Abstract
The three-dimensional structure of glutamate-1-semialdehyde aminomutase (EC 5.4.3.8), an α2-dimeric enzyme from Synechococcus, has been determined by x-ray crystallography using heavy atom derivative phasing. The structure, refined at 2.4-Å resolution to an R-factor of 18.7% and good stereochemistry, explains many of the enzyme’s unusual specificity and functional properties. The overall fold is that of aspartate aminotransferase and related B6 enzymes, but it also has specific features. The structure of the complex with gabaculine, a substrate analogue, shows unexpectedly that the substrate binding site involves residues from the N-terminal domain of the molecule, notably Arg-32. Glu-406 is suitably positioned to repel α-carboxylic acids, thereby suggesting a basis for the enzyme’s reaction specificity. The subunits show asymmetry in cofactor binding and in the mobilities of the residues 153–181. In the unliganded enzyme, one subunit has the cofactor bound as an aldimine of pyridoxal phosphate with Lys-273 and, in this subunit, residues 153–181 are disordered. In the other subunit in which the cofactor is not covalently bound, residues 153–181 are well defined. Consistent with the crystallographically demonstrated asymmetry, a form of the enzyme in which both subunits have pyridoxal phosphate bound to Lys-273 through a Schiff base showed biphasic reduction by borohydride in solution. Analysis of absorption spectra during reduction provided evidence of communication between the subunits. The crystal structure of the reduced form of the enzyme shows that, despite identical cofactor binding in each monomer, the structural asymmetry at residues 153–181 remains.
Keywords: x-ray structure analysis, chlorophyll biosynthesis, pyridoxal-5′-phosphate, asymmetric dimer, reaction specificity
Enzymes using pyridoxal-5′-phosphate (PLP) as cofactor provide an excellent example of the evolutionary development of catalytic versatility in homologous enzymes. Thus the α-family of PLP-dependent enzymes (1) contains aminotransferases, racemases, decarboxylases, mutases, and synthetases. Understanding how the related proteins confer separate reaction specificities on the coenzyme requires knowledge of the three-dimensional structures of representative enzymes that catalyze the different reactions.
The tetrapyrrole nucleus in compounds such as haem and chlorophyll is assembled from eight molecules of 5-aminolevulinate (2). In plants and at least some bacteria, this compound is made from glutamate 1-semialdehyde (GSA) in a reaction catalyzed by GSA aminomutase (GSA-AT, EC 5.4.3.8), an α2-dimeric enzyme with 433 residues per chain (3, 4) which is clearly related in sequence to the aminotransferases of the α-family of B6-dependent enzymes (5). The reaction mechanism (refs. 6 and 7; Fig. 1) is also closely analogous to that of the aminotransferases (8). The first step is imine formation between pyridoxamine-5′-phosphate (PMP) of the enzyme and the aldehyde function of GSA. The double bond of this imine 2 is shifted, by proton transfer from C4′ of the coenzyme to C5 of the substrate, to yield an imine 3 of PLP with the 5-amino group of 4,5-diaminovalerate 4. Next, by a mechanism which is uncertain, this intermediate is isomerized to an aldimine 5 with the 4-amino group of 4,5-diaminovalerate. The remainder of the reaction is analogous to that normally presented as the first half of a classical transamination reaction with proton transfers now being conducted from C4 of diaminovalerate to C4′ of the cofactor. The similarity between this reaction and that of the true aminotransferases is reinforced by the observation that significant amounts of the pyridoxaldimine form of the enzyme and free diaminovalerate are formed (6, 9). However, unlike the aminotransferases, the enzyme converts substrate to product by an intramolecular exchange of amino and oxo functions and has no requirement for α-carboxylic amino or oxo acids to complete the reaction (6, 7). This ability to reject potential alternative substrates is pivotal in providing the enzyme with the reaction specificity of a mutase. One reason for determining the crystal structure of GSA-AT was to provide a structural explanation of this property.
Figure 1.
Intermediates in the conversion of GSA to 5-aminolevulinate. R, (CH2)2COO−; Py, the 5′-phosphopyridoxyl group of the cofactor.
