Abstract
ClpA, a newly discovered ATP-dependent molecular chaperone, remodels bacteriophage P1 RepA dimers into monomers, thereby activating the latent specific DNA binding activity of RepA. We investigated the mechanism of the chaperone activity of ClpA by dissociating the reaction into several steps and determining the role of nucleotide in each step. In the presence of ATP or a nonhydrolyzable ATP analog, the initial step is the self-assembly of ClpA and its association with inactive RepA dimers. ClpA-RepA complexes form rapidly and at 0°C but are relatively unstable. The next step is the conversion of unstable ClpA-RepA complexes into stable complexes in a time- and temperature-dependent reaction. The transition to stable ClpA-RepA complexes requires binding of ATP, but not ATP hydrolysis, because nonhydrolyzable ATP analogs satisfy the nucleotide requirement. The stable complexes contain approximately 1 mol of RepA dimer per mol of ClpA hexamer and are committed to activating RepA. In the last step of the reaction, active RepA is released upon exchange of ATP with the nonhydrolyzable ATP analog and ATP hydrolysis. Importantly, we discovered that one cycle of RepA binding to ClpA followed by ATP-dependent release is sufficient to convert inactive RepA to its active form.
Keywords: protein folding, plasmid P1, ATP-dependent proteolysis, molecular chaperones
It was first suggested that the universal and highly conserved Clp family of proteins might be molecular chaperones because they possess several characteristics of known chaperones (1). Many chaperones are heat-shock proteins involved during heat stress in the refolding of nonnative proteins and the degradation of irreversibly denatured proteins. Similarly, many Clp proteins are induced by heat shock, and several are regulatory components of two component ATP-dependent proteases. Recently, Escherichia coli ClpA and ClpX and yeast Hsp104 have been demonstrated to reactivate and disaggregate proteins, defining functions of Hsp60 and Hsp70 chaperones (2–5).
Yeast Hsp104 has been shown to function as a chaperone in vivo (2). Mutant cells defective in Hsp104 have lost the ability to dissolve protein aggregates formed by heat shock and are unable to reactivate heat-inactivated luciferase (2). Hsp104 is also involved in reactivating mRNA splicing after heat inactivation (6) and in altering the conformation of a yeast prion protein in vivo (7).
E. coli ClpA was first identified as the regulatory component of ClpAP protease (protease Ti) (8, 9). ClpP is the peptidase component and is unrelated to the Clp family. ClpA has two ATP binding sites and is an ATPase. It forms an oligomer with 6-fold symmetry in the presence of MgATP (10). ClpAP protease complexes contain a tetradecamer of ClpP with a ClpA hexamer at one or both ends, forming structures strikingly similar to the eukaryotic 26S proteasome (10).
Studies from our laboratory showed that ClpA, independent of ClpP, performs the ATP-dependent chaperone function of DnaJ/DnaK/GrpE in activating the plasmid P1 initiator protein, RepA, in vitro (3). The chaperones stimulate the specific DNA binding activity of RepA by about 100-fold but are themselves not part of the RepA-DNA complex (11). Gel filtration (12) and velocity sedimentation experiments (unpublished work) showed that activation is the conversion of inactive RepA dimers to active monomers. Other chaperones, including ClpB, ClpX, Hsp104, and GroEL/GroES, do not activate RepA (3).
Additional evidence that ClpA is a chaperone is the ability of ClpA to protect RepA from heat inactivation and to prevent firefly luciferase from irreversible heat inactivation in vitro (3). ClpA also targets RepA for in vitro degradation by ClpP in the presence of ATP (3). ClpA is directly required for degradation and not simply for generating RepA monomers, because RepA activated by DnaJ/DnaK/GrpE is not degraded by ClpP without ClpA.
ClpX functions as a molecular chaperone in bacteriophage Mu DNA replication and transposition (5, 13, 14). It mediates the ATP-dependent disassembly in vitro of stable complexes of MuA tetramers and Mu DNA, releasing MuA monomers (5). In another in vitro reaction, ClpX prevents and reverses heat-induced aggregation of bacteriophage λ O protein (4). ClpX is an alternative regulator of ClpP, forming ClpXP protease (15, 16) and two ClpXP substrates are MuA (5) and λ O protein (15, 16).
Thus, the Clp family of proteins has emerged as a new class of ATP-dependent molecular chaperones able to prevent aggregation, refold and remodel proteins, and participate in degradation. In this report, we have investigated the mechanism of ClpA chaperone activity using the ClpA activation of RepA. We have isolated several intermediate complexes in the reaction and found that ClpA-RepA complexes committed to activating RepA are formed in a time-, temperature-, and nucleotide-dependent reaction. We discovered that one round of RepA binding to ClpA and release from ClpA is sufficient to activate RepA.
