Abstract
A hammerhead ribozyme [R(-)] targeting the minus strand RNA of potato spindle tuber viroid (PSTVd) and a mutated nonfunctional ribozyme [mR(-)] were designed, cloned, and transcribed. As predicted, both monomer and dimer transcripts of the active R(-) ribozyme gene could cleave the PSTVd minus strand dimer RNA into three fragments of 77, 338, and 359 bases in vitro at 25 and 50°C. The tandem dimer genes of R(-) and mR(-) were subcloned separately into the plant expression vector pROK2. Transgenic potato plants (cultivar Desirée) were generated by Agrobacterium tumefaciens-mediated transformation. Twenty-three of 34 independent transgenic plant lines expressing the active ribozyme R(-) resulted in having high levels of resistance to PSTVd, being free of PSTVd accumulation after challenge inoculation with PSTVd, but the remaining lines showed weaker levels of resistance to PSTVd with low levels of PSTVd accumulation. In contrast, 59 of 60 independent transgenic lines expressing the mutated ribozyme mR(-) were susceptible to PSTVd inoculation and had levels of PSTVd accumulation similar to that of the control plants transformed with the empty vector. The resistance against PSTVd replication was stably inherited to the vegetative progenies.
Keywords: hammerhead/minus RNA/disease resistance
Ribozymes are small RNA molecules capable of highly specific catalytic cleavage of RNA (1–3); therefore, they have enormous potential to inhibit gene expression. Recent examples include reduction in viral RNA replication of HIV-1 (4, 5) and arena virus (6), as well as inhibition of gene expression for C-fos (7), tumor necrosis factor (8), and α-lactalbumin (9) in mammalian cells, U7 snRNA (10), and 28S rRNA (11) in Xenopus oocytes and a complete reduction of neomycin phosphotransferase activity in tobacco protoplasts (12). Although much success has been achieved in vitro for ribozyme-mediated gene inhibition, progress in vivo has been markedly slower, especially with RNA viruses. In transgenic tobacco plants expressing low levels of ribozymes targeting tobacco mosaic virus (TMV) RNA, symptom development after TMV infection was delayed (13). Recently, De Feyter et al. (14) reported that a ribozyme gene and an antisense gene are equally effective in conferring resistance to tobacco mosaic virus in transgenic tobacco. Also, in turnip, an active cis-hairpin ribozyme incorporated into cauliflower mosaic virus (CaMV) was shown to delay systemic viral symptoms significantly compared with a nonfunctional ribozyme control (15).
So far, ribozymes mainly have been used in attempts to inhibit RNA viruses that have an exclusively cytoplasmic replication cycle (4–6, 13, 14). Although it is assumed that ribozymes move from the nucleus into the cytoplasm to cleave the RNAs, we postulated that ribozymes could be more effective in cleaving the RNA of viruses and viroids that have a nuclear replication phase, such as, for example, potato spindle tuber viroid (PSTVd) (16). Viroids (17) are small (246–375 nt), single-stranded, covalently closed circular RNA molecules that do not encode any protein. PSTVd, the best characterized viroid, contains 359 nt (18, 19) and, like most viroids, shows intramolecular base pairing and forms a rod-like structure under native conditions (20). The secondary structure of PSTVd is thought to contain five structural domains (21). Viroid replication is totally dependent on host enzymes, probably normally DNA-dependent RNA polymerase II in the nucleus (22, 23). Replication occurs through a rolling circle-type mechanism via oligomeric minus-sense replication intermediates, which serve as templates for the transcription of plus-sense oligomers (24).
Here we show that transgenic potato (Solanum tuberosum) plants expressing a ribozyme directed against PSTVd minus-strand RNA possess a high resistance against PSTVd replication. Our results provide the first example of a ribozyme suppressing a viroid pathogen to an undetectable level in planta.
MATERIALS AND METHODS
Design and Construction of Ribozyme Genes.