The formation and dissociation of diaminovalerate from the enzyme appears to be mechanistically unnecessary but the existence of this step makes the enzyme susceptible, through its pyridoxaldimine form, to mechanism-based inhibitors such as gabaculine (10). Because no counterpart of this enzyme exists in animals, it is a promising target for safe, selective herbicides and the rational design of such compounds has provided part of the driving force for the work presented in this paper. An explanation for the gabaculine tolerance of a Met-248 → Ile mutant, produced when photosynthetic bacteria are grown in a medium containing this compound (10), is a further aim of this work.
MATERIALS AND METHODS
Crystallization and Data Collection.
Synechococcus GSA-AT, expressed in Escherichia coli, was purified and crystallized as described (10, 11). The gabaculine complex was formed by soaking a crystal with dimensions of 0.1 × 0.1 × 0.5 mm3 at 4°C in Na-cacodylate (50 mM, pH 7.0) containing 19.5% polyethylene glycol 10,000, 100 mM Mg-acetate, and 3 mM gabaculine for 3 days. GSA-AT, in which both subunits contained the cofactor as an internal aldimine (7), was converted to the fully reduced form with 10 mM NaBH3CN at pH 7.8. Crystals of reduced GSA-AT were grown by macroseeding (11, 12).
GSA-AT crystallizes in the orthorhombic space group P212121 with cell dimensions a = 68.6 Å, b = 108.0 Å, and c = 122.4 Å. Except for derivative MMC(b) (Table 1), for which data were collected at the Deutsches Elektronen Synchrotron (Hamburg, Germany; EMBL beamline X31) with wavelength 0.95 Å, all x-ray data were collected with CuKα radiation from the laboratory rotating anode, recorded on a MAR-Research (Hamburg, Germany) image plate area detector and evaluated with mosflm (13).
Table 1.
Data collection statistics for native and heavy metal derivative data of GSA-AT
| Soaking conditions | Maximum resolution, Å | Total reflections | Unique reflections | Completeness, % | Rsym,* % | Riso,† % | No. of sites | Rc,‡ % | Phasing power | |
|---|---|---|---|---|---|---|---|---|---|---|
| Native | — | 2.4 | 110,780 | 34,132 | 94.6 | 6.8 | ||||
| Reduced form | — | 2.5 | 91,742 | 31,141 | 96.8 | 7.3 | 20.3 | |||
| Gabaculine | 3 mM | 3.0 | 57,653 | 18,662 | 98.0 | 11.4 | 13.9 | |||
| Hg-acetate | 0.1 mM; 3 days | 4.0 | 27,969 | 7,898 | 98.1 | 4.5 | 29.8 | 4 | 0.86 | 0.68 |
| EMTS§ | 0.01 mM; 14 days | 3.5 | 36,865 | 11,229 | 98.3 | 7.9 | 17.4 | 4 | 0.71 | 1.08 |
| Pt¶ | Saturated; 7 days | 4.0 | 20,946 | 7,885 | 96.3 | 6.0 | 20.5 | 6 | 0.87 | 0.65 |
| MMC(a)¶ | 10 mM; 3 h | 2.8 | 64,746 | 22,369 | 97.0 | 4.9 | 37.0 | 9 | 0.77 | 1.00 |
| MMC(b)¶ | 1 mM; 3 h | 3.5 | 30,545 | 10,405 | 86.7 | 5.0 | 30.5 | 9 | 0.76 | 0.98 |
All derivatives were prepared by soaking crystals in heavy atom solutions in stabilizing buffer (Na-cacodylate buffer: 50 mM, pH 7.0/200 mM Mg-acetate/19.5% polyethylene glycol 10,000).
Rsym = Σhkl Σi |I(hkl)i−〈I(hkl)〉|/Σhkl Σi 〈I(hkl)i〉.
Riso = Σhkl ∥FPH(hkl) − FP(hkl)∥/Σhkl FP(hkl).
Rc = Σhkl |∥FPH(hkl)| ± |FP(hkl)∥ − |FHcalc(hkl)∥/Σhkl ∥FPH(hkl)| ± |FP(hkl)∥ for centric reflections.
Ethylmercury thiosalicylate;
Pt, platinum ethylene diamine dichloride; MMC(a), methylmercury chloride, data collected in the laboratory, and MMC(b), methylmercury chloride, data collected using synchrotron radiation and a wavelength of 0.95 Å.
