Abstract
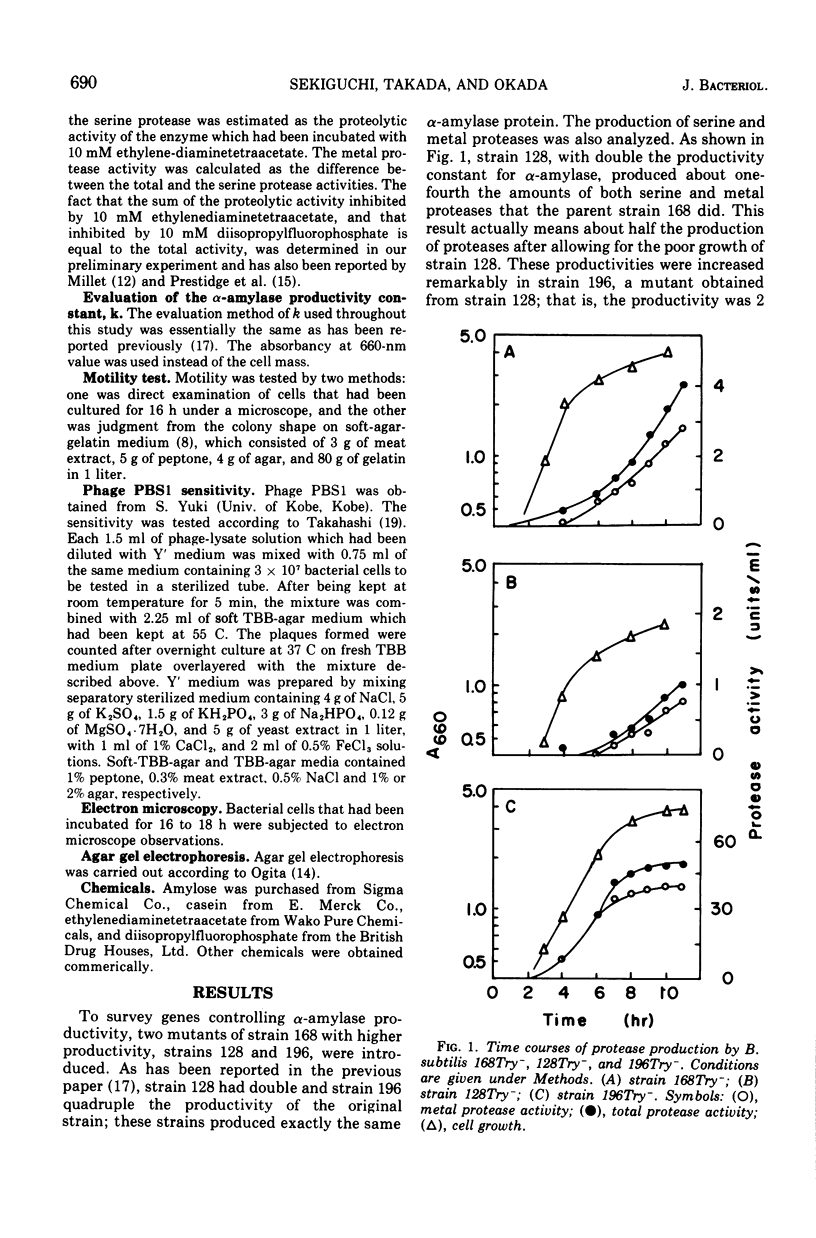
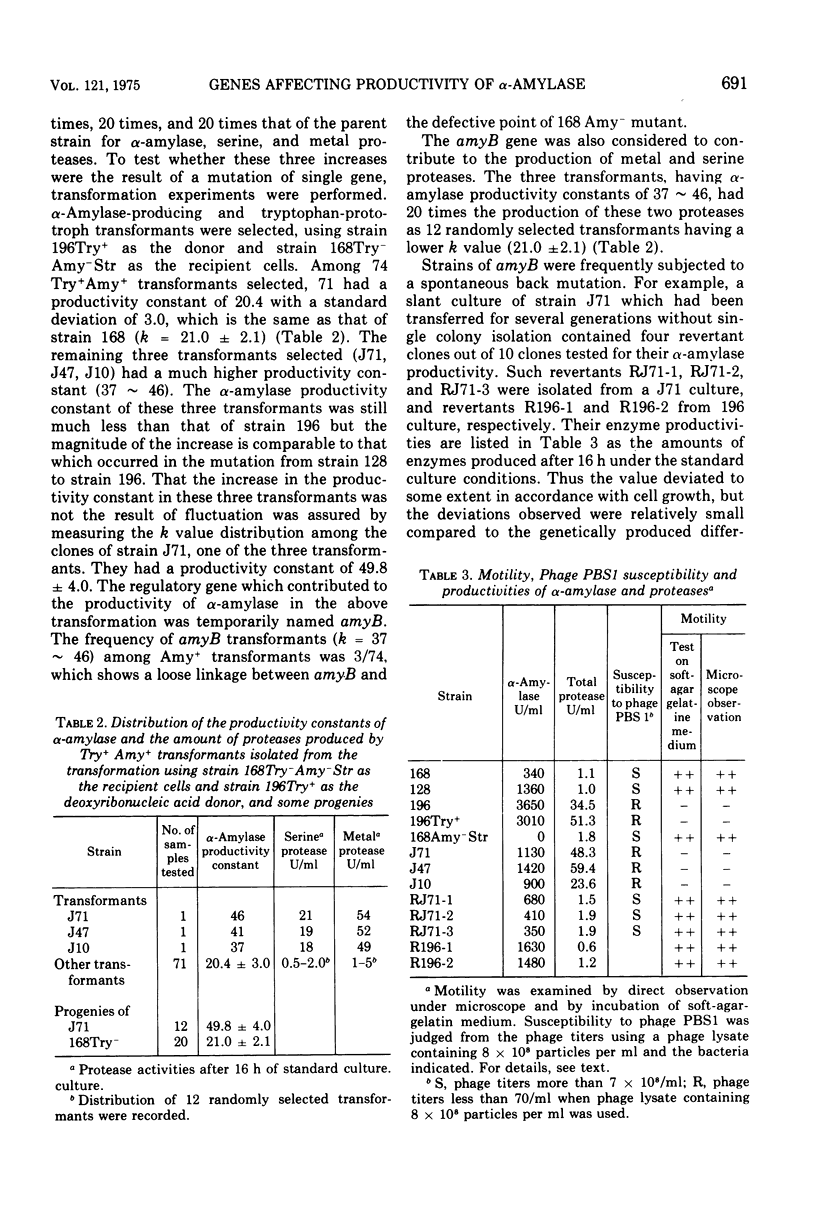
Genetic control of alpha-amylase (alpha-1,4-glucan glucanohydrolase, EC 3.2.1.1.) production by Bacillus subtilis 168 was studied from the standpoint that alpha-amylase production by bacteria is dependent on a long-lived messenger ribonucleic acid and obeys the following equation: E = kappa integral of X-DT where x = cell mass at time t, E = alpha amylase produced, t = culture time, and kappa = productivity constant. So a productivity constand (kappa) is obtained from the slope of the straight line plot of alpha-amylase formed versus the total mass of cells accumulated over that time during the culture process. The following results were obtained. (i) Two sequential mutants, derived from the 168(kappa = 20) strain and having improved alpha-amylase productivity (168 leads to 196), were analyzed for their serine and metal protease production. Strain 128 (kappa = 40) produced half the amount of both proteases, but strain 196 (kappa = 60 similar to 80) produced 20 times that in the original strain. (ii) Amy+ transformants, using the 196 strain as the other three had higher productivity (kappa = 37 similar to 46). These transformants (J71, J47, groups. Seventy-one of 74 Amy+ transformants had a kappa value of 21.0 plus or minus 2.1 and the other three had higher productivity (kappa = 37 similar to 46). These transformants (J71,J47, and J10) produced levels of serine and metal proteases 20 times higher than the other transformants. (iii) Strains 196, J71, J47, and J10 were found to be nonmotile and resistant to phage PBS1, whereas other strains, including strains 168, 128, 3 revertants