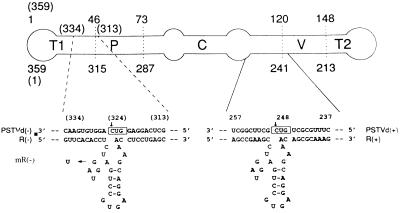
Based on the model for ribozyme-mediated catalytic cleavage of RNA put forward by Haseloff and Gerlach (3), a hammerhead ribozyme [R(−)] was designed to target the PSTVd minus strand RNA and a mutated, nonfunctional ribozyme [mR(−)] was constructed as a control. As shown in Fig. 1, a region of the PSTVd minus strand RNA containing a GUC trinucleotide (positions 322–324) was chosen for construction of a ribozyme [R(−)] cleaving the minus strand. The R(−) ribozyme is 49 nt long and is aimed at a putative binding site of DNA-dependent RNA polymerase II within the T1 domain of the PSTVd secondary structure (21, 25–26). The mutant, nonfunctional ribozyme [mR(−)] contains U instead of G in the conserved region of the ribozyme’s catalytic domain. For comparative purposes, a hammerhead ribozyme [R(+)] also was designed to target the PSTVd plus strand RNA. The R(+) ribozyme is 50 nt long and targets a GUC trinucleotide at positions 246–248 of the PSTVd plus strand RNA toward the C–V domain, which is a postulated binding site of DNA-dependent RNA polymerase III and a region involved in the processing of viroid replicative intermediates (21, 27–29). Short recognition antisense sequences (9–11 bases) were used in this study because longer sequences generally result in a strong binding and slow product release thus limiting the maximal rate of turnover (30–32) (Fig. 1).
Figure 1.
Base pairing of three hammerhead ribozymes, R(−), mR(−), and R(+), and their target sites within the respective T1 and C–V domains of minus (−) and plus (+) strands of the PSTVd RNA. A schematic map of the PSTVd RNA is shown at the top of the figure together with the numbering of the nucleotides. The numbering for PSTVd (−) is in brackets. T1, left-hand terminal domain; P, pathogenic domain; C, conserved central domain; V, variable domain; and T2, right-hand terminal domain. Locations of the PSTVd domains are from Keese and Symons (21).
Three single-stranded oligodeoxy nucleotides corresponding to the ribozymes R(−), mR(−), and R(+) were synthesized in a DNA synthesizer model 381 (Applied Biosystem). At the same time, two primers complementary to the 3′ end of R(−) and R(+) oligonucleotides also were synthesized. Both single-stranded oligonucleotides and primers were phosphorylated. The double-stranded DNAs of ribozymes R(−), mR(−), and R(+) were synthesized by Klenow treatment and cloned into the transcription vector pGEM-3Zf(+) (Promega) at its SmaI site. The recombinants containing R(−), mR(−), or R(+) genes were selected by hybridization with 32P-labeled ribozyme cDNA probes. The monomers (M) or dimers (D) of these ribozyme genes were identified by DNA sequencing.
In Vitro Transcription and Cleavage Testing.
PSTVd cDNA clone pST-B19 was kindly supplied by R. A. Owens (Molecular Plant Pathology Laboratory, Agricultural Research Service, United States Department of Agriculture, Beltsville, MD), and the PSTVd isomonomer was prepared by subcloning a BamHI fragment from the PSTVd cDNA dimer into pGEM-3Zf(+). The PSTVd(−) dimer transcript containing 772 nt (including 54 nt from the vector) was synthesized in vitro using SP6 RNA polymerase and pST-B19 linearized with HindIII as template. The PSTVd(+) isomonomer transcript containing 425 nt (including 66 nt from the vector) was synthesized in vitro using SP6 RNA polymerase and pGEM–PSTVd DNA linearized with EcoRI as template. These substrate RNAs were radiolabeled by inclusion of −[32P]GTP in the synthesis reactions. The transcribed RNAs of the ribozyme monomer and dimer were separately synthesized in vitro using pGEM R(−)M, R(−)D, R(+)M, or R(+)D linearized with EcoRI as template for SP6 RNA polymerase. All transcriptions were performed with an RNA transcription kit (Boehringer Mannheim).
In vitro cleavage reactions contained ribozyme and 32P-labeled substrate in a 1:1 molar ratio in 50 mM Tris⋅HCl, pH 8.0/20 mM MgCl2. The reactions were carried out at either 50°C (2 h) or 25°C (4 h) and were stopped by adding an equal volume of termination solution (98% formamide/10 mM EDTA/0.1% bromophenol blue/0.1% xylene cyanol) and incubating at 80°C for 30 s. Samples were analyzed on denaturing 6% polyacrylamide gels containing 8 M urea. Radiolabeled RNA bands were detected and quantified by autoradiography.
Potato Transformation and Inoculation.