Structure Determination.
An extensive search resulted in five useful isomorphous heavy atom derivatives (Table 1). The heavy atom locations were determined by a difference Patterson synthesis for derivative MMC(a) and by difference Fourier methods for the others. All calculations were performed using the CCP4 package (13). The heavy atom parameter refinement and multiple isomorphous replacement phasing resulted in an overall figure of merit of 0.40 for data between 10.0 Å and 2.8 Å resolution. A 4.0-Å resolution electron density map clearly showed the solvent–protein boundary but was not of sufficient quality for tracing. From equivalent heavy atom sites in the two subunits, orientation and position of the molecular 2-fold axis were established. A molecular envelope was determined using rave (14). Combined solvent flattening, histogram matching, and molecular 2-fold averaging, as implemented in the program dm (13), was used to improve the phases. The resulting electron density map, calculated at 2.8-Å resolution, was of sufficient quality to trace the polypeptide chain. An atomic model was built from the “bones” coordinates with the graphics package “o” (15). The region corresponding to residues 153–181 showed very weak electron density and could not be built.
The atomic model was refined using the x-plor package (16) and the atomic restraint parameters of Engh and Huber (17). Initially, the model was subjected to a full simulated annealing refinement procedure at 2.8 Å with strict 2-fold noncrystallographic symmetry and the resulting model was reappraised by “o” (15). This reduced the R-factor from 42.5% to 28.1% (free R-factor, 34.3%) for data between 10.0- and 2.8-Å resolution. Further coordinate and temperature factor refinement was performed with relaxed 2-fold symmetry restraints. This improved the electron density for residues 153–181 of one subunit (A) sufficiently to include this stretch in the molecular model. This part of the structure was transferred to the second subunit (B) using the noncrystallographic symmetry. Inclusion of these additional residues of subunit B in the refinement increased their B-factors to values around 100 Å2. Thereafter, these residues were excluded from refinement by setting their occupancies to zero. Further regular manual updates, during which water molecules were also built into the structure, finally produced an R-factor of 18.3% for the working data set and 25.0% for the test data set (free R-factor, no σ cutoff) in the resolution range of 10.0 to 2.4 Å (Table 2).
Table 2.
Refinement statistics
| Wild type | Reduced form | Gabaculine complex | |
|---|---|---|---|
| No. of protein atoms | 6,255 | 6,255 | 6,255 |
| No. of water molecules | 347 | 285 | 166 |
| rms deviation from | |||
| Ideal distances (1-2), Å | 0.009 | 0.011 | 0.012 |
| Ideal bond angles, ° | 1.76 | 1.85 | 1.98 |
| Residues in most favored | |||
| regions of Ramachandran plot, % | 89.4 | 89.3 | 88.8 |
| Mean B-factor | |||
| Protein atoms, Å2 | 29.6 | 28.4 | 33.2 |
| Water molecules, Å2 | 41.6 | 37.2 | 28.0 |
| R-factor* | |||
| Working set | 0.183 | 0.187 | 0.159 |
| Test set, 5% of data | 0.250 | 0.264 | 0.250 |
R-factor = Σhkl |F(hkl)o − F(hkl)c|/ΣhklF(hkl)o.
Soaking in gabaculine of crystals grown from native GSA-AT changed the cell dimensions to a = 68.6 Å, b = 108.6 Å, and c = 123.5 Å. GSA-AT reduced with BH3CN has cell dimensions of a = 68.6 Å, b = 108.3 Å, and c = 122.4 Å. Both structures were analyzed by difference Fourier methods using the refined native GSA-AT structure as reference and subsequently refined by x-plor (Table 2).
Kinetic Analyses.
Consecutive absorption spectra were analyzed by global least-squares fitting and singular value decomposition using the program specfit (Spectrum Software Associates, Chapel Hill, NC). Rapid scanning experiments were carried out with a stopped-flow diode-array spectrophotometer (Hi-Tech, Salisbury, UK).
RESULTS AND DISCUSSION
Monomer Structure.