MATERIALS AND METHODS
Proteins and DNA.
P1 RepA (17) and ClpA (18) were purified as described. Throughout, both ClpA and RepA are expressed as mol of subunits. Plasmid DNA carrying the P1 origin of replication (100 μg) was labeled by incubating it for 1.5 hr at 37°C in 200 μl of HhaI buffer containing 125 units of HhaI methylase (New England Biolabs) and 0.5 mCi (80 Ci/mmol; 1 Ci = 37 GBq) of S-adenosyl-l-[methyl-3H]methionine (DuPont/NEN). The DNA was purified by phenol extraction and Sepharose 6B column chromatography.
In Vivo Labeling of RepA Protein.
Overnight cultures of E. coli MZ1 carrying pLRepA, a plasmid encoding RepA under the control of the λ PL promoter, were grown at 30°C in M63 minimal media supplemented with 2% glucose, 0.05% B1, 1% casamino acids, and 50 μg/ml ampicillin. Cells were then diluted into 1 liter of M63 media containing the same supplements, grown to an OD600 of 0.8, collected by centrifugation, resuspended in 1 liter of M63 media containing all supplements except casamino acids and ampicillin, and incubated at 30°C for 1 hr. RepA expression was induced by raising the temperature to 42°C for 30 min. Cells were labeled with either 5 mCi of Tran35S-label or 1 mCi of l-[14C]leucine (both from ICN) for 5 min at 42°C. Cells were immediately harvested, and RepA was purified. The specific activities of the purified 35S- and 14C-labeled RepA were 2 Ci/mmol and 0.3 Ci/mmol, respectively. Both unlabeled and labeled RepA were activated by either DnaK/DnaJ/GrpE or ClpA to similar extents.
RepA Activation Reactions.
Reaction mixtures (20 μl) for RepA activation contained buffer A (20 mM Tris⋅HCl, pH 7.5/100 mM KCl/0.01 mM EDTA/5 mM DTT/10 mM MgCl2), 1 mM ATP, 100 μg/ml BSA, 300 ng of ClpA and RepA as indicated. The proteins were diluted in 20 mM Tris⋅HCl, pH 7.5/100 mM KCl/0.01 mM EDTA/5 mM DTT/0.01% Triton X-100 (vol/vol). After 15 min at 23°C, the mixtures were chilled to 0°C. One microgram of calf thymus DNA and 20 to 40 fmol of 3H-labeled plasmid DNA containing the P1 origin were added. After 5 min at 0°C, the mixtures were filtered through nitrocellulose filters, and the retained radioactivity, indicating RepA binding, was measured.
RESULTS
One Cycle of Binding and Release of RepA to ClpA Activates RepA.
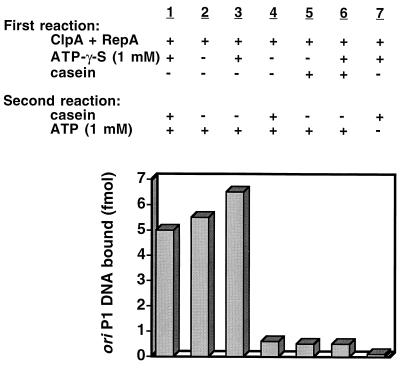
We dissected the reaction into several steps and determined the role of nucleotide in each step to understand the mechanism of the chaperone activity of ClpA. Previous work showed that ClpA both assembles into hexamers (10) and interacts with inactive RepA dimers to form stable ClpA-RepA complexes (3) in reactions requiring ATP, but not ATP hydrolysis. We used this information to determine whether one or multiple rounds of binding and release of RepA to ClpA are necessary to refold RepA. First, excess ClpA was incubated with RepA and adenosine 5′-O-(3-thiotriphosphate) (ATP[γ-S]), a poorly hydrolyzable ATP analog, at 23°C to form ClpA-RepA complexes. Free ClpA was then trapped by adding a 10-fold molar excess of casein, which binds with high affinity to ClpA (18). Next, knowing that the half-time for ATP exchange with ATP[γ-S] bound to ClpA is fast (less than a minute, unpublished observation), we added ATP and measured RepA activation. We discovered that ClpA efficiently activated RepA despite the casein trap (Fig. 1). The level of RepA activation was similar to that reached with the standard RepA activation conditions or with casein omitted during the exchange of ATP[γ-S] and ATP (Fig. 1). As controls, the omission of ATP[γ-S] during the first reaction or the addition of casein, either without or with ATP[γ-S], blocked RepA activation (Fig. 1). Similarly, RepA was not activated when the second incubation lacked ATP, because ATP is required for activation (Fig. 1) (3). These experiments show that ClpA binding to RepA requires nucleotide binding but that RepA activation and release requires ATP hydrolysis. Because it is known that the active RepA is free of ClpA, these experiments demonstrate that a single cycle of ClpA binding and release activates RepA.