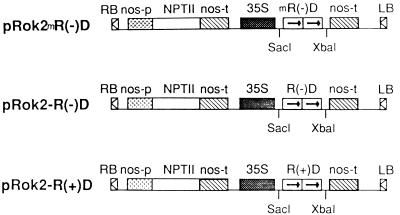
The tandem dimer sequences of R(−), mR(−), and R(+) were excised from pGEM-3Zf(+) with SacI/XbaI and subcloned separately into pROK2, a plant expression vector kindly provided by D. Baulcombe (Sainsbury Laboratory, John Innes Center, Norwich, UK) containing a 35S promotor from CaMV and terminator from the nopaline synthase in addition to a selectable marker, neomycin phosphotransferase conferring kanamycin resistance (Fig. 2). Recombinant vectors pROK2–R(−) D, pROK2–mR(−)D, and pROK2–R(+)D were transferred from Escherichia coli MC1022 into Agrobacterium tumefaciens LBA 4404 by triparental mating and were used to transform virus-free slices of potato tuber (cultivar Desirée) (33). As a control, the empty vector pROK2 was used to transform potato. Transformed potato seedlings exhibiting kanamycin resistance were regenerated and planted in an insect-proof greenhouse kept at 25–30°C. Each seedling was inoculated at the 7- to 8-leaf stage with 20 ng of PSTVd (type strain) by dusting carborundum on all of the leaves and gently rubbing carborundum-dusted leaves with a gloved finger. Infectious PSTVd was isolated from PSTVd-inoculated tomato (Lycopersicon esculentum Mill cv. Rutgers) leaves.
Figure 2.
Construction of expression vectors for potato transformation containing ribozyme genes under the control of 35S promotor. R(−)D, mR(−)D, and R(+)D, tandem R(−), mR(−), and R(+) ribozymes, respectively; nos-p, nos promotor; nos-t, nos terminator; NPTII, neomycin phosphotransferase II; RB, right border; LB, left border.
Nucleic Acid Extraction.
Total nucleic acids were extracted by the phenol–chloroform method (34) from each PSTVd-inoculated plant transformed with the ribozyme gene or with the empty vector 1 month after inoculation. Leaf tissue (1–1.5 g) was ground in a mortar containing 5 ml of extraction buffer (100 mM Tris⋅HCl, pH 9.0/100 mM NaCl/10 mM Na2 EDTA/2% SDS/2% bentonite/1% 2-mercaptoethanol) and treated with an equal volume of phenol–chloroform (1:1, vol/vol). RNAs were further purified by DEAE–cellulose column chromatography, precipitated by three volumes of ethanol, washed with 70% ethanol, vacuum dried, and suspended in 100 μl of TE buffer (10 mM Tris⋅HCl/1 mM EDTA, pH 8.0). Total RNA concentration was determined by UV spectrophotometry, one OD260 unit corresponding to 40 μg/ml RNA.
Return Gel Electrophoresis and Northern Blotting.
Total (10 μg) RNA from individual transgenic plants was applied onto 5% polyacrylamide gels (acrylamide-to-bis = 29:1) in TBE buffer (89 mM Tris/89 mM boric acid/2.5 mM Na2 EDTA, pH 8.3) and analyzed by return gel electrophoresis (35). The first electrophoresis was performed at room temperature at 150 V for 3–4 h until the xylene cyanol had migrated to the bottom of the gel. The second electrophoresis was carried out from bottom to top under denaturing condition at 70°C and 200 V for 1 h in fresh TBE buffer preheated to 70°C until the xylene cyanol migrated to the top of the gel. The gel was silver-stained (35). For Northern blot analysis, 4 μg of total RNAs extracted from each plant was analyzed by gel electrophoresis without the second run as described above. The nucleic acids were transferred to S & S Nytran Nylon membranes (Schleicher & Schuell) by capillary blotting and were cross-linked to the membrane by UV irradiation according to the manufacturer’s instructions. The membranes were treated at 80°C for 2 h and used for Northern hybridization with a 32P-labeled probe—either PSTVd cDNA or a ribozyme cDNA. All membranes were prehybridized in 20 ml of hybridization solution [50% vol/vol deionized formamide/5 × standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA)/0.1% wt/vol SDS/5 × Denhardt’s solution/100 μg/ml sheared denatured herring sperm DNA] in a plastic bag at 42°C for 8 h. 32P-Labeled DNA probes were prepared using the gel-purified DNA restriction fragment of the PSTVd clone or the R(−) DNA clone as a probe template in the oligonucleotide primer extension reaction. Hybridization was carried out at 42°C for 20 h. The membranes were washed twice at 37°C with a solution of 5 × standard saline phosphate/EDTA (0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA)/0.1% SDS (15 min per change) and then exposed by autoradiography at −70°C for 48 h.