The GSA-AT fold (Figs. 2 and 3) can be divided into three domains. The N-terminal domain consists of about 70 residues that form an α-helix followed by a three-stranded antiparallel β-sheet. The main, cofactor-binding domain, residues 70–326, contains a central seven-stranded β-sheet with six parallel strands, a–f, and one antiparallel strand, g, the order being agfedbc, as first observed in aspartate aminotransferase (19). This structural feature is surrounded by several α-helices of different length. The C-terminal domain, comprising residues 327–433, folds into a three-stranded antiparallel β-sheet covered on the outer surface with four helices, including one at the C terminus. This topology corresponds to that of the other enzymes from subgroup II of the α-family of vitamin B6 enzymes (1), namely dialkylglycine decarboxylase (20, 21) and ornithine aminotransferase (22). The folds of the cofactor-binding domains of all three structures are similar, but there are major differences in the N-terminal part of the polypeptide chain and in several loop regions. If GSA-AT is compared with dialkylglycine decarboxylase, the largest deviations are found in the first 40 amino acid residues which in GSA-AT line the substrate binding site (see below). Further major deviations include the loops 153–181 (different conformation), 189–193 (deletion in GSA-AT), 365–385 (insertion), and 400–410 (deletion). The GSA-AT-dimer, which has dimensions of about 90 Å × 45 Å × 45 Å, is compact and has a large subunit interface area (≈5,000 Å2).
Figure 2.
Schematic drawing of the secondary structure of GSA-AT. Rectangles and arrows represent α-helices and β-strands, respectively. Secondary structure definitions were made with the program dssp (18).
Figure 3.
Stereoview of GSA-AT showing the overall fold and the secondary structure. The cofactor is shown in ball and stick. The view is down the 2-fold symmetry axis of the dimer. Subunit A is in yellow and subunit B is in green. Residues 153–181 are shown as blue in subunit A and as red in subunit B, but it should be noted that in subunit B, this part of the chain is disordered and does not have the structure indicated.
Dimer Structure.
The refined structure comprises the polypeptide chain from Phe-7 to the C terminus, Leu-433, for both subunits except for residues 153–181 which are disordered in subunit B. Superposition of the subunits using 397 Cα atoms results in an rms deviation of 0.48 Å2. The backbone conformations of the two subunits of GSA-AT are identical with two exceptions. The first is a 29-residue length from His-153 to Thr-181, which is well defined in subunit A but disordered in subunit B, with mean atomic B-factors of 34.4 Å2 and 103.4 Å2, respectively, compared with 29.6 Å2 for the overall structure. A second, smaller, deviation concerns the residues Pro-30 to Ile-42. The electron density for this region is well defined in both subunits, but the conformations differ somewhat in the two subunits. The largest differences in Cα position (Fig. 4) occur at Ala-33 (1.4 Å) and Val-31 (1.2 Å). The average B-factors for atoms in subunit B (31.8 Å2) are slightly lower than those for the same atoms in subunit A (35.7 Å2).
Figure 4.
Stereoviews of superpositions of the active sites of the two subunits. The bonds of subunits A and B are shown in yellow and cyan, respectively, except for residues 303–305 which are shown in green (subunit A) and blue (subunit B). (A) Wild-type enzyme. (B) The enzyme after reduction of the Schiff base double bond by cyanoborohydride.
Cofactor Binding.
Unless preparations of GSA-AT are deliberately treated to convert the enzyme completely into either the PLP or PMP form, solutions invariably contain both forms in approximately equal amounts (6, 7, 23). This finding is confirmed by the crystal structure and extended significantly by the observation that the distribution of the two forms of the cofactor is asymmetric within each dimer. Fig. 4 shows the location and conformation of the cofactor and Lys-273 in both subunits by superposition. While the location of the phosphate group, which behaves like an anchor, is the same in both subunits, the orientations of the pyridine rings differ by about 30° (Fig. 4). In subunit B there is continuous electron density between the cofactor and the lysine side chain, consistent with the protonated aldimine between Lys-273 and PLP that is responsible for the 418-nm chromophore in the absorption spectrum (6). In subunit A there is no continuous electron density between the cofactor and Lys-273, thus demonstrating the absence of a covalent bond and suggesting that, in this subunit, the cofactor is PMP. If this suggestion is correct, the PMP and Lys-273 amino group nitrogens are 2.7 Å apart, and the amino group of PMP points away from Lys-273 so that it can make a hydrogen bonded salt bridge with the ionized 3-hydroxyl group of the cofactor.