of strain J71 and 2 revertants of strain 196, were all motile and sensitive to the phage. (iv) Strains 196 and J71 were nonflagellated under electron microscopic observation but strain 168, 128 and a revertant of J71 were flagellated. From the above experimental results, the existence of a quality controlling gene (amyB) was deduced, which is loosely linked to the structural gene and controls productivities of alpha-amylase and proteases, and flagellation. The probable existence of another regulatory gene, amyC, is also discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both G. W., McInnes J. L., Hanlon J. E., May B. K., Elliott W. H. Evidence for an accumulation of messenger RNA specific for extracellular protease and its relevance to the mechanism of enzyme secretion in bacteria. J Mol Biol. 1972 Jun 20;67(2):199–217. doi: 10.1016/0022-2836(72)90236-7. [DOI] [PubMed] [Google Scholar]
- COHEN G. N., MONOD J. Bacterial permeases. Bacteriol Rev. 1957 Sep;21(3):169–194. doi: 10.1128/br.21.3.169-194.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel R. W., Joys T. M. Adsorption Specificity of Bacteriophage PBS1. J Bacteriol. 1966 Aug;92(2):388–389. doi: 10.1128/jb.92.2.388-389.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GREEN D. M., COLARUSSO L. J. THE PHYSICAL AND GENETIC CHARACTERIZATION OF A TRANSFORMABLE ENZYME: BACILLUS SUBTILIS ALPHA-AMYLASE. Biochim Biophys Acta. 1964 Aug 26;89:277–290. doi: 10.1016/0926-6569(64)90216-0. [DOI] [PubMed] [Google Scholar]