RESULTS
In Vitro Cleavage Activity of Ribozymes.
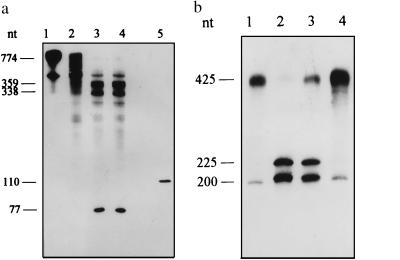
The ribozyme DNA monomers [pGEM–R(−)M and pGEM R(+)M] and dimers [pGEM–R(−)D, pGEM–mR(−)D, and pGEM–R(+)D] designed to specifically target PSTVd minus and plus strands, respectively, were obtained after cloning and evaluated by sequencing. The transcripts of PSTVd(−) dimer and isomonomer and pGEM–R(+)M, pGEM–R(+)D, pGEM–R(−)M, and pGEM–R(−)D were prepared in large scale. Cleavage activity tests conducted at 50°C for 2 h followed by denaturing PAGE showed that, after incubation of R(−) monomer and dimer with PSTVd(−) dimer, ≈95% of the latter was cleaved into three RNA fragment bands with 77, 336, and 359 bases as predicted by ribozyme design (Fig. 3A). When R(+) monomer and dimer were incubated with PSTVd(+) isomonomer, ≈70–95% of the latter cleaved into two RNA fragments containing 200 and 225 bases (Fig. 3B). Some ribozyme cleavage activity was observed at 25°C for 4 h although it was far lower than that observed during incubation at 50°C.
Figure 3.
In vitro cleavage of PSTVd transcripts by ribozymes R(−) and R(+). R(−) and R(+) were reacted with substrate PSTVd at 50°C for 2 h, followed by 6% PAGE containing 8 M urea and autoradiography. (a) Lanes: 1, PSTVd(−) dimer; 2, PSTVd(−) dimer incubated alone; 3 and 4, PSTVd(−) dimer incubated with R(−) monomer and R(−) dimer, respectively; and 5, R(−) monomer (110 nt). (b) Lanes: 1, PSTVd(+) isomonomer incubated alone; and 2–4, PSTVd(+) isomonomer incubated with R(+) monomer, R(+) dimer, and R(−) monomer, respectively.
High Resistance Against PSTVd Infection in Transgenic Potato Plants Expressing R(−).
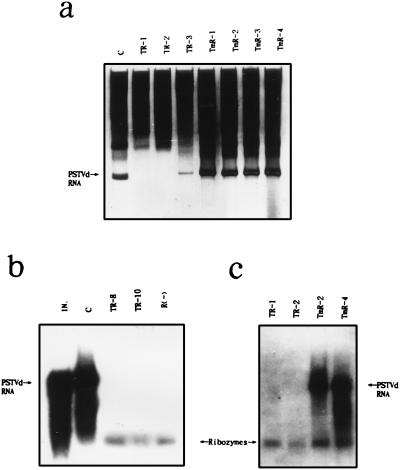
A total of 34 independent transgenic potato plants expressing ribozyme R(−) were generated. Analysis of RNA extracted from the transgenic plants 1 month after challenge inoculation by return PAGE showed that 23 lines were free of PSTVd accumulation (Table 1; see Fig. 4a, TR-1 and TR-2 for representative data). Although variable, the remaining 11 plants accumulated PSTVd to ≈1/20 of the control plants transformed with pROK2 alone (Fig. 4a, compare lanes TR-3 and C). Sixty independent transgenic potato plants expressing mutant ribozyme mR(−) were regenerated. Return PAGE analysis indicated that all but one line had levels of PSTVd accumulation similar to those of the control (Table 1; Fig. 4a, compare lanes TmR-1 and TmR-4). These results were confirmed by Northern hybridization analysis of total RNA extracted from the transgenic lines using 32P-labeled, full length PSTVd cDNA as a probe (Fig. 4, b and c). The results also showed that ribozyme RNAs were present in the 23 PSTVd-free lines, when in vitro R(−) was used as marker [Fig. 4b. TR-8, TR-10, and R(−)]. Lines transformed by the nonfunctional mR(−) ribozyme showed both PSTVd and mutant ribozyme (Fig. 4c, lanes TmR-2 and TmR-4). Although mR(−) was expressed in transgenic plants, no cleavage activity was observed.
