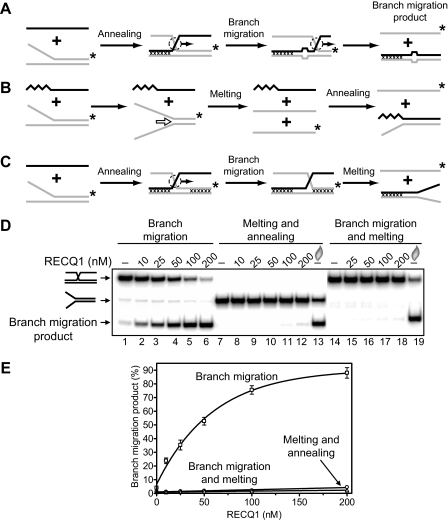
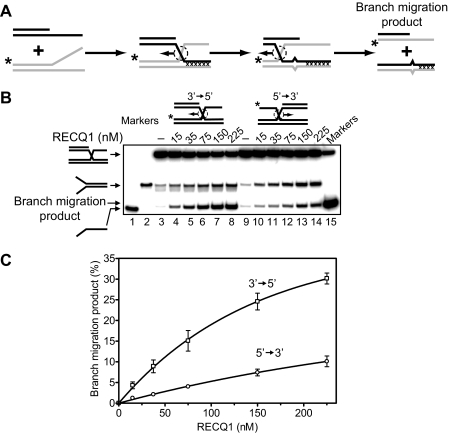
RECQ1 Promotes Three- and Four-stranded Branch Migration in the 3′ → 5′ Direction—It is known that two human RecQ helicases, BLM and WRN, promote branch migration of Holliday junctions (29, 30). Here we investigated whether RECQ1 may also catalyze DNA branch migration. To examine RECQ1 branch migration activity, we used synthetic DNA substrates. Previously, we used similar synthetic DNA substrates to analyze branch migration activity of RAD54 protein (24, 25, 50). We first tested the three-stranded branch migration activity of RECQ1 helicase. A pair of DNA substrates was constructed in a such way that one could support branch migration only in the 3′ → 5′ direction and another only in the 5′ → 3′ direction (Fig. 2, A and B, top). We also incorporated four mismatches in DNA branches to minimize spontaneous branch migration (Fig. 2A). The data show that RECQ1 efficiently catalyzes three-stranded branch migration (Fig. 2, B and C). The enzyme demonstrates a strong polarity bias: RECQ1 promoted branch migration in the 3′ → 5′ direction, but not in the 5′ → 3′ direction. To confirm the identity of the branch migration DNA products, we applied another labeling scheme for the DNA substrates, in which ssDNA (oligonucleotide 201 or 379) was 32P-labeled. Branch migration was monitored by incorporation of the ssDNA substrate into the mismatched dsDNA product (supplemental Fig. S1A). The rate and the polarity of RECQ1 branch migration were the same as with DNA substrates in which forked DNA (strand 169/71 or 193/117) was 32P-labeled (supplemental Fig. S1, B and C). In contrast, as expected RAD54 promotes the three-stranded branch migration on both these substrates with similar efficiency (supplemental Fig. S2). Branch migration activity of RECQ1 showed dependence on ATP hydrolysis (Fig. 3); no branch migration was observed in the absence of nucleotide cofactor (Fig. 3B, lane 2) or in the presence of ADP or ATPγS, a nonhydrolyzable ATP analog (Fig. 3B, lanes 3 and 4). Among other cofactors tested, only dATP supported the reaction.
FIGURE 3.
Effect of a different nucleotide cofactors on the DNA branch migration activity of RECQ1 helicase. A, scheme of branch migration assay. Mutually heterologous terminal region is marked by crosses. Unpaired ssDNA regions (four bases) that arise during branch migration are shown by widening in DNA duplex. The asterisks indicate 32P label at the 5′-end DNA. B, the reactions were carried out in the presence of indicated nucleotide cofactor (lanes 2–12) or in the absence of nucleotide cofactor (lane 2). Branch migration was initiated by the addition of 100 nm RECQ1 followed by incubation for 30 min at 37 °C. The DNA products of the branch migration reactions were separated by electrophoresis in a 8% polyacrylamide gel. In control, storage buffer was added instead of RECQ1 (lane 1). In lane 13 melting of DNA substrate was carried out by heating and followed by thermal reannealing.
It was previously shown that RECQ1 has a helicase activity, although it is weak (42). Therefore, we examined the possibility that DNA product observed in Fig. 2B could be formed through the helicase activity of RECQ1, not branch migration, as a result of unwinding of DNA substrates followed by cross-reannealing of the resultant ssDNA strands (Fig. 4B). We tested this possibility by using two types of DNA substrates for RECQ1. First, we use a mixture of the fork DNA (strand 169/71, 63 bp of dsDNA region) and ssDNA (Fig. 4B). The ssDNA was complementary to the ssDNA strands that were expected to be produced by RECQ1 helicase unwinding of the forked DNA. However, we found that in this case the expected DNA product was not produced (Fig. 4D, lanes 7–12); in contrast, it was detected after the forked DNA was dissociated by heat followed by annealing of DNA strands after cooling of the reaction mixture (Fig. 4D, lane 13). Then we used partially movable three-stranded molecule to confirm that RECQ1 acts in the three-stranded reaction (Fig. 2B) as a branch migration protein, not a helicase (Fig. 4C). This substrate was designed in a such way that it can migrate through only 13 bp, but complete strands separation would require melting of the terminal 50 bp through the helicase activity. The data show that RECQ1 cannot dissociate this partially movable three-stranded structure (Fig. 4, D, lanes 14–18, and E), even though RECQ1 can disrupt partially movable four-stranded substrates with shorter dsDNA arms (25 bp) under favorable conditions (low free Mg2+ concentration) for its helicase activity (27). Taken together, the results indicate that RECQ1 promotes efficient three-stranded reaction (Fig. 2B) through the branch migration but not the helicase mechanism.