Figure 1.
Casein trap experiment showing that one cycle of ClpA binding and ATP-dependent release activates RepA. ClpA-RepA complexes were formed by incubating 4.3 pmol of ClpA and 0.3 pmol of RepA in 20 μl of buffer A containing 1 mM ATP[γ-S] for 20 min at 23°C. Then 42.4 pmol of casein was added, followed by 1 mM ATP where indicated. After an additional 15 min at 23°C, activated RepA was measured by adding 1 μg of calf thymus DNA and 20 fmol of 3H-labeled oriP1 plasmid DNA. After 5 min at 0°C, the mixtures were filtered through nitrocellulose filters, and the retained radioactivity was measured. In control experiments ATP[γ-S] was omitted from the first reaction, casein was added to the first reaction with or without ATP[γ-S], or ATP was omitted from the second reaction, as indicated.
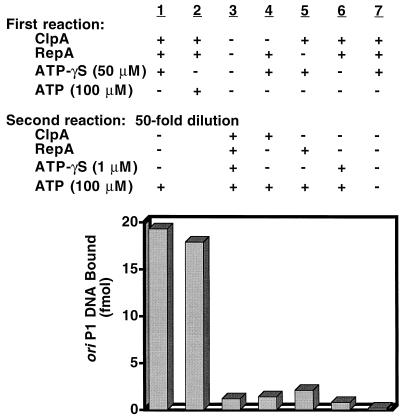
We performed dilution experiments to corroborate these results. ClpA was incubated with RepA and ATP[γ-S] as described above. Then the reaction mixtures were diluted 50-fold to prevent further assembly or reassembly of ClpA and RepA. Lastly, ATP was added. We found that after dilution and ATP exchange, RepA was fully activated, and the extent of RepA activation was similar to that obtained when the reaction was performed with ATP without dilution (Fig. 2). When the entire reaction was carried out in the dilute conditions of the second incubation, activation was prevented, indicating that dilution restricts activation of RepA to a single cycle (Fig. 2). When ClpA, RepA, or ATP[γ-S] was omitted from the first reaction and added back during the second incubation after dilution, no activation was observed (Fig. 2). The omission of ATP from the second reaction also prevented activation. When the experiments were performed using adenylyl imidodiphosphate, a nonhydrolyzable ATP analog, in place of ATP[γ-S], similar results were obtained (results not shown). Thus, these results confirm that ClpA activates RepA in one cycle of nucleotide-dependent binding and ATP-hydrolysis-dependent release. The requirement for RepA in the first reaction shows that the assembly reaction involves not just ClpA hexamerization but also binding to RepA. Nucleotide binding presumably induces an allosteric change in the chaperone to allow substrate binding, but a second transition requiring ATP hydrolysis is needed to complete the cycle and release the active RepA monomers.
Figure 2.
Dilution experiment showing that one cycle of ClpA binding and ATP-dependent release activates RepA. ClpA-RepA complexes were formed by incubating 6.0 pmol of ClpA and 1.1 pmol of RepA in 15 μl of buffer A containing 50 μM ATP[γ-S]. After 20 min at 23°C, 3 μl was diluted to 160 μl with the same buffer plus 100 μg/ml BSA and 100 μM ATP and incubated 10 min at 23°C. Activated RepA was determined as described in Fig. 1. In column 2, RepA and ClpA were incubated without nucleotide in 15 μl. Then 3 μl was diluted to 20 μl in buffer A containing 100 μg/ml BSA and 100 μM ATP and incubated as above. In column 3, the entire reaction was carried out in 160 μl with 1.2 pmol of ClpA and 0.2 pmol of RepA. In other control experiments, RepA, ClpA, or ATP[γ-S] was omitted from the first reaction where indicated and was added back in the second reaction. In the last control, ATP was omitted from the second reaction.
Formation of Uncommitted ClpA-RepA Complexes and Their Conversion to Committed Complexes.