Table 1.
Resistance of transgenic potatoes containing anti-PSTVd ribozymes against high concentrations of PSTVd inoculum
| Expression vector | Transformed plants, n* | Inoculated plants, n†
|
||
|---|---|---|---|---|
| PSTVd not detected | PSTVd detected as in control | PSTVd decreased | ||
| pRok2-R(−)D | 34 | 23 | 0 | 11 |
| pRok2-mR(−)D | 60 | 0 | 59 | 1 |
| pRok2-R(+)D | 6 | 2 | 4 | 0 |
| pRok2 | 8 | 0 | 8 | 0 |
Integration of the neomycin phosphotransferase II and ribozymes was detected by Southern hybridization analysis. Copy numbers of the ribozymes within the genome varied from one to five (data not shown).
Three or four leaves of transgenic potato plants were mechanically inoculated with 20 ng of PSTVd RNA per plant. After 1 month, PSTVd was detected by return gel electrophoresis and Northern hybridization analysis (see Fig. 4).
Figure 4.
Expression of ribozyme R(−) and nonfunctional mutant mR(−) in transgenic potato plants. (a) Detection of PSTVd isolated from transgenic potato lines. Lanes: c, transformed with empty vector pROK2; TR-1–3, transformed with pROK2–R(−)D; and TmR-1–4, transformed with pROK2–mR(−)D. Total RNA (10 μg) isolated from stems and leaves of individual plant was loaded in each lane of 5% polyacrylamide gel, using return gel electrophoresis. Gel was stained by silver staining (35). (b and c) Northern blot analysis of RNA isolated from transgenic potato lines. Lanes: IN, PSTVd inoculum (0.1 μg); c, control plant transformed with empty vector pROK2; TR-1, -2, -8, and -10, transgenic plants transformed with pROK2–R(−)D; TmR-2 and -4, transgenic plants transformed with pROK2–mR(−)D; and R(−), in vitro-transcripted ribozyme R(−)M as a marker. Total RNA (4 μg) isolated from the transgenic plant was loaded in each lane of 5% polyacrylamide gel and electroblotted onto an S & S Nytron Nylon membrane (Schleicher & Schuell). Blotted RNAs were hybridized with a 32P-labeled PSTVd cDNA fragment probe. The PSTVd, ribozyme R(−), and mutant mR(−) are indicated by arrows.
Resistance Against PSTVd of Vegetative Progeny Plants Expressing R(−).
All of the transgenic lines of R(−) showing high resistance to PSTVd replication were individually propagated vegetatively from tubers. No PSTVd was detected by RNA analyses in progeny plants. These results suggest that the resistance against PSTVd replication was stably transmitted to the progenies. In contrast, levels of PSTVd accumulation similar to those of control plants were found in vegetative progenies of mR(−) transgenic plant lines (Table 2). Taken together, our results show that R(−) is capable of inhibiting the formation of PSTVd(−) and preventing PSTVd(+) synthesis, resulting in no detectable levels of PSTVd in the majority of transgenic lines.
Table 2.
Resistance of first generation vegetative progeny plants from transgenic potatoes containing anti-PSTVd ribozymes against PSTVd infection
| Expression vector | Progeny tubers from transgenic plants | Plants, n | Plants, n
|
|
|---|---|---|---|---|
| PSTVd not detected | PSTVd detected as in control | |||
| pRok2-R(−)D | PSTVd not detected | 20 | 20 | 0 |
| pRok2-mR(−)D | PSTVd detected as in control | 18 | 0 | 18 |
| pRok2 | PSTVd detected | 10 | 0 | 10 |
Transgenic potatoes expressing R(−) do not display any growth abnormalities. The tubers and plants of the control transgenic plants showed very mild symptoms of PSTVd infection after one generation of vegetative propagation post-PSTVd infection. Furthermore, the PSTVd-free R(−) transgenic plants did not develop any PSTVd-associated symptoms after one generation of vegetative propagation. Results obtained from the experimental cultivar Desirée were confirmed by transformation of a commercial potato cultivar, Favorita, with pROK2–R(−)D (data not shown).