FIGURE 4.
RECQ1 helicase catalyzes the three-stranded branch migration more efficiently than DNA unwinding. Shown are experimental schemes to distinguish between the three-stranded branch migration and the helicase activity of RECQ1 using mobile Kappa structure (A), forked DNA plus ssDNA (B), and partially mobile Kappa structures (C) as substrates. The asterisk indicates 32P label at the 5′-end DNA. The arrows indicate the direction of branch migration or DNA unwinding. Mutually heterologous terminal regions (in A and C) are marked by crosses. Unpaired DNA regions that arise during branch migration (A) are shown by widening in DNA duplex (four bases). The zig-zag line (B) denotes a nonhomologous DNA sequence that was introduced into the 3′-region of ssDNA to prevent formation of the PX junction by annealing to the forked DNA, which could be processed by branch migration. D, the DNA products were visualized by electrophoresis in a 8% polyacrylamide gel. The reactions were initiated by the addition of indicated amount of RECQ1 to DNA substrates followed by incubation for 30 min at 37 °C. In lanes 14–19, oligonucleotide 2 was added to prevent reannealing of the Kappa structure after melting by RECQ1. In controls, storage buffer was added instead of RECQ1. In lanes 13 and 19, melting of DNA substrates was carried out by heating and followed by thermal reannealing. E, the data from D presented as a graph. The error bars indicate S.E.
We next tested whether RECQ1 can promote branch migration on long DNA substrates. The substrates were prepared by carrying out RecA-mediated DNA strand exchange between circular pBSK(-) ssDNA and linearized pBSK(+) dsDNA (Fig. 5A). pBSK(+) and pBSK(-) plasmids (2958 bp) are fully homologous, except for the f1 ori (306 bp) that is present in these plasmids in the inverted orientations. Inversion of the f1 ori creates a region of heterology between linear pBSK(+) dsDNA and pBSK(-) ssDNA, which caused termination of DNA strand exchange, preventing formation of the nicked circular product but permitting formation of joint molecules (Fig. 5A). Joint molecules were then deproteinized and used a substrate for RECQ1 (Fig. 5B, lane 2). RECQ1 efficiently promoted three-stranded branch migration through a 2243-bp DNA region, as evidenced by formation of linear dsDNA (Fig. 5, B and C). In contrast, it was previously shown that RECQ1 cannot efficiently unwind DNA duplexes longer than 30 bp by the helicase mechanism (42).
Then, we tested RECQ1 branch migration activity in the four-stranded reaction using synthetic PX junctions (Fig. 6A). We found that RECQ1 can promote the four-stranded reaction. In this reaction, RECQ1 also shows a preference toward branch migration in the 3′ → 5′ direction. However, in this case the bias was less significant; branch migration occurred approximately three times more efficiently in this direction than in the 5′ → 3′ direction (Fig. 5, B and C).
FIGURE 6.
RECQ1 helicase promotes the four-stranded DNA branch migration in the 3′ → 5′ direction. A, schemes depict the four-stranded branch migration. Mutually heterologous terminal regions are marked by crosses. Unpaired ssDNA regions (one base) that arise during branch migration are shown as a mismatch. These nonhomologous regions were introduced to block spontaneous branch migration. The asterisks indicate 32P label at the 5′-end DNA. B, the DNA products of the branch migration reactions were visualized by electrophoresis in a 8% polyacrylamide gel. Branch migration was initiated by the addition of indicated amount of RECQ1 to appropriate DNA substrates followed by incubation for 30 min at 37 °C. In controls, storage buffer was added instead of RECQ1. Lanes 1, 2, and 15 show migration of DNA markers. C, the data from B presented as a graphs. The error bars indicate S.E.
It should be noted that the efficiency of the four-stranded branch migration was much weaker than three-stranded reaction. Thus, the four-stranded reaction proceeded efficiently only at low concentrations of Mg2+ (less than 1 mm) that are favorable for RECQ1 helicase activity (27) and facilitate formation of the open PX junction conformation, which is required for branch migration. At the high Mg2+ concentration (5 mm) that we used for the three-stranded reaction, RECQ1 is unable to promote efficient four-stranded branch migration on PX or X junctions (data not shown). The efficiency of the four-stranded branch migration promoted by RECQ1 was comparable with the protein helicase activity, because we also observed appearance of the forked DNA product (reversal of the annealing), which resulted from the helicase activity of RECQ1 on a short 31-bp region (Fig. 6B). In contrast, in the three-stranded reaction we did not observe formation of the fork DNA product (Figs. 2B and 4D, lanes 1–6), because the efficiency of the three-stranded branch migration is significantly higher than that of the helicase activity of RECQ1.
In contrast to RECQ1, RAD54 catalyzes efficient four-stranded branch migration without significant polarity bias (supplemental Fig. S3). We also tested the polarity of four-stranded branch migration of another human RecQ helicase, BLM. Similar to RAD54 BLM promotes efficient four-stranded branch migration of the PX junction and shows only a slight preference for migration in the 3′ → 5′ direction (supplemental Fig. S3).
Thus, the results indicate that RECQ1 is a three-stranded branch migration protein that shows a significant preference for branch migration in the 3′ → 5′ direction. In contrast, RECQ1 shows low efficiency in promoting the four-stranded branch migration.