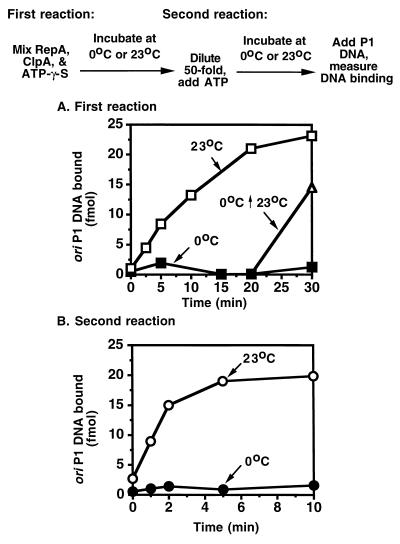
The dilution protocol readily divided the activation reaction into discrete stages. Interestingly, we discovered that the first step of the dilution experiment (complex formation) is relatively slow, requiring at least 20 to 30 min for completion at 23°C (Fig. 3A). Moreover, the extent of the reaction was much less at 0°C than at 23°C (Fig. 3A). Both of these results were surprising because many protein–protein interactions occur rapidly and relatively independent of temperature.
Figure 3.
Time course of dilution experiment suggesting that RepA-ClpA complexes assemble in a time- and temperature-dependent reaction. (A) Reaction mixtures identical to those described in Fig. 2 containing ClpA, RepA, and ATP[γ-S] were incubated at 0°C (▪) or 23°C (□) for the times indicated and then diluted 50-fold into buffer A containing ATP as described in Fig. 2. After a second incubation of 10 min at 23°C, activated RepA was determined as in Fig. 1. In one set of reaction mixtures, after incubating at 0°C for 20 min, the mixtures were further incubated at 23°C for 10 min before dilution (▵). (B) Reaction mixtures were identical to those described in Fig. 2. After a first incubation of 20 min at 23°C, reactions were diluted, and ATP was added as in Fig. 2. The second reaction was incubated at 0°C (•) or 23°C (○) for the times indicated.
To determine if ClpA hexamer assembly was the slow and temperature-sensitive step, we incubated ClpA and ATP[γ-S] for 20 min in the absence of RepA to allow hexamerization and then added RepA 2 min before dilution. The amount of RepA activated after dilution was still very low (data not shown), indicating that the slow first reaction must involve RepA.
We measured complexes directly to further characterize the ClpA-RepA complex formation reaction. Using radiolabeled RepA and a Microcon 100 filtration assay, we were able to separate free RepA from ClpA-RepA complexes and quantitate the amount of RepA associated with ClpA. With this technique, less than 10% of the [14C]RepA, both inactive dimers and active monomers, was retained on the filters (Table 1). Greater than 95% of the ClpA, incubated alone or with ATP[γ-S], was retained on the filters, as determined by SDS/PAGE (data not shown). When ClpA was incubated with [14C]RepA with or without ATP[γ-S] at 0°C or 23°C, 47% to 98% of the RepA was retained on the filters with ClpA (Table 1). Thus, these complexes, unlike the complexes formed in the first step of the dilution experiment, assembled independent of temperature and nucleotide binding.
Table 1.
Requirements for committed and uncommitted ClpA-RepA complex formation
| Additions to complex formation reactions | RepA in complex with ClpA, % RepA retained on Microcon 100 filters
|
|||
|---|---|---|---|---|
| No NaCl
|
1 M NaCl
|
|||
| 0°C | 23°C | 0°C | 23°C | |
| RepA + ClpA + ATP[γ-S] | 75 | 98 | 5 | 85 |
| RepA + ClpA | 47 | 65 | 1 | 3 |
| RepA | 10 | 10 | 8 | 7 |
| RepA + ClpA + ATP | 10 | 12* | 5 | 4* |
Reaction mixtures (50 μl) containing buffer A, 100 μg/ml BSA, 0.02% Triton X-100 (vol/vol), 72 pmol of ClpA, 10 pmol of [14C]RepA, and 1 mM ATP[γ-S] or 1 mM ATP, where indicated, were incubated at 23°C or 0°C for 20 min. Mixtures were adjusted to 100 μl with buffer A containing 100 μg/ml BSA, 0.02% Triton X-100 (vol/vol), and adjusted to 1 M NaCl, as indicated. Reactions were centrifuged in Microcon 100 filtering devices (Amicon) at 3,200 × g for 5 min. Retentates containing ClpA-RepA complexes were recovered with 50 μl of 10% SDS, and the radioactivity was measured. The radioactivity in the filtrates was also measured.