Transgenic Potato Plants Expressing R(+).
Only six transgenic plants expressing R(+) were obtained even though the same number of potato tuber slices were used for transformation as with pROK2–R(−)D. Possibly, the pROK2–R(+)D RNA transcript interferes with the growth of transgenic plants. RNA analyses showed that two lines had no PSTVd accumulation after infection, but four lines contained PSTVd levels similar to those in the control plants (Table 1).
DISCUSSION
On the bases of structural and replication features of viroid, useful models to develop viroid-resistance have been studied. Matousek et al. (36) showed that antisense RNA mediated an inhibitory effect on PSTVd replication in transgenic potato plants expressing antisense RNA against minus-strand as well as plus-strand viroid replication intermediates. However, there was a high degree of variability among different plant genotypes and even between different plants transformed by the same clone. Atkins et al. reported (37) that citrus exocortis viroid accumulation in tomato plants expressing antisense constructs targeting the negative-strand RNA molecule was only slightly reduced. In contrast, transgenic plants expressing constructs targeting the positive-strand citrus exocortis viroid molecule resulted in an increase in the accumulation of citrus exocortis viroid.
Our results demonstrate that ribozymes aimed at the minus-strand replicative intermediate of PSTVd can efficiently block viroid replication and confer high levels of resistance to viroid replication when expressed in transgenic plants. In comparison, lower levels of resistance were observed in plants transformed with ribozyme genes targeting the positive-strand PSTVd. This phenomenon is in accord with the results of Atkins et al. (37) with respect to the effects of the transgenic RNAs targeting the (+) or (−) strand of PSTVd. Thus, minus-strand viroid RNA is apparently more accessible to transgenic antisense or hammerhead ribozyme RNA. Faustmann et al. (38) and Matousek et al. (39) identified differences in the accumulation of plus- and minus-strand PSTVd. Specifically, there are lower levels of minus-strand intermediates in infected plant cells than circular plus-strand viroid progeny perhaps because (−) strand intermediaries are either synthesized at lower rate or are degraded much faster than the plus strand monomers. This situation may permit the establishment of a more effective ratio of transgenic RNA-to-target (−) strand intermediates. In addition, the minus-strand RNA may be more accessible to the transgenic RNA transcripts (39).
As mentioned by Atkins et al. (37), the addition of hammerhead ribozyme sequences to both classes of antisense constructs resulted in lower resistance levels in comparison to antisense expression alone. The authors suggest that the additional hammerhead sequences reduced the net effect of the antisense RNA interaction with citrus exocortis viroid, by reducing its association with either a host factor(s) or the viroid RNA. In our work, R(−) constructs with short antisense recognition sequences (9–11 bases) were used, possibly providing a higher rate of turnover. In other eukaryotic systems, similar short arm ribozymes also were used to cleave virus pathogen genes in vivo (40). Another factor influencing the ribozyme effectiveness could be the type of test plant used. In our laboratory, transgenic tomato plants expressing R(−) ribozyme did not show resistance to PSTVd replication (unpublished data). The inefficiency of anti-PSTVd ribozymes in transgenic tomato may be the result of the ability of PSTVd to replicate much faster in tomato than in its natural host, potato.
It should be pointed out that effective expression of catalytically active ribozymes targeting PSTVd(−) in vivo may be the direct result of the nuclear replication of the viroid. To further explore this possibility, we concentrated our efforts on CaMV, a plant DNA virus, whose transcription processes are located within nuclei. Transgenic rape (Brassica napus L.) plants expressing a ribozyme specifically targeting the CaMV [35S]RNA transcript were created and shown to confer high levels of resistance to CaMV infection (41). On the contrary, transgenic tobacco plants expressing a ribozyme targeting the TMV 54-KDa coding region failed to display resistance against TMV infection (unpublished data), perhaps because TMV replication occurs in the cytoplasm.