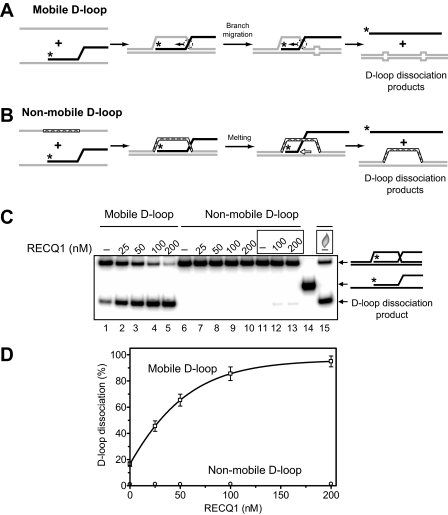
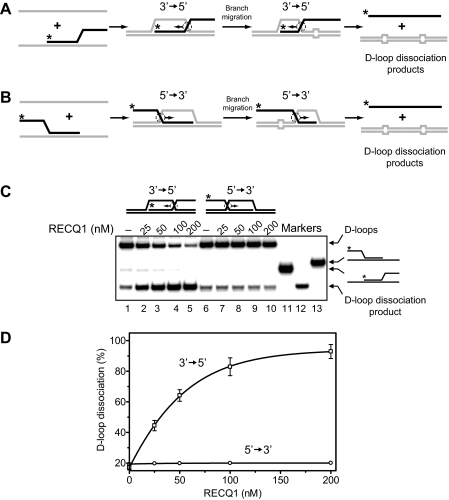
RECQ1 Promotes D-loops Dissociation by Branch Migration, not by DNA Unwinding—D-loops, key intermediates of HR, can be either extended by the four-stranded branch migration or disrupted through the three-stranded branch migration or the helicase mechanism (Fig. 1). Previously, we showed that RAD54 dissociates D-loops by branch migration (24, 25). BLM helicase can also promote efficient disruption of D-loops (35), but the mechanism of D-loop dissociation by BLM is not completely understood because BLM has both DNA unwinding (51, 52) and DNA branch migration activities (29). Here, we examined whether RECQ1 can promote D-loop disruption through its three-stranded branch migration activity that we described above (Figs. 2B and 4D). We used synthetic mobile D-loop substrate, which was prepared by annealing of ssDNA to the branched DNA molecule (Fig. 7A). To minimize spontaneous dissociation of resulting D-loops, we introduced four mismatches from each side of the D-loops that lead to significant reduction of spontaneous branch migration (Fig. 7C, lane 1). Addition of RECQ1 helicase to the reactions results in efficient dissociation of mobile D-loops (Fig. 7, C, lanes 2–5, and D). We examined whether RECQ1 dissociates D-loops by the branch migration (Fig. 7A) or by the helicase mechanism (Fig. 7B). For this purpose, we used synthetic nonmobile D-loops as a substrate, which can be dissociated only by the helicase mechanism. We found that RECQ1 cannot disrupt synthetic nonmobile D-loops, which would require melting of a 63-bp heteroduplex region (Fig. 7, C, lanes 6–13, and D), even though it can disrupt shorter nonmobile D-loops (21 bp of a heteroduplex region) under optimal conditions (low free Mg2+ concentration) for RECQ1 helicase activity (27). Therefore, we conclude that D-loop dissociation occurs through the branch migration mechanism (Fig. 7A). We then examined the directionality bias in D-loop disruption by RECQ1 using synthetic mobile D-loops, which can be dissociated in either the 3′ → 5′ (Fig. 8A) or the 5′ → 3′ (Fig. 8B) directions. We found that RECQ1 specifically disrupts synthetic D-loops in the 3′ → 5′ direction but is unable to promote efficient D-loop dissociation in the 5′ → 3′ direction. In this respect RECQ1 helicase is different from another branch migration protein, RAD54, which does not show distinct polarity in branch migration (25) and can efficiently dissociate D-loops in the 5′ → 3′ direction (24).
FIGURE 7.
RECQ1 helicase promotes dissociation of D-loops by the branch migration but not the helicase mechanism. A and B, the schemes depict the structures of mobile and nonmobile oligonucleotide-based D-loops and the expected mechanisms of their dissociation by the branch migration (A) or helicase (B) activities. Unpaired DNA regions that arise during branch migration are shown by widening in DNA duplex (four bases from each side of mobile D-loops), and the nonhomologous displaced strand of nonmobile D-loops is indicated by the cross-hatched segment. The arrows indicate the direction of branch migration and DNA unwinding. The asterisks indicate 32P label at the 5′-end DNA. C, the DNA products of D-loop dissociation were visualized by electrophoresis in a 6% polyacrylamide gel. The dissociation was initiated by the addition of RECQ1 in indicated concentration to the mobile or nonmobile D-loops followed by incubation for 30 min at 30 °C. In controls, storage buffer was added instead of RECQ1. In reactions in lanes 11–13 and 15 (marked by the rectangles), an oligonucleotide complementary to one DNA strand of D-loops was added to prevent their possible reannealing after melting. In lane 15, D-loop melting was carried out by heating followed by thermal reannealing. Lane 14 shows the migration of DNA markers. D, the data from C presented as a graph. The error bars indicate S.E.
FIGURE 8.
RECQ1 catalyzes dissociation of mobile D-loops in the 3′ → 5′ direction. A and B, the schemes depict the structures of mobile oligonucleotide-based D-loops that migrates in either 3′ → 5′ (A) or 5′ → 3′ (B) direction. Unpaired DNA regions that arise during branch migration are shown by widening in DNA duplex (four bases from each side of mobile D-loops). The arrows indicate the direction of DNA branch migration. The asterisks indicate 32P label at the 5′-end DNA. C, the DNA products of branch migration were visualized by electrophoresis in a 6% polyacrylamide gel. The reactions were initiated by the addition of RECQ1 in indicated concentration to the mobile D-loops, which can be dissociated by branch migration either in the 3′ → 5′ (lanes 1–5) or in the 5′ → 3′ (lanes 6–10) direction. In controls (lanes 1 and 6), storage buffer was added instead of RECQ1. Lanes 11–13 show the migration of DNA markers. D, the data from C presented as a graph. The error bars indicate S.E.