With these two conditions, the RepA recovered in the filtrate was active for oriP1 DNA binding.
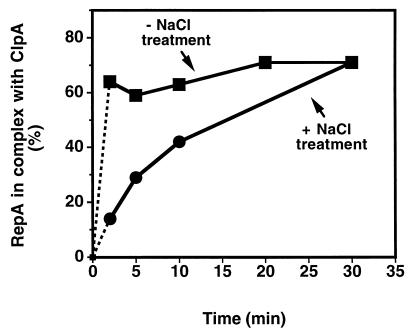
We then looked at the stability of the ClpA-RepA complexes. By varying reaction conditions, less stable ClpA-RepA complexes could be distinguished from very stable complexes. When we challenged complexes with a high concentration of salt, we found that only ClpA-RepA complexes formed in the presence of ATP[γ-S] at 23°C persisted in 1 M NaCl (Table 1). Stable ClpA-RepA complexes were also formed with adenylyl imidodiphosphate at 23°C (data not shown). In contrast, complexes formed at 0°C with or without ATP[γ-S] and at 23°C without ATP[γ-S], were dissociated by the NaCl treatment. Similar results were obtained when the complexes were challenged by diluting the reaction mixtures 30-fold (data not shown). Consistent with the dilution experiments, ClpA-RepA complexes that were stable to NaCl treatment formed slowly over a 30-min time course at 23°C (Fig. 4). In contrast, less stable complexes formed rapidly, reaching completion by 2 min, the earliest time point possible using this technique (Fig. 4).
Figure 4.
Stable ClpA-RepA complexes assemble slowly, although unstable ClpA-RepA complexes form rapidly. ClpA (76 pmol) and 9 pmol of [14C]RepA were incubated together in 50 μl of buffer A containing 100 μg/ml BSA, 0.02% Triton X-100 (vol/vol), and 100 μM ATP[γ-S] for 0 to 30 min at 23°C. Reactions were adjusted to 1 M NaCl (•) in a final volume of 100 μl of the above buffer or were adjusted to 100 μl in the above buffer without NaCl (▪). Reactions were centrifuged in Microcon 100 filtering devices, and the radioactivity in the filtrates and retentates was determined as in Table 1. In the absence of ClpA 8% of the [14C]RepA was retained on the filters and has been subtracted.
Thus, complexes formed in the presence of ATP[γ-S] at 23°C are distinguished from those formed at 0°C in the presence or absence of ATP[γ-S] or at 23°C in the absence of ATP[γ-S] in that (i) they are more stable, and (ii) they are committed† to activate RepA. The less stable uncommitted complexes could be converted into committed complexes by further incubating at 23°C with ATP[γ-S] (Fig. 3A). A time course of the second step of the dilution experiment, after dilution and ATP addition, showed that release of active RepA from committed complexes is relatively fast and complete in 5 min (Fig. 3B).
Stoichiometry of ClpA to RepA in Committed ClpA-RepA Complexes.
We measured the stoichiometry of RepA to ClpA in committed complexes by incubating increasing amounts of 14C-labeled RepA at 23°C with a fixed concentration of ClpA in the presence of ATP[γ-S]. The amount of RepA bound to ClpA was determined by measuring 14C retained on Microcon 100 filters and the amount of free RepA, by measuring 14C in the filtrate (Fig. 5). The results extrapolated to a molar ratio of 1.2 RepA dimers per ClpA hexamer. In a similar set of experiments using unlabeled RepA, the amount of free RepA and RepA in the ClpA-RepA complex was determined by the activation assay (Fig. 5). The results from these experiments also extrapolated to a molar ratio of about one RepA dimer per ClpA hexamer, suggesting close to equimolar binding of RepA dimers to ClpA hexamers in the committed complex.
Figure 5.
Determination of the molar ratio of ClpA to RepA in ClpA-RepA complexes. Reaction mixtures (100 μl) containing buffer A, 1 mM ATP[γ-S], 210 pmol of ClpA, and 7 to 280 pmol of [35S]RepA or unlabeled RepA were incubated for 10 min at 23°C. Reactions were then centrifuged in Microcon 100 filtering devices. In the experiments with [35S]RepA, radioactivity recovered in the retentates and filtrates was measured as in Table 1 (•). In the experiments with unlabeled RepA, RepA activity in the filtrates and retentates was assayed as described in Materials and Methods (▪).
Reversibility of the RepA Activation Reaction.