In recent years, many efforts have been devoted to the search for new strategies to increase ribozyme cleavage activity in vivo. Sullenger et al. (42) used a retroviral vector-mediated gene transfer system to substantially increase the effectiveness of a ribozyme. Michienzi et al. (43) reported that small nuclear RNA chimeric ribozymes targeting the regulator of expression of virion protein pre-mRNA of HIV-1 displayed efficient cleavage activity in Xenopus laevis oocytes. From these experimental results, it has been demonstrated that the effectiveness of ribozymes is closely related to colocalization with their target substrate (42, 43). In our study, transgenic plants expressing ribozymes targeted to PSTVd showed high levels of resistance, possibly reflecting the principle that useful ribozyme activity depends on the compartmentalization of both ribozyme and target inside the nucleus.
Acknowledgments
We are grateful to Yongbiao Xue for useful discussion and for help in the preparation of the manuscript. We thank Drs. R. A.Owens and J. W. Davies for critical reading of the manuscript. This work was supported by a grant from the Chinese National Natural Science Foundation and was partially supported by United Nations Environment Programme through Microbiological Resources Center-China.
ABBREVIATIONS
- TMV
tobacco mosaic virus
- CaMV
cauliflower mosaic virus
- PSTVd
potato spindle tuber viroid
- R(−)
ribozyme (−)
- mR(−)
mutated ribozyme (−)
- R(+)
ribozyme(+)
- TR
transgenic ribozyme
- TmR
transgenic mutated ribozyme
- M
monomer
- D
dimer
- C
control
- IN
inoculum
References
- 1.Zaug A J, Been M D, Cech T R. Nature (London) 1986;324:429–433. doi: 10.1038/324429a0. [DOI] [PubMed] [Google Scholar]
- 2.Uhlenbeck O C. Nature (London) 1987;328:596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- 3.Haseloff J, Gerlach W L. Nature (London) 1988;334:585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- 4.Sarver N, Cantin E M, Chang P S, Zaia J A, Ladne P A, Stephens D A, Rossi J J. Science. 1990;247:1222–1225. doi: 10.1126/science.2107573. [DOI] [PubMed] [Google Scholar]
- 5.Ojwang J O, Hampel A, Looney D J, Wong-Staal F, Rappaport J. Proc Natl Acad Sci USA. 1992;89:10802–10806. doi: 10.1073/pnas.89.22.10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xing Z, Whitton J L. J Virol. 1992;66:1361–1369. doi: 10.1128/jvi.66.3.1361-1369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scanlon K J, Jiao L, Funato T, Wang W, Tone T, Rossi J J, Kashani-Saber M. Proc Natl Acad Sci USA. 1991;88:10591–10595. doi: 10.1073/pnas.88.23.10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sioud M, Natvig J B, Forre O. J Mol Biol. 1992;223:831–835. doi: 10.1016/0022-2836(92)90244-e. [DOI] [PubMed] [Google Scholar]
- 9.Huillier P J L, Davis S R, Bellamy A R. EMBO J. 1992;11:4411–4418. doi: 10.1002/j.1460-2075.1992.tb05541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotten M, Birnstiel M L. EMBO J. 1989;8:3861–3866. doi: 10.1002/j.1460-2075.1989.tb08564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saxena S K, Ackerman E J. J Biol Chem. 1990;265:17106–17109. [PubMed] [Google Scholar]
- 12.Steinecke P, Herger T, Schreier P H. EMBO J. 1992;11:1525–1530. doi: 10.1002/j.1460-2075.1992.tb05197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edington B V, Dixon R A, Nelson R S. In: Transgenic Plants. Hiatt A, editor. New York: Dekker; 1992. pp. 301–323. [Google Scholar]
- 14.De Feyter R, Young M, Schroeder K, Dennis E S, Gerlach W. Mol Gen Genet. 1996;250:329–338. doi: 10.1007/BF02174391. [DOI] [PubMed] [Google Scholar]
- 15.Borneman J, Tritz R, Hampel A, Altschuler M. Gene. 1995;159:137–142. doi: 10.1016/0378-1119(95)00173-4. [DOI] [PubMed] [Google Scholar]