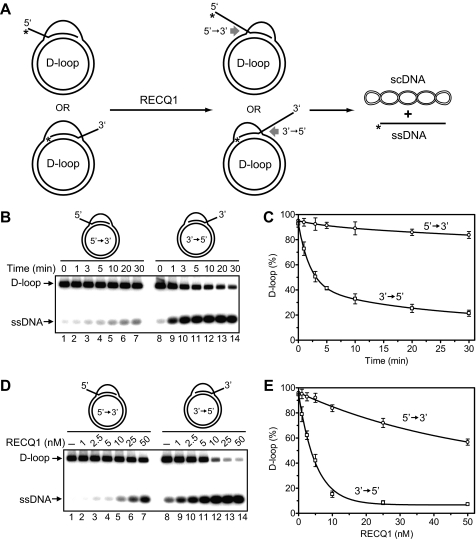
RECQ1 Dissociates D-loops Formed on Plasmid DNA by the 5′-Invading ssDNA—The data described above suggest that the 3′ → 5′ polarity preference in branch migration would make RECQ1 specifically able to dissociate D-loops formed with a 5′-invading but not with a 3′-invading DNA strand (Fig. 9A). To test this prediction we prepared plasmid-based D-loops with either 3′- or 5′-invading DNA strands using RAD51-promoted DNA strand exchange. After deproteinization and purification, the D-loops were used as substrates for RECQ1 helicase (Fig. 9A). We found that RECQ1 promotes efficient dissociation of D-loops formed by the 5′-invading ssDNA (Fig. 9, B, lanes 8–14, and C). In contrast, RECQ1 poorly dissociated D-loops formed by the 3′-invading ssDNA (Fig. 9, B, lanes 1–7, and C). We next determined the dependence of D-loop disruption on the RECQ1 concentration. We found that 50% dissociation of D-loops with the 3′-invading ssDNA required more than 10-fold higher RECQ1 concentration than dissociation of 50% of D-loops with the 3′-invading ssDNA, 5 nm versus more than 50 nm RECQ1, respectively (Fig. 9, D and E).
FIGURE 9.
RECQ1 helicase catalyzes specific dissociation of 5′-invaded joint molecules (D-loops). A, scheme representing dissociation of D-loops formed by 3′- and 5′-invaded ssDNA. The asterisks indicate 32P label at the 5′-end DNA. scDNA indicates supercoiled dsDNA. B, the time course of D-loop dissociation by RECQ1. The reaction was initiated by adding RECQ1 (7.5 nm) to purified D-loops, carried out for the indicated periods of time, and analyzed by electrophoresis in a 1% agarose gel. C, the data from B, shown as a graph. D, effect of the RECQ1 concentrations on dissociation of D-loops (150 pm). RECQ1 was added in the indicated concentrations; the reactions were carried out for 30 min, and the DNA products were analyzed in a 1% agarose gel. E, the data from D are presented as a graph. The error bars in C and E indicate S.E.
We then examined whether dissociation of plasmid-based D-loops by RECQ1 occurs through the branch migration or the helicase mechanism. Because ssDNA-binding proteins RPA and SSB are known to enhance DNA unwinding activity of the RecQ family helicases including RECQ1 (28, 42, 53–55), we performed D-loop disruption by RECQ1 in the presence of RPA or SSB. We found that neither SSB nor RPA stimulated RECQ1-dependent D-loop dissociation (supplemental Fig. S4). Thus, these results are inconsistent with the helicase mechanism and indicate that branch migration activity of RECQ1 is responsible instead for D-loop disruption.
The observed efficiency of dissociation of the 5′-invaded D-loop was comparable with the efficiency reported previously for BLM (35); however, in contrast to RECQ1, BLM showed only a small preference toward disruption of the 5′-invaded D-loops (supplemental Fig. S5), which is consistent with previously published data (35). Thus, RECQ1 is unique among known branch migration proteins in that it shows preferential dissociation of the 5′-invaded D-loops.
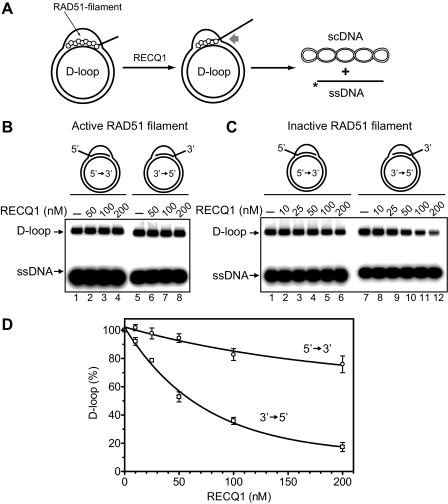
Effect of RAD51 on D-loop Dissociation by RECQ1—In vivo, D-loops are generally formed by Rad51 recombinase. Afterward, Rad51 likely remains associated with D-loops and therefore may represent a barrier for their dissociation. Here we asked whether the branch migration activity of RECQ1 can dissociate native D-loops (nondeproteinized D-loops with RAD51 protein bound to them), which mimic in vivo recombination intermediates (Fig. 10A). Previously, we have shown that dissociation of native D-loops by RAD54 or BLM depends on the conformation of the RAD51 (15, 24), which exists in an active (ATP-bound) or in an inactive (ADP-bound) form (48). To promote DNA strand exchange, RAD51 has to be in an active form but can be converted to an inactive conformation during ATP hydrolysis in the presence of Mg2+. In vitro, an active form of RAD51 can be mimicked by the RAD51 K133R ATPase mutant, which can bind but not hydrolyze ATP. RAD51 in an active form inhibits D-loop dissociation by both RAD54 and BLM; RAD51 inactivation is required for D-loop dissociation (15, 24). Here, we found that RECQ1 can promote dissociation of native D-loops. Similar to RAD54 and BLM (15, 24), RECQ1 could only dissociate native D-loops when RAD51 bound to the D-loops was in an inactive form (Fig. 10, B–D). As with deproteinized D-loops (Fig. 9, B–E), RECQ1 preferentially dissociates native D-loops formed by the 5′-invading ssDNA (Fig. 10, C and D).