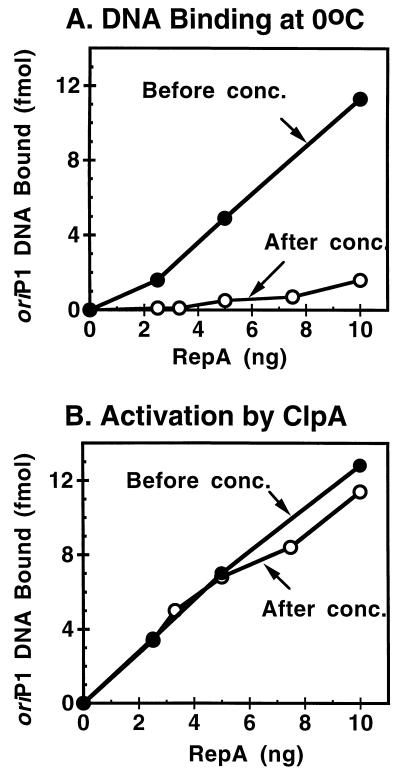
To show that activation of RepA by ClpA is simply a conversion of a dimer to a monomer and does not entail covalent modification of RepA, we demonstrated the reversibility of the reaction. First we activated RepA by incubating it with ClpA and ATP. Activated RepA was isolated by centrifuging the reaction mixture through a Microcon 100 filter. The filtrate contained activated RepA, as determined by its ability to bind oriP1 DNA at 0°C in the absence of ClpA (Fig. 6A) and to not be further stimulated after another incubation with ClpA and ATP (Fig. 6B). The filtrate was then concentrated about 100-fold by centrifugation through a Microcon 30 filter. The concentrated material was inactive in binding to oriP1 DNA because the monomerization was reversed (Fig. 6A). The RepA was not irreversibly inactivated, because it was activated again by incubation with ClpA and ATP (Fig. 6B). These results demonstrate that activation by ClpA is reversible and does not involve covalent modification of RepA.
Figure 6.
Reversibility of ClpA-activated RepA. RepA (50 pmol) and 180 pmol ClpA were incubated in 1.1 ml with 1 mM ATP in buffer A for 15 min at 23°C. The reaction mixture was centrifuged in Microcon 100 filtering devices at 3,200 × g for 8 min. One milliliter of the filtrate was then concentrated to 0.05 ml by centrifuging in a Microcon 30 filtering device at 14,000 × g for 8 min. Samples were assayed for activated RepA in A by measuring DNA binding at 0°C before (•) and after (○) concentration. In B they were assayed for total RepA by incubating them with ClpA and ATP and then measuring DNA binding as described in Materials and Methods before (•) and after (○) concentration.
DISCUSSION
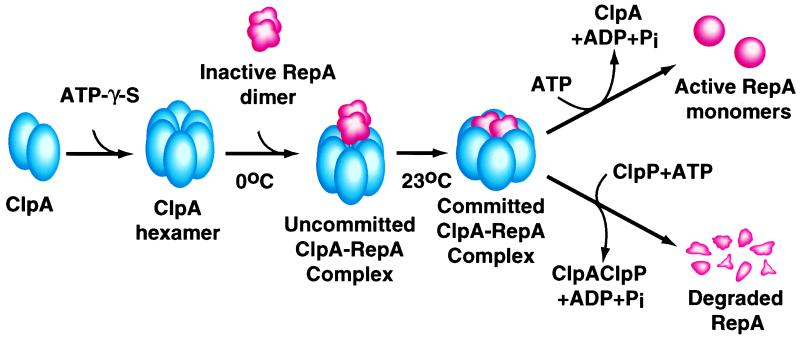
Fig. 7 summarizes the pathway of protein remodeling by ClpA implied by our results identifying several intermediates in the reaction. First, ClpA assembles into six-membered rings in the presence of ATP or nonhydrolyzable analogs (10). The ClpA hexamers interact with the substrate, in this case RepA dimers, forming ClpA-RepA complexes. These complexes form rapidly at 0°C. Because they can be dissociated readily into the original components, we refer to them as uncommitted complexes. A slow, temperature-dependent reaction converts the uncommitted ClpA-RepA complexes into very stable ClpA-RepA complexes, containing equimolar amounts of ClpA hexamers and RepA dimers. The molecular basis of this time- and temperature-dependent reaction is not known yet, but we imagine that in this slow reaction, RepA dimers are transformed from loose to tight complexes with ClpA. We refer to these complexes as committed complexes, because they are “committed,” or predetermined, to release active RepA monomers upon ATP hydrolysis. It is possible that on ATP hydrolysis ClpA undergoes a conformational change causing the dissociation of RepA dimers and the release of RepA monomers. Alternatively, ClpA may convert RepA dimers to monomers during the slow first reaction and then release the monomers upon ATP hydrolysis. It will be necessary to combine single turnover experiments with measurements of time-dependent conformational changes in the RepA substrate to decide between these alternatives. The RepA monomerization reaction carried out by ClpA is reversed by increasing the concentration of active monomers as is RepA activation by urea-treatment (12). Fig. 7 also shows our current model for the alternate fate for RepA, degradation by ClpAP protease (3). Although the intermediates in this reaction remain to be characterized, we have isolated complexes of ClpA, ClpP, and RepA that require ATP[γ-S] for formation (unpublished result).