- Joys T. M. Correlation between susceptibility to bacteriophage PBS1 and motility in Bacillus subtilis. J Bacteriol. 1965 Dec;90(6):1575–1577. doi: 10.1128/jb.90.6.1575-1577.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Michel J. F., Millet J. Physiological studies on early-blocked sporulation mutants of Bacillus subtilis. J Appl Bacteriol. 1970 Mar;33(1):220–227. doi: 10.1111/j.1365-2672.1970.tb05246.x. [DOI] [PubMed] [Google Scholar]
- Millet J. Characterization of proteinases excreted by Bacillus subtilis Marburg strain during sporulation. J Appl Bacteriol. 1970 Mar;33(1):207–219. doi: 10.1111/j.1365-2672.1970.tb05245.x. [DOI] [PubMed] [Google Scholar]
- NESTER E. W., LEDERBERG J. Linkage of genetic units of Bacillus subtilis in DNA transformation. Proc Natl Acad Sci U S A. 1961 Jan 15;47:52–55. doi: 10.1073/pnas.47.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OGITA Z. I. IMPROVED AGAR GEL MEDIA FOR THIN LAYER ELECTROPHORESIS. Med J Osaka Univ. 1964 Dec;15:141–153. [PubMed] [Google Scholar]
- Prestidge L., Gage V., Spizizen J. Protease activities during the course of sporulation on Bacillus subtilis. J Bacteriol. 1971 Sep;107(3):815–823. doi: 10.1128/jb.107.3.815-823.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKAHASHI I. Transducing phages for Bacillus subtilis. J Gen Microbiol. 1963 May;31:211–217. doi: 10.1099/00221287-31-2-211. [DOI] [PubMed] [Google Scholar]
- Yoneda Y., Yamane K., Maruo B. Membrane mutation related to the production of extracellular -amylase and protease in bacillus subtilis. Biochem Biophys Res Commun. 1973 Feb 5;50(3):765–770. doi: 10.1016/0006-291x(73)91310-7. [DOI] [PubMed] [Google Scholar]
- Yuki S. On the gene controlling the rate of amylase production in Bacillus subtilis. Biochem Biophys Res Commun. 1968 Apr 19;31(2):182–187. doi: 10.1016/0006-291x(68)90727-4. [DOI] [PubMed] [Google Scholar]