FIGURE 10.
Dissociation of native D-loops by RECQ1 helicase required inactivation of RAD51 filament. A, The experimental scheme. The asterisks indicate the 32P label at the 5′-DNA end. scDNA indicates supercoiled dsDNA. B, RECQ1 cannot disrupt native D-loops in the presence of an active RAD51 filament (formed by the RAD51 K133R ATPase-deficient mutant). The reactions were initiated by the addition of RECQ1 in indicated concentrations. C, RECQ1 disrupts D-loops formed by RAD51 after filament inactivation (Ca2+ removal). The protein concentrations are indicated above the gel. In lanes 1 and 7, the protein was replaced by RECQ1 storage buffer. D, the data from C presented as a graph. The error bars indicate S.E.
DISCUSSION
Previously, it was shown that RECQ1 is important for the maintenance of genome stability in mice and human cells (38, 39). Here we investigated the biochemical properties of RECQ1 that may contribute to its biological function. We found that RECQ1 possesses an ATPase-dependent DNA branch migration activity. We also demonstrated that RECQ1 may employ its DNA branch migration activity to disrupt recombination intermediates, D-loops, indicating a possible role for RECQ1 in HR.
HR is essential for DSB repair, segregation of homologous chromosomes, propagation of genetic diversity, and maintenance of telomeres (20, 23, 56–61). However, inappropriate or untimely initiation of HR can generate harmful genome rearrangements and lead to loss of heterozygosity (62). Therefore, cells have evolved specific mechanisms to control recombination and to coordinate it with other cellular functions. Previous work implicated RecQ helicases in regulation of HR events. Mutations in the RecQ helicase genes show a hyper-recombination phenotype causing genome instability that leads to inherited human disorders, Werner, Bloom, and Rothmund-Thomson syndromes (5, 6). We recently found that BLM can prevent untimely initiation of HR by disrupting the RAD51 filament (15). Moreover, it was also reported by Hu et al. (14) that RECQ5 has similar RAD51-filament disrupting activity. At a later stage, HR can also be inhibited by disrupting joint molecules (Fig. 1B), important HR intermediates. It was shown that BLM helicase can promote dissociation of HR joint molecules, D-loops (15, 35, 52). However, D-loop disruption activity of BLM can also drive HR forward, channeling the HR intermediates into the synthesis-dependent strand annealing (SDSA) pathway, in which D-loop disruption represents a necessary step toward completion of the recombination process, e.g. displacement of the invaded ssDNA end after extension by DNA polymerase is required for annealing to the second ssDNA end (Fig. 11A) (15, 63, 64).
FIGURE 11.
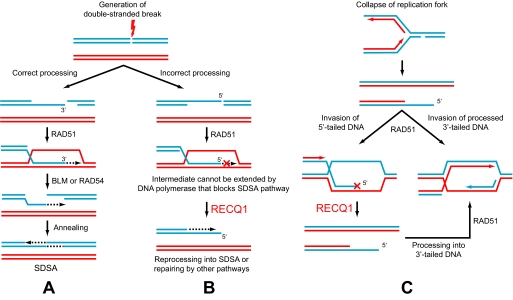
Possible role of RECQ1 helicase in repair of nonproductive HR intermediates. A, The SDSA mechanism of DSB repair. Processing of DSB results in formation of the 3′-tailed dsDNA, which can invade the homologous chromosome forming D-loops. To retrieve lost information, the 3′-end of tailed DNA is extended by DNA polymerase extended joint molecules are then dissociated by branch migration proteins (e.g. by BLM or RAD54) followed by reannealing of the extended tailed dsDNA with the second end of broken chromosome. B, unconventional processing of DSB results in generation of the 5′-tailed dsDNA. D-loops resulted from invasion of the 5′-tailed DNA cannot be readily extended by DNA polymerase along the SDSA mechanism. These unproductive intermediates can be disrupted by RECQ1 helicase. After disruption, tailed DNA with the 5′-protruding end can be reprocessed into tailed DNA with the 3′-protruding ends and channeled into the SDSA mechanism or can be repaired by another mechanism. C, restart of collapsed replication forks by HR. Invasion of 5′-tailed DNA creates nonproductive intermediates, which may be dissociated by RECQ1 (left). After dissociation, 5′-tailed DNA are processed by exonucleases into 3′-tailed DNA followed by Rad51-mediated invasion to initiate replication restart.