- 16.Diener T O. Virology. 1971;45:411–428. doi: 10.1016/0042-6822(71)90342-4. [DOI] [PubMed] [Google Scholar]
- 17.Diener T O. The Viroids. New York: Plenum; 1987. [Google Scholar]
- 18.Gross H J, Domdey H, Lossow C, Jank P, Raba M, Alberty H. Nature (London) 1978;273:203–208. doi: 10.1038/273203a0. [DOI] [PubMed] [Google Scholar]
- 19.Sanger H L. In: The Microbe. Mahy B W J, Pathison J R, editors. London: Cambridge Univ. Press; 1984. pp. 281–334. [Google Scholar]
- 20.Sanger H L, Klotz G, Riesner D, Gross H J, Kleinschmidt A K. Proc Natl Acad Sci USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keese P, Symons R H. Proc Natl Acad Sci USA. 1985;82:4582–4585. doi: 10.1073/pnas.82.14.4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanger H L. In: The Viroids. Diener T O, editor. New York: Plenum; 1987. pp. 117–166. [Google Scholar]
- 23.Schumacher J, Sanger H L, Riesner D. EMBO J. 1983;2:1549–1555. doi: 10.1002/j.1460-2075.1983.tb01622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Branch A D, Benefeld B T, Robertson H D. Proc Natl Acad Sci USA. 1988;85:9128–9132. doi: 10.1073/pnas.85.23.9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodman T C, Nagel L, Rappold W, Klotz G, Riesner D. Nucleic Acids Res. 1984;12:6231–6246. doi: 10.1093/nar/12.15.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rackwitz J R, Rohde W, Sanger H L. Nature (London) 1981;291:297–301. doi: 10.1038/291297a0. [DOI] [PubMed] [Google Scholar]
- 27.Semancik J S. Viroids and Viroid-Like Pathogens. Boca Raton, FL: CRC; 1987. [Google Scholar]
- 28.Diener T O. Proc Natl Acad Sci USA. 1989;86:9370–9374. doi: 10.1073/pnas.86.23.9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Candresse T, Diener T O, Owens R A. Virology. 1990;175:232–237. doi: 10.1016/0042-6822(90)90203-4. [DOI] [PubMed] [Google Scholar]
- 30.Herschlag D, Cech T R. Biochemistry. 1990;29:10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- 31.Perrotta A T, Been M D. Biochemistry. 1992;31:16–21. doi: 10.1021/bi00116a004. [DOI] [PubMed] [Google Scholar]
- 32.Fedor M J, Uhlenbeck O C. Biochemistry. 1992;31:12042–12054. doi: 10.1021/bi00163a012. [DOI] [PubMed] [Google Scholar]
- 33.Edwards G A, Hepher A, Clerk S P, Boulter D. Plant Mol Biol. 1991;17:89–100. doi: 10.1007/BF00036809. [DOI] [PubMed] [Google Scholar]
- 34.Hadidi A, Yang X. J Virol Methods. 1990;30:261–270. doi: 10.1016/0166-0934(90)90068-q. [DOI] [PubMed] [Google Scholar]
- 35.Schumacher J, Meyer N, Riesner D, Weidemann H L. J Phytopathol. 1986;115:332–343. [Google Scholar]
- 36.Matoušek J, Schröder A R W, Trněná L, Reimers M, Baumstark T, Dědič P, Vlasák J, Becker I, Kreuzaler F, Fladung M, Riesner D. Biol Chem Hopper-Seyler. 1994;375:765–777. doi: 10.1515/bchm3.1994.375.11.765. [DOI] [PubMed] [Google Scholar]
- 37.Atkins D, Young M, Uzzell S, Kelly L, Fillatti J, Gerlach W L. J Gen Virol. 1995;76:1881–1796. doi: 10.1099/0022-1317-76-7-1781. [DOI] [PubMed] [Google Scholar]
- 38.Faustmann O, Kern R, Sänger H L, Mühlbach H P. Virus Res. 1986;4:213–227. [Google Scholar]
- 39.Matoušek J, Rakouský S, Trněná L, Riesner D. Biol Plant. 1993;35:131–135. [Google Scholar]
- 40.Castanotto D, Rossi J J, Deshler J D. Crit Rev Eukaryotic Gene Expression. 1992;2:331–339. [PubMed] [Google Scholar]
- 41.Wang S Y, Yie Y, Zhao S Z, Tien P. Abstracts of 15th Annual Meeting of American Society for Virology. London, Ontario, Canada: University of Western Ontario; 1996. p. 105. [Google Scholar]
- 42.Sullenger B A, Cech T R. Science. 1993;262:1566–1569. doi: 10.1126/science.8248806. [DOI] [PubMed] [Google Scholar]
- 43.Michienzi A, Prislei S, Bozzoni I. Proc Natl Acad Sci USA. 1996;93:7219–7224. doi: 10.1073/pnas.93.14.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]