It is apparent that asymmetry of coenzyme binding coincides with asymmetry in the conformation of residues 153–181. In subunit A, which contains PMP, this segment is structured. However, in subunit B, which contains PLP bound as aldimine to Lys-273, the corresponding segment is disordered.
The possibility that the asymmetry is related to the catalytic action of the enzyme prompted further experiments aimed at revealing its origins. The enzyme can be converted completely into the PLP form, but we have not succeeded in growing crystals good enough for x-ray analysis, because this form is unstable in solution, approximately half of it reverting to the PMP form within a few days. Consequently, the absorption spectrum and the structure of a crystal grown from enzyme converted to the double PLP form correspond to those of the native enzyme (M.H. and J.N.J., unpublished data). So far, the only form of the enzyme to yield useful crystals in which it is certain that the cofactor is present in the same form in both monomers, is that obtained by reducing the double PLP form as soon as it is made. The cofactor in this sample was entirely in the 330-nm absorbing form and electro-spray mass spectroscopy confirmed the covalent single bond between Lys-273 and PLP in both monomers (R.C. and R.A.J. unpublished data). The same covalent bond was evident in the crystal structure in both monomers but the conformational asymmetry of residues Gly-153 to Thr-181 remained, demonstrating that it persists even when cofactor binding is symmetric.
Asymmetry in Solution.
Many oligomeric enzymes show asymmetric kinetic behavior. Binding of a ligand to one subunit in, for example, an α2 dimer may cause a conformational change that influences the binding affinity of the second subunit for the ligand in a positive (positive cooperativity) or negative way (negative cooperativity). The most extreme form of negative cooperativity corresponds to “half of the sites reactivity” (24). Niefind et al. (25) described negative cooperativity in binding of NAD+, coupled to strong deviations from 222 symmetry for the α4 tetramer l-2-hydroxyisocaproate dehydrogenase from Lactobacillus confusus. There appears, however, to be no example of an oligomer in which crystallographically demonstrated asymmetry in the absence of ligands can be linked with asymmetric kinetic behavior in solution.
In the symmetric dimer aspartate aminotransferase, reduction of the PLP form by NaBH3CN produces a single first order reduction of the 430-nm absorbing protonated imine to colorless product (26). The course of reduction of GSA-AT in its double PLP form does not fit to a single first order process but is biphasic. When the kinetics of the reduction are observed by measuring the amount of remaining unreduced PLP, two first order processes are seen and the amplitudes of the two phases show that half of the enzyme is reduced rapidly and the other half slowly, consistent with the existence of an asymmetric dimer in which one active site is less accessible than the other. When the reduction is followed by observing changes in the absorption spectrum as the reaction proceeds, further complexity is evident in that the spectral changes of the fast phase are different from those of the slow phase (Fig. 5).
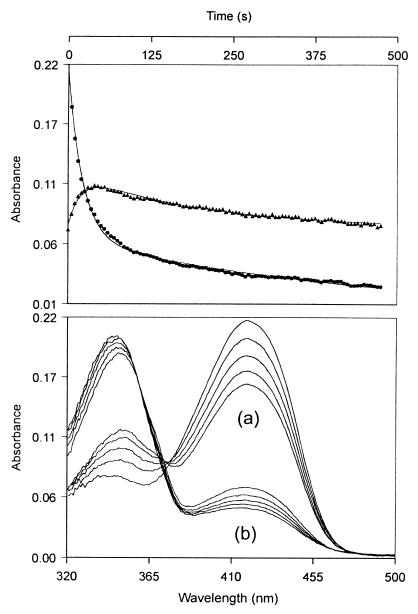
Figure 5.
Changes in absorption spectrum during reduction. The enzyme (37 μM) was reacted with NaBH3CN (0.1 M) in 0.1 M tricine (pH 7.8, 37°C). Absorption spectra were taken at 5-s intervals. (Upper) Absorbance changes observed at single wavelengths of 418 nm (•) and 370 nm (▴). (Lower) Spectra taken (a) at 5-s intervals from the beginning of the reaction and (b) at 50-s intervals 250 s after the start of reaction.