Here, we found that another member of the RecQ helicase family, RECQ1, catalyzes dissociation of D-loops. Our analysis revealed that RECQ1 dissociates D-loops by the branch migration but not the helicase mechanism. We found that RECQ1 preferentially disrupts D-loops with 5′-invaded ssDNA (Figs. 9 and 10). By analogy with yeast, it is generally thought that in human cells during DSB repair by HR, the exonucleases mainly generate tailed intermediates with the 3′-protruding ends (Fig. 11A), although the mechanism(s) of DNA recession is not yet fully elucidated (22, 23). However, given a broad spectrum of nucleases and helicases in human cells, it seems very likely that at least some fraction of 5′-ssDNA can be generated during DNA repair (Fig. 11B). Furthermore, in vitro RAD51 protein can utilize tailed DNA with the 5′-protruding ends with similar or even greater efficiency than tailed DNA with the 3′-protruding ends (65). Because such D-loops cannot prime DNA synthesis, they may represent a dead end product of HR (Fig. 11B). The RECQ1 activity described in this work may serve to specifically disrupt these potentially toxic HR intermediates (Fig. 11B). After disruption of dead end D-loops, tailed DNA with the 5′-protruding end either can be reprocessed into tailed DNA with the 3′-protruding ends, e.g. by action of DNA polymerase and/or of 5′ to 3′ exonucleases, and channeled into the SDSA mechanism or can be repaired by another mechanism (Fig. 11B).
RECQ1 may also play a role in a HR-dependent restart of collapsed replication forks. A one-ended DSB with the 5′-protruding ends can be generated when incoming replication forks encounter nicks on the DNA template (Fig. 11C). They can also be produced by endonucleolytic cleavage of stalled replication forks. In both cases, invasion of nonprocessed 5′-tailed DNA into homologous duplex DNA may result in failed replication restart or at least may require additional activities, e.g. four-stranded branch migration activity, to switch 5′-invaded D-loop into productive replication fork. RECQ1 may specifically dissociate nonproductive 5′-invaded D-loops, allowing procession of 5′-tailed DNA ends into 3′-tailed ones by exonucleases followed by their invasion into homologous dsDNA by RAD51 (Fig. 11C).
All of the proteins of the RecQ family show the 3′ → 5′ polarity in their helicase activity (66). Consistent with this polarity, BLM, for instance, disrupts nonmobile D-loops formed by 5′-invaded ssDNA, which only can be disrupted by the action of a helicase, with a significantly higher efficiency than D-loops formed by 3′-invading ssDNA (see scheme in Fig. 7B) (52). However, surprisingly, the helicase polarity does not seem to “automatically” translate into the same polarity in branch migration. Thus, BLM promotes disruption of mobile D-loops, which likely proceeds through the branch migration mechanism, with similar efficiency regardless of whether they are formed by 3′-or5′-invaded ssDNA (35) (also supplemental Fig. S3). In contrast, RECQ1 shows the same polarity bias in disrupting of nonmobile (27) and mobile D-loops (our current data), indicating an identical polarity preference of its helicase and branch migration activities. The mechanistic basis for these differences in the behavior of RECQ1 and BLM remains to be elucidated.
Similar to other branch migration proteins, branch migration activity of RECQ1 showed dependence on ATP hydrolysis. Typical for the RecQ family helicases (26), apart from ATP, the RECQ1 activity was also supported by dATP. Less selective are the RECQ5α (67), WRN, and BLM (68) helicases, which in addition to ATP and dATP can utilize most of the nucleotide cofactors to promote DNA unwinding. Interesting to note, RAD54 protein shows even higher selectivity than RECQ1 with respect to nucleotide cofactors; it promotes DNA branch migration only in the presence of ATP.3
In summary, our current results show that RECQ1 has a unidirectional 3′ → 5′ branch migration activity and promotes preferential dissociation of model recombination intermediates that result from invading of the 5′-ssDNA into homologous dsDNA (Fig. 11). In vivo, this activity of RECQ1 may help to dissolve nonproductive and potentially toxic HR intermediates and facilitate the restart of collapsed replication forks.
Supplementary Material
Acknowledgments
We thank P. Sung and W. Holloman for human RAD51 and BLM expression vectors and M. Rossi and O. Mazina for the comments and discussion.
This work was supported, in whole or in part, by National Institutes of Health Grant CA100839 and by a grant from the Intramural Research Program of NIA, National Institutes of Health (to R. M. B., Jr.). This work was also supported by Leukemia and Lymphoma Society Scholar Award 1054-09 (to A. V. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and supplemental Figs. S1–S5.
Footnotes
The abbreviations used are: HR, homologous recombination; dsDNA, double-stranded DNA; ssDNA, single-stranded DNA; BSA, bovine serum albumin; DTT, dithiothreitol; DSB, double-stranded break; ATPγS, adenosine 5′-O-(thiotriphosphate); SDSA, synthesis-dependent strand annealing; SSB, single-strand binding protein.