Figure 7.
Mechanism of the ClpA chaperone activity and model for ClpAP protease activity. (See text for discussion.)
Our results have shown that all of the RepA released from committed ClpA-RepA complexes is in the active monomeric form. This observation leads to the interesting question of whether or not the ability of ClpA to fully remodel substrates in a single round of binding and release is a property of the ClpA chaperone. The folding process may be processive such that once a substrate is bound, it goes through multiple processive cycles of ATP-dependent protein remodeling, but is not released until the native form is reached. This mechanism would be consistent with the known processive degradation mechanism of the ClpAP protease, where it has been shown that protein substrates are cleaved at multiple sites without the release of intermediate degradation products (20). A processive mechanism of chaperone action is not generally observed with other ATP-dependent chaperones. One-cycle experiments have been performed with DnaK/DnaJ/GrpE and luciferase, and the observation was that only about 30% of the released luciferase was reactivated (21). Similarly, for many GroEL/ES substrates, the majority of the polypeptides generated after one round of substrate binding and release are in nonnative forms. This has been seen with rhodanese (22–24), ribulose bisphosphate carboxylase (25), dihydrofolate reductase (26), and mitochondrial malate dehydrogenase (27). However, with ornithine transcarbamylase (28), a large fraction of the molecules fold after a single round of release. It has been suggested that by binding and releasing substrates GroEL/GroES kinetically partitions nonnative polypeptides (22). Thus, with each round of binding and release some polypeptides reach their native form. Other nonnative forms have the opportunity to rebind GroEL, bind other chaperones, aggregate, or interact with proteolytic components.
We do not know whether ClpA remodels substrates other than RepA in one cycle of binding and release. RepA may be a rather unique substrate in that it is virtually always remodeled into an active conformation with few nonnative forms arising. It will be interesting to learn if RepA is activated by other chaperones, namely DnaJ/DnaK/GrpE, in one cycle or if other substrates are remodeled by ClpA in one cycle.
The specificity of both ClpA and ClpX for their respective chaperone substrates is paralleled by the specificity of ClpAP and ClpXP proteases for their degradation substrates. That is, ClpA but not ClpX activates RepA; and, ClpAP but not ClpXP degrades RepA (3). Similarly, ClpX dissociates MuA tetramers and disaggregates λ O protein, and ClpXP degrades λ O and MuA (4, 5, 15, 16). These observations imply that ClpA and ClpX may directly determine the fate of their respective protein substrates by either releasing native proteins or presenting nonnative proteins to the proteolytic component, ClpP. Very likely, the relative concentrations of ClpA/ClpX and ClpAP/ClpXP as well as the relative kinetic parameters for remodeling or degrading a substrate are also critical in determining the fate of a substrate.
In summary, we have explored the mechanism of the ClpA chaperone activity in protein remodeling by dissociating the reaction into stages. We found that ClpA forms unstable complexes with RepA that are converted to stable complexes in a time-, temperature-, and nucleotide-dependent fashion. Stable complexes are committed to release active RepA upon ATP hydrolysis. Understanding the mechanism of remodeling by ClpA should provide important clues about both the early steps of substrate presentation to ClpAP protease and the coupling mechanism of ClpA chaperone function and ClpAP protease function.
Acknowledgments
We thank Susan Gottesman, Michael Maurizi, Joel Hoskins, and Keith McKenney for helpful discussions and Joel Hoskins and Keith McKenney for reading the manuscript. We thank Joel Hoskins for preparing the labeled DNA.
ABBREVIATION
- ATP[γ-S]
adenosine-5′-O-(3-thiotriphosphate)
Footnotes
We have used the phrase “committed complex” to refer to ClpA-RepA complexes that are destined to release RepA in its active conformation upon ATP hydrolysis. The phrase “committed conformation” is used in the literature to refer to a conformation of a polypeptide that is released from a chaperone and is destined to refold to the native form (19).