Singular value decomposition of these results, based on analysis of absorbance changes at all wavelengths, showed that they were consistent with two consecutive reactions and inconsistent with two parallel reactions, thus demonstrating the presence of a spectrally distinct intermediate. The analysis showed the intermediate to contain a significant chromophore with λmax = 360 nm. This cannot be the reduced cofactor (λmax = 330 nm), and its spectrum suggests that it is the unprotonated aldimine. Importantly, it must have arisen in the subunit which has not been reduced and therefore provides direct evidence of communication between the subunits. Specifically, in this interpretation, reduction of the cofactor aldimine in one subunit lowers the pK of the aldimine in the other subunit.
Substrate Binding Site.
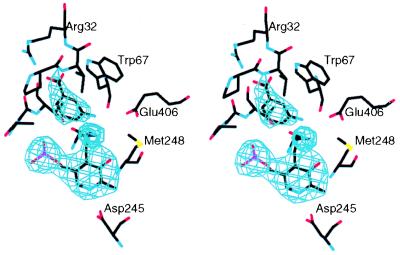
The substrate binding site was identified using gabaculine (3-amino-2,3-dihydrobenzoic acid), a substrate analogue and mechanism-based inhibitor of GSA-AT (10). The electron density of a crystal soaked in 3 mM gabaculine shows unambiguously (Fig. 6) that the molecule binds in a pocket which is positioned differently from those of all other members of the α-family of aminotransferases studied so far. In subunit A, which in the native structure contains noncovalently bound cofactor, gabaculine is bound noncovalently with its carboxylate group at hydrogen bonding distance from the guanidinium group of Arg-32 and its amino group near that of PMP and the ɛ-group of Lys-273. Thus Arg-32, an invariant residue in GSA-AT from all species, binds the carboxylate of gabaculine, and, by inference therefore, the carboxylate of the substrate, GSA. In subunit B there is only very weak electron density for gabaculine. This finding is puzzling in that the expected covalent binding of gabaculine to PLP in subunit B is not apparent. A recent experiment (M. Moser and J.N.J., unpublished data) in which native GSA-AT was cocrystallized with gabaculine, gave crystals isomorphous with those of the native enzyme. In subunit A gabaculine binds as in the soaking experiment, but in subunit B gabaculine is covalently bound, as expected for the PLP form of the enzyme.
Figure 6.
Active site of subunit A with gabaculine noncovalently bound, superimposed on a 3.0-Å resolution omit map, contoured at 4 σ.
The funnel-shaped, gabaculine-binding pocket is about 12 Å long with the internal, narrow (6 Å) end near Arg-32 and the least buried, wider (9 Å) end between the side chains of Tyr-150 and Glu-406. The side chains of the following residues line the pocket: Ser-29, Val-31, Arg-32, Trp-67, Tyr-150, Ser-163, Asn-217, Met-248, Glu-406, Ala-303*, Gly-304*, and Thr-305* (the ∗ indicates residues from the neighboring subunit). All these residues are invariant in the 12 sequences of GSA-AT known so far except Met-248 which is valine in Xanthomonas campestris (27). Glu-406 is likely to have an important role. It is well placed to bind to the 5-amino group of diaminovalerate and could hydrogen bond with the aldehyde of GSA. However, perhaps its most important role is the exclusion of glutamate from the pocket by charge repulsion. It may therefore be the residue that contributes most to the differentiation of this enzyme from the aminotransferases in the α-family which bind the α-carboxylate with an invariant arginine (1, 5, 8).
Gabaculine inhibition is much relieved in a mutant in which Met-248 is replaced by isoleucine (10). The side chain of Met-248 has an extended conformation (Fig. 4), approximately perpendicular to the plane of the pyridine ring of the cofactor. In subunit A the distances of the Cɛ atom of the Met-248 side chain to Nζ of K273 and O3 of the cofactor are 3.6 Å and 3.2 Å, respectively, while in subunit B these distances are 3.8 Å and 3.6 Å. It is unlikely therefore that Met-248 plays an important role in reactions with the true substrate but its substitution by isoleucine may impose restrictions on movements of the cofactor and gabaculine because of the extra bulk of the inhibitor.