D. V. Bugreev and A. V. Mazin, unpublished observation.
References
- 1.Brosh, R. M., Jr., and Bohr, V. A. (2007) Nucleic Acids Res. 35 7527-7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hickson, I. D. (2003) Nat. Rev. Cancer 3 169-178 [DOI] [PubMed] [Google Scholar]
- 3.Opresko, P. L., Cheng, W. H., and Bohr, V. A. (2004) J. Biol. Chem. 279 18099-18102 [DOI] [PubMed] [Google Scholar]
- 4.Wu, L., and Hickson, I. D. (2006) Annu. Rev. Genet. 40 279-306 [DOI] [PubMed] [Google Scholar]
- 5.Hanada, K., and Hickson, I. D. (2007) Cell Mol. Life Sci. 64 2306-2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrigan, J. A., and Bohr, V. A. (2003) Biochimie. (Paris) 85 1185-1193 [DOI] [PubMed] [Google Scholar]
- 7.German, J. (1995) Dermatol. Clin. 13 7-18 [PubMed] [Google Scholar]
- 8.Ellis, N. A., Groden, J., Ye, T. Z., Straughen, J., Lennon, D. J., Ciocci, S., Proytcheva, M., and German, J. (1995) Cell 83 655-666 [DOI] [PubMed] [Google Scholar]
- 9.Epstein, C. J., Martin, G. M., Schultz, A. L., and Motulsky, A. G. (1966) Medicine (Baltimore) 45 177-221 [DOI] [PubMed] [Google Scholar]
- 10.Yu, C. E., Oshima, J., Fu, Y. H., Wijsman, E. M., Hisama, F., Alisch, R., Matthews, S., Nakura, J., Miki, T., Ouais, S., Martin, G. M., Mulligan, J., and Schellenberg, G. D. (1996) Science 272 258-262 [DOI] [PubMed] [Google Scholar]
- 11.Wang, L. L., Levy, M. L., Lewis, R. A., Chintagumpala, M. M., Lev, D., Rogers, M., and Plon, S. E. (2001) Am. J. Med. Genet. 102 11-17 [DOI] [PubMed] [Google Scholar]
- 12.Kitao, S., Shimamoto, A., Goto, M., Miller, R. W., Smithson, W. A., Lindor, N. M., and Furuichi, Y. (1999) Nat. Genet. 22 82-84 [DOI] [PubMed] [Google Scholar]
- 13.Lindor, N. M., Furuichi, Y., Kitao, S., Shimamoto, A., Arndt, C., and Jalal, S. (2000) Am. J. Med. Genet. 90 223-228 [DOI] [PubMed] [Google Scholar]
- 14.Hu, Y., Raynard, S., Sehorn, M. G., Lu, X., Bussen, W., Zheng, L., Stark, J. M., Barnes, E. L., Chi, P., Janscak, P., Jasin, M., Vogel, H., Sung, P., and Luo, G. (2007) Genes Dev. 21 3073-3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bugreev, D. V., Yu, X., Egelman, E. H., and Mazin, A. V. (2007) Genes Dev. 21 3085-3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bjergbaek, L., Cobb, J. A., and Gasser, S. M. (2002) Swiss Med. Wkly. 132 433-442 [DOI] [PubMed] [Google Scholar]
- 17.Stewart, E., Chapman, C. R., Al-Khodairy, F., Carr, A. M., and Enoch, T. (1997) EMBO J. 16 2682-2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.German, J. (1993) Medicine (Baltimore) 72 393-406 [PubMed] [Google Scholar]
- 19.Wyman, C., and Kanaar, R. (2006) Annu. Rev. Genet. 40 363-383 [DOI] [PubMed] [Google Scholar]
- 20.West, S. C. (2003) Nat. Rev. Mol. Cell Biol. 4 435-445 [DOI] [PubMed] [Google Scholar]
- 21.McGlynn, P., and Lloyd, R. G. (2002) Nat. Rev. Mol. Cell Biol. 3 859-870 [DOI] [PubMed] [Google Scholar]
- 22.Pâques, F., and Haber, J. E. (1999) Microbiol. Mol. Biol. Rev. 63 349-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krogh, B. O., and Symington, L. S. (2004) Annu. Rev. Genet. 38 233-271 [DOI] [PubMed] [Google Scholar]
- 24.Bugreev, D. V., Hanaoka, F., and Mazin, A. V. (2007) Nat. Struct. Mol. Biol. 14 746-753 [DOI] [PubMed] [Google Scholar]
- 25.Bugreev, D. V., Mazina, O. M., and Mazin, A. V. (2006) Nature 442 590-593 [DOI] [PubMed] [Google Scholar]
- 26.Bachrati, C. Z., and Hickson, I. D. (2003) Biochem. J. 374 577-606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma, S., Sommers, J. A., Choudhary, S., Faulkner, J. K., Cui, S., Andreoli, L., Muzzolini, L., Vindigni, A., and Brosh, R. M., Jr. (2005) J. Biol. Chem. 280 28072-28084 [DOI] [PubMed] [Google Scholar]
- 28.Cui, S., Arosio, D., Doherty, K. M., Brosh, R. M., Jr., Falaschi, A., and Vindigni, A. (2004) Nucleic Acids Res. 32 2158-2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karow, J. K., Constantinou, A., Li, J. L., West, S. C., and Hickson, I. D. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 6504-6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Constantinou, A., Tarsounas, M., Karow, J. K., Brosh, R. M., Bohr, V. A., Hickson, I. D., and West, S. C. (2000) EMBO Rep. 1 80-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheok, C. F., Wu, L., Garcia, P. L., Janscak, P., and Hickson, I. D. (2005) Nucleic Acids Res. 33 3932-3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macris, M. A., Krejci, L., Bussen, W., Shimamoto, A., and Sung, P. (2006) DNA Repair (Amst.) 5 172-180 [DOI] [PubMed] [Google Scholar]
- 33.Garcia, P. L., Liu, Y., Jiricny, J., West, S. C., and Janscak, P. (2004) EMBO J. 23 2882-2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Machwe, A., Xiao, L., Groden, J., Matson, S. W., and Orren, D. K. (2005) J. Biol. Chem. 280 23397-23407 [DOI] [PubMed] [Google Scholar]
