Abstract
Interactions among transcription factors that bind to separate sequence elements require bending of the intervening DNA and juxtaposition of interacting molecular surfaces in an appropriate orientation. Here, we examine the effects of single amino acid substitutions adjacent to the basic regions of Fos and Jun as well as changes in sequences flanking the AP-1 site on DNA bending. Substitution of charged amino acid residues at positions adjacent to the basic DNA-binding domains of Fos and Jun altered DNA bending. The change in DNA bending was directly proportional to the change in net charge for all heterodimeric combinations between these proteins. Fos and Jun induced distinct DNA bends at different binding sites. Exchange of a single base pair outside of the region contacted in the x-ray crystal structure altered DNA bending. Substitution of base pairs flanking the AP-1 site had converse effects on the opposite directions of DNA bending induced by homodimers and heterodimers. These results suggest that Fos and Jun induce DNA bending in part through electrostatic interactions between amino acid residues adjacent to the basic region and base pairs flanking the AP-1 site. DNA bending by Fos and Jun at inverted binding sites indicated that heterodimers bind to the AP-1 site in a preferred orientation. Mutation of a conserved arginine within the basic regions of Fos and transversion of the central C:G base pair in the AP-1 site to G:C had complementary effects on the orientation of heterodimer binding and DNA bending. The conformational variability of the Fos–Jun–AP-1 complex may contribute to its functional versatility at different promoters.
The regulation of transcription initiation in eukaryotic organisms relies on the concerted action of multiple transcription control proteins at the promoter. The regulatory information encoded in the linear array of control elements is interpreted through the assembly of a multiprotein transcription factor complex at the promoter (1–3). Assembly of this complex requires DNA bending as well as juxtaposition of complementary molecular surfaces in the correct orientation. DNA can be either actively bent by proteins that alter DNA structure, or it can passively adopt a bent conformation as a result of looping mediated by proteins that bind to separate recognition elements. Likewise, the orientation of protein binding can be determined by asymmetric recognition of the DNA-binding site or by interactions with other proteins at the promoter. Both active DNA bending and oriented protein binding can facilitate assembly of the transcription complex by promoting transcription factor interactions. Thus, identification of the molecular mechanisms of DNA bending and asymmetric DNA recognition are important for understanding the structural basis of transcription complex assembly.
Structural studies of the basic leucine zipper (bZIP) domain have revealed a simple and elegant mechanism for cooperative DNA recognition. The leucine zipper forms a coiled–coil dimerization interface and the basic regions form α helices that contact base pairs within the major groove of the DNA recognition site (4–12). DNA bending by Fos and Jun has been investigated using many different methods. Phasing analysis, a method that is based on the phase-dependent interaction between two closely spaced bends, first indicated that Fos–Jun heterodimers and Jun homodimers induce DNA bending and that the directions of DNA bending induced by the complexes were opposite (13, 14). However, x-ray crystallographic analysis of peptides encompassing the minimal basic DNA binding and leucine zipper dimerization domains of Fos and Jun bound to the AP-1 site detected little DNA bending (12). Whereas phase-sensitive detection and ligase catalyzed cyclization experiments failed to detect significant bending (15), subsequent analysis of the effects of probe geometry on phasing analysis and cyclization demonstrated that, using appropriate probes, DNA bending can be readily detected by both methods (16). DNA bending by Fos and Jun has also been visualized directly by atomic force microscopy (17). X-ray crystallographic analysis and solution studies of intrinsic DNA bending have also reached diametrically opposite conclusions (18–21). Crystal packing forces as well as agents used to promote crystallization can influence DNA bending (22–24). Thus, additional methods are required for analysis of the structural basis of protein-induced DNA bending.
MATERIALS AND METHODS
Plasmid Construction and Protein Purification.
Phasing analysis plasmids pNR412–26..-36, pNR421–28..-38, pNR414–26..36, pNR441–28..-38, and pNR502–26..-36 (where the suffix indicates the separation in base pairs between the centers of the AP-1 site and the intrinsic bend and .. indicates a series spaced by two base pairs), containing sites M, X, M-6T, X-6G, and W, respectively, were constructed by cloning the oligonucleotides shown in Fig. 2A between the XbaI and SalI sites of plasmids pTK401-26 and pTK401-28 (13). To generate plasmids with different separations between the centers of the AP-1 sites and the intrinsic DNA bends, oligonucleotides of different lengths containing an additional TGAC or TGACTGAC sequence inserted between the AP-1 site and the intrinsic bend were used. The plasmid vectors for expression of Fos and Jun deletion derivatives encompassing the leucine zipper dimerization and basic DNA binding domains were constructed by PCR amplification of the regions encoding residues 139–200 of Fos and 257–318 of Jun from plasmids encoding the full-length proteins, and insertion of these fragments into the same expression vectors with hexahistidine purification tags (13). Mutations in residues adjacent to the basic regions were constructed by insertion of oligonucleotides between unique restriction sites engineered into the coding regions. All of these proteins contained a Factor Xa cleavage site preceding the native coding sequence. All of the constructs were verified by sequencing of the cloned regions. The proteins were expressed in Escherichia coli as hexahistidine fusions and were purified by nickel chelate affinity chromatography to greater than 90% homogeneity as described (13).
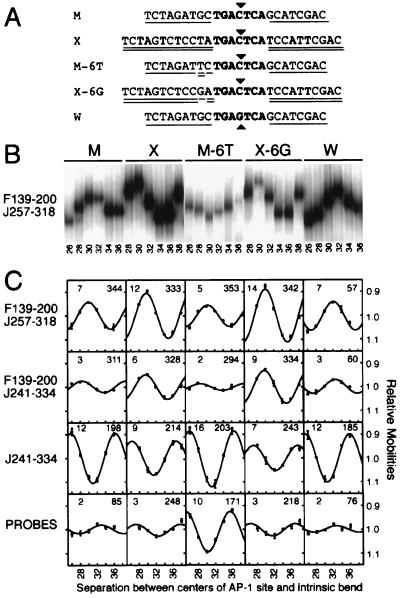
Figure 2.
Effects of sequences flanking the AP-1 site on DNA bending by the bZIP domains of Fos and Jun. (A) Sequences of AP-1 sites analyzed. Flanking sequences shared with the M site are single underlined whereas flanking sequences shared with the X site are double underlined. The core AP-1 recognition element is shown in boldface. A central C:G base pair is indicated by a triangle above the sequence whereas a central G:C base pair is indicated by a triangle below the sequence. (B) Phasing analysis of Fos–Jun bZIP domain heterodimers bound to different AP-1 sites. Fos139–200 and Jun257–318 were incubated with phasing analysis probes containing the sites shown above, and the complexes were analyzed by PAGE. The complexes were run on the same gel and the lanes were rearranged for a more concise figure. (C) The average mobilities of complexes formed by Fos139–200—Jun257–318, Fos139–200—Jun241–334, and Jun241–334 on different binding sites were normalized for differences in probe mobilities (shown at the bottom of the figure) and were plotted as a function of the separation between the centers of the AP-1 site and the intrinsic bend. The DNA bend angle and direction calculated based on the mobility variation are indicated in the upper left and upper right hand corners of each plot. Standard deviations of the mobilities in multiple independent experiments are plotted for each complex. Multivariate analysis of the variance in DNA bending for all of the complexes indicated that the differences in DNA bending were highly significant (probability