References
- 1.Squires C, Squires C L. J Bacteriol. 1992;174:1081–1085. doi: 10.1128/jb.174.4.1081-1085.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parsell D A, Kowal A S, Singer M A, Lindquist S. Nature (London) 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 3.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi M R. Proc Natl Acad Sci USA. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M. EMBO J. 1995;14:1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levchenko I, Luo L, Baker T A. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 6.Vogel J L, Parsell D A, Lindquist S. Curr Biol. 1995;5:306–317. doi: 10.1016/s0960-9822(95)00061-3. [DOI] [PubMed] [Google Scholar]
- 7.Chernoff Y O, Lindquist S L, Ono B, Inge-Vechtomov S G, Liebman S W. Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 8.Chung C H. Science. 1993;262:372–373. doi: 10.1126/science.8211156. [DOI] [PubMed] [Google Scholar]
- 9.Gottesman S. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 10.Kessel M, Maurizi M R, Kim B, Kocsis E, Trus B L, Singh S K, Steven A C. J Mol Biol. 1995;250:587–594. doi: 10.1006/jmbi.1995.0400. [DOI] [PubMed] [Google Scholar]
- 11.Wickner S, Hoskins J, McKenney K. Nature (London) 1991;350:165–167. doi: 10.1038/350165a0. [DOI] [PubMed] [Google Scholar]
- 12.Wickner S, Hoskins J, McKenney K. Proc Natl Acad Sci USA. 1991;88:7903–7907. doi: 10.1073/pnas.88.18.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mhammedi-Alaoui A, Pato M, Gama M J, Toussaint A. Mol Microbiol. 1994;11:1109–1116. doi: 10.1111/j.1365-2958.1994.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 14.Kruklitis R, Welty D J, Nakai H. EMBO J. 1996;15:935–944. [PMC free article] [PubMed] [Google Scholar]
- 15.Gottesman S, Clark W P, de Crecy-Lagard V, Maurizi M R. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 16.Wojtkowiak D, Georgopoulos C, Zylicz M. J Biol Chem. 1993;268:22609–22617. [PubMed] [Google Scholar]
- 17.Wickner S H. Proc Natl Acad Sci USA. 1990;87:2690–2694. doi: 10.1073/pnas.87.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maurizi M R, Thompson M W, Singh S K, Kim S H. Methods Enzymol. 1994;344:314–331. doi: 10.1016/0076-6879(94)44025-5. [DOI] [PubMed] [Google Scholar]
- 19.Martin J, Langer T, Boteva R, Schramel A, Horwich A L, Hartl F U. Nature (London) 1991;352:36–42. doi: 10.1038/352036a0. [DOI] [PubMed] [Google Scholar]
- 20.Thompson M W, Singh S K, Maurizi M R. J Biol Chem. 1994;269:18209–15. [PubMed] [Google Scholar]
- 21.Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl F U. Proc Natl Acad Sci USA. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burston S G, Weissman J S, Farr G W, Fenton W A, Horwich A L. Nature (London) 1996;383:96–99. doi: 10.1038/383096a0. [DOI] [PubMed] [Google Scholar]
- 23.Smith K E, Fisher M T. J Biol Chem. 1995;270:21517–21523. doi: 10.1074/jbc.270.37.21517. [DOI] [PubMed] [Google Scholar]
- 24.Weissman J, Kashi Y, Fenton W, Horwich A. Cell. 1994;78:693–702. doi: 10.1016/0092-8674(94)90533-9. [DOI] [PubMed] [Google Scholar]
- 25.Todd M J, Viitanen P V, Lorimer G H. Science. 1994;265:659–666. doi: 10.1126/science.7913555. [DOI] [PubMed] [Google Scholar]
- 26.Mayhew M, da Silva A C R, Martin J, Erdjument-Bromage H, Tempst P, Hartl F U. Nature (London) 1996;379:420–426. doi: 10.1038/379420a0. [DOI] [PubMed] [Google Scholar]
- 27.Ranson N L, Dunster N J, Burston S G, Clarke A R. J Mol Biol. 1995;250:581–586. doi: 10.1006/jmbi.1995.0399. [DOI] [PubMed] [Google Scholar]
- 28.Weissman J S, Hohl C M, Kovalenko O, Kashi Y, Chen S, Braig K, Saibil H R, Fenton W A, Horwich A L. Cell. 1995;83:577–587. doi: 10.1016/0092-8674(95)90098-5. [DOI] [PubMed] [Google Scholar]