- 35.Bachrati, C. Z., Borts, R. H., and Hickson, I. D. (2006) Nucleic Acids Res. 34 2269-2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, L., and Hickson, I. D. (2003) Nature 426 870-874 [DOI] [PubMed] [Google Scholar]
- 37.Kawabe, T., Tsuyama, N., Kitao, S., Nishikawa, K., Shimamoto, A., Shiratori, M., Matsumoto, T., Anno, K., Sato, T., Mitsui, Y., Seki, M., Enomoto, T., Goto, M., Ellis, N. A., Ide, T., Furuichi, Y., and Sugimoto, M. (2000) Oncogene 19 4764-4772 [DOI] [PubMed] [Google Scholar]
- 38.Sharma, S., Stumpo, D. J., Balajee, A. S., Bock, C. B., Lansdorp, P. M., Brosh, R. M., Jr., and Blackshear, P. J. (2007) Mol. Cell Biol. 27 1784-1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma, S., and Brosh, R. M., Jr. (2007) PLoS ONE 2 e1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puranam, K. L., and Blackshear, P. J. (1994) J. Biol. Chem. 269 29838-29845 [PubMed] [Google Scholar]
- 41.Seki, M., Miyazawa, H., Tada, S., Yanagisawa, J., Yamaoka, T., Hoshino, S., Ozawa, K., Eki, T., Nogami, M., Okumura, K., Taguchi, H., Hanaoka, F., and Enomoto, T. (1994) Nucleic Acids Res. 22 4566-4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cui, S., Klima, R., Ochem, A., Arosio, D., Falaschi, A., and Vindigni, A. (2003) J. Biol. Chem. 278 1424-1432 [DOI] [PubMed] [Google Scholar]
- 43.Muzzolini, L., Beuron, F., Patwardhan, A., Popuri, V., Cui, S., Niccolini, B., Rappas, M., Freemont, P. S., and Vindigni, A. (2007) PLoS Biol. 5 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doherty, K. M., Sharma, S., Uzdilla, L. A., Wilson, T. M., Cui, S., Vindigni, A., and Brosh, R. M., Jr. (2005) J. Biol. Chem. 280 28085-28094 [DOI] [PubMed] [Google Scholar]
- 45.Johnson, F. B., Lombard, D. B., Neff, N. F., Mastrangelo, M. A., Dewolf, W., Ellis, N. A., Marciniak, R. A., Yin, Y., Jaenisch, R., and Guarente, L. (2000) Cancer Res. 60 1162-1167 [PubMed] [Google Scholar]
- 46.Seki, T., Tada, S., Katada, T., and Enomoto, T. (1997) Biochem. Biophys. Res. Commun. 234 48-53 [DOI] [PubMed] [Google Scholar]
- 47.Mazina, O. M., and Mazin, A. V. (2004) J. Biol. Chem. 279 52042-52051 [DOI] [PubMed] [Google Scholar]
- 48.Bugreev, D. V., and Mazin, A. V. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 9988-9993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bugreev, D. V., Mazina, O. M., and Mazin, A. V. (2006) Nature Protocols, doi: 2010.1038/nprot.2006.2217 [DOI] [PubMed]
- 50.Mazina, O. M., Rossi, M. J., Thomaa, N. H., and Mazin, A. V. (2007) J. Biol. Chem. 282 21068-21080 [DOI] [PubMed] [Google Scholar]
- 51.Karow, J. K., Chakraverty, R. K., and Hickson, I. D. (1997) J. Biol. Chem. 272 30611-30614 [DOI] [PubMed] [Google Scholar]
- 52.van Brabant, A. J., Ye, T., Sanz, M., German, I. J., Ellis, N. A., and Holloman, W. K. (2000) Biochemistry 39 14617-14625 [DOI] [PubMed] [Google Scholar]
- 53.Shereda, R. D., Bernstein, D. A., and Keck, J. L. (2007) J. Biol. Chem. 282 19247-19258 [DOI] [PubMed] [Google Scholar]
- 54.Brosh, R. M., Jr., Li, J. L., Kenny, M. K., Karow, J. K., Cooper, M. P., Kureekattil, R. P., Hickson, I. D., and Bohr, V. A. (2000) J. Biol. Chem. 275 23500-23508 [DOI] [PubMed] [Google Scholar]
- 55.Brosh, R. M., Jr., Orren, D. K., Nehlin, J. O., Ravn, P. H., Kenny, M. K., Machwe, A., and Bohr, V. A. (1999) J. Biol. Chem. 274 18341-18350 [DOI] [PubMed] [Google Scholar]
- 56.Helleday, T., Lo, J., van Gent, D. C., and Engelward, B. P. (2007) DNA Repair (Amst.) 6 923-935 [DOI] [PubMed] [Google Scholar]
- 57.Sung, P., and Klein, H. (2006) Nat. Rev. Mol. Cell Biol. 7 739-750 [DOI] [PubMed] [Google Scholar]
- 58.Agarwal, S., Tafel, A. A., and Kanaar, R. (2006) DNA Repair (Amst.) 5 1075-1081 [DOI] [PubMed] [Google Scholar]
- 59.Whitby, M. C. (2005) Biochem. Soc. Trans. 33 1451-1455 [DOI] [PubMed] [Google Scholar]
- 60.Neale, M. J., and Keeney, S. (2006) Nature 442 153-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoeijmakers, J. H. (2001) Nature 411 366-374 [DOI] [PubMed] [Google Scholar]
- 62.Branzei, D., and Foiani, M. (2007) Genes Dev. 21 3019-3026 [DOI] [PubMed] [Google Scholar]
- 63.Adams, M. D., McVey, M., and Sekelsky, J. J. (2003) Science 299 265-267 [DOI] [PubMed] [Google Scholar]
- 64.McVey, M., Larocque, J. R., Adams, M. D., and Sekelsky, J. J. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 15694-15699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mazin, A. V., Zaitseva, E., Sung, P., and Kowalczykowski, S. C. (2000) EMBO J. 19 1148-1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harmon, F. G., and Kowalczykowski, S. C. (2001) J. Biol. Chem. 276 232-243 [DOI] [PubMed] [Google Scholar]
- 67.Ozsoy, A. Z., Sekelsky, J. J., and Matson, S. W. (2001) Nucleic Acids Res. 29 2986-2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brosh, R. M., Jr., Majumdar, A., Desai, S., Hickson, I. D., Bohr, V. A., and Seidman, M. M. (2001) J. Biol. Chem. 276 3024-3030 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.