Inspection of space filling models reveals that, in subunit A, there is no opening through which the substrate may enter its binding pocket. In contrast, in subunit B with its flexible region 153–181, a gate to the substrate pocket would be opened by the displacement of the short helical section Ser-163 to Leu-168 which closes the entrance (Fig. 3). Thus flexibility in this region would readily permit substrate entry and product release. The proposal that this region is functionally important, is supported by the fact that residues Ala-161, Gly-162, Ser-163, and Gly-164 are invariant in all 12 sequences of GSA-AT which are known so far and for residues Thr-167 and Leu-168 there is only one substitution. The gate is delimited by the structurally well-defined residues Val-31, Tyr-150, Asn-217, Tyr-301, Tyr-375, Ser-403, and Phe-405. Thus the crystal structure of native GSA-AT reveals a dimer in which one subunit (B) has the cofactor PLP bound as a Schiff-base to K273 and a flexible region 153–181 which allows the entry of the substrate. The other subunit, A, has the noncovalently bound form of the cofactor, PMP and a well-defined structure in the region 153–181 which restricts entry and release of substrates and products.
Conclusions.
Both the crystallography and the kinetics of reduction in solution point strongly to asymmetry in the GSA-AT dimer. The fact that the asymmetry in the polypeptide chain is confined to a loop, the position of which controls access to the active site and which is of clearly different mobility in each monomer, suggests that the asymmetry is related to the catalytic mechanism. The crystallographic observation that access to the active site of subunit A is much less than to subunit B must be reconciled with the fact that the reaction must begin with entry of the substrate to the PMP-containing site. Thus, the closed subunit cannot present a total kinetic barrier to substrate binding. Although its preferred conformation is closed it must open sufficiently frequently to allow entry of the substrate and release of the product. The enzyme that has both subunits in the PLP form reacts very much more slowly with GSA (9). Consequently, if both subunits were converted to the PLP form during the normal reaction, the enzyme would be essentially inactive. The asymmetry may provide a mechanism to prevent this eventuality by ensuring (i) that when one subunit is open the other is closed and, (ii) that when one subunit is in the PLP form the other subunit cannot open. Such a mechanism would prevent the enzyme from being converted into the almost inactive form in which both monomers contain PLP.
Acknowledgments
We thank M. Jenny for excellent technical support in purification and crystallization. We thank the staff of the European Molecular Biology Laboratory outstation at Deutsches Elektronen Synchrotron (Hamburg, Germany) for synchrotron data collection facilities. This research was supported by the European Community Human Capital and Mobility Program 1991–1995 (contract CHRX-CT93-0179) with Swiss contributions by the “Bundesamt für Bildung und Wissenschaft,” Berne, Switzerland (Contract 93.0305). It was also supported in part by Grant 31-36432.92 of the Swiss National Science Foundation (to J.N.J.).
ABBREVIATIONS
- GSA
glutamate 1-semialdehyde
- GSA-AT
GSA aminomutase
- PLP
pyridoxal-5′-phosphate
- PMP
pyridoxamine-5′-phosphate.
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Chemistry Department, Brookhaven National Laboratory, Upton, NY 11973 (accession codes 2GSA, 3GSA, 4GSA).
References
- 1.Alexander F W, Sandmeier E, Mehta P K, Christen P. Eur J Biochem. 1994;219:953–960. doi: 10.1111/j.1432-1033.1994.tb18577.x. [DOI] [PubMed] [Google Scholar]
- 2.Jordan P M, Shemin D. In: The Enzymes. 3rd Ed. Boyer P D, editor. Vol. 7. New York: Academic; 1972. pp. 339–356. [Google Scholar]
- 3.Kannangara G, Gough S P, Bryant P, Hoober J K, Kahn A, von Wettstein D. Trends Biochem Sci. 1988;13:139–143. doi: 10.1016/0968-0004(88)90071-0. [DOI] [PubMed] [Google Scholar]
- 4.Grimm B, Bull A, Breu V. Mol Gen Genet. 1991;225:1–10. doi: 10.1007/BF00282635. [DOI] [PubMed] [Google Scholar]
- 5.Mehta P K, Christen P. Biochem Biophys Res Commun. 1994;198:138–143. doi: 10.1006/bbrc.1994.1020. [DOI] [PubMed] [Google Scholar]
- 6.Smith M A, Kannangara C G, Grimm B, Smith M. Eur J Biochem. 1991;202:749–757. doi: 10.1111/j.1432-1033.1991.tb16429.x. [DOI] [PubMed] [Google Scholar]
- 7.Pugh C, Harwood J L, John R A. J Biol Chem. 1992;267:1584–1588. [PubMed] [Google Scholar]
- 8.Christen P, Metzler D E, editors. Transaminases. New York: Wiley; 1985. [Google Scholar]
- 9.Tyacke R J, Harwood J L, John R A. Biochem J. 1993;293:697–701. doi: 10.1042/bj2930697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimm B, Smith A J, Kannangara C G, Smith M. J Biol Chem. 1991;19:12495–12501. [PubMed] [Google Scholar]
- 11.Hennig M, Grimm B, Jenny M, Müller R, Jansonius J N. J Mol Biol. 1994;242:591–594. doi: 10.1006/jmbi.1994.1606. [DOI] [PubMed] [Google Scholar]
- 12.Thaller C, Weaver L H, Eichele G, Wilson E, Karlsson R, Jansonius J N. J Mol Biol. 1981;147:465–469. doi: 10.1016/0022-2836(81)90496-4. [DOI] [PubMed] [Google Scholar]
- 13.Science and Engineering Research Council. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 14.Kleywegt G J, Jones T A. Acta Crystallogr D. 1996;52:826–828. doi: 10.1107/S0907444995014983. [DOI] [PubMed] [Google Scholar]
- 15.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 16.Brünger A T. x-plor. New Haven, CT: Yale Univ. Press; 1992. , Version 3.1. [Google Scholar]
- 17.Engh R A, Huber R. Acta Crystallogr A. 1991;47:392–400. [Google Scholar]
- 18.Kabsch W, Sander Ch. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 19.Ford G C, Eichele G, Jansonius J N. Proc Natl Acad Sci USA. 1980;77:2559–2563. doi: 10.1073/pnas.77.5.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toney M D, Hohenester E, Cowan S W, Jansonius J N. Science. 1993;261:756–759. doi: 10.1126/science.8342040. [DOI] [PubMed] [Google Scholar]
- 21.Toney M D, Hohenester E, Keller J W, Jansonius J N. J Mol Biol. 1995;245:151–179. doi: 10.1006/jmbi.1994.0014. [DOI] [PubMed] [Google Scholar]
- 22.Jansonius J N, Genovesio-Taverne J-C, Hennig M, Hohenester E, Jenny M, Malashkevich V N, Moser M, Shen B W, Stark W, von Stosch A, Toney M D. In: Biochemistry of Vitamin B6 and PQQ. Marino G, Sannia G, Bossa F, editors. Basel: Birkhäuser; 1994. pp. 29–35. [Google Scholar]
- 23.Brody S, Andersen J S, Kannangara C G, Meldgaard M, Roepstorff P, von Wettstein D. Biochemistry. 1995;34:15918–15924. doi: 10.1021/bi00049a006. [DOI] [PubMed] [Google Scholar]
- 24.Koshland D E., Jr Curr Opin Struct Biol. 1996;6:757–761. doi: 10.1016/s0959-440x(96)80004-2. [DOI] [PubMed] [Google Scholar]
- 25.Niefind K, Hecht H-J, Schomburg D. J Mol Biol. 1995;251:256–281. doi: 10.1006/jmbi.1995.0433. [DOI] [PubMed] [Google Scholar]
- 26.Garnier A L, John R A. Eur J Biochem. 1993;216:763–768. doi: 10.1111/j.1432-1033.1993.tb18196.x. [DOI] [PubMed] [Google Scholar]
- 27.Murakami K, Korbsrisate S, Asahara N, Hashimoto Y, Murooka Y. Appl Microbiol Biotechnol. 1993;38:502–506. doi: 10.1007/BF00242945. [DOI] [PubMed] [Google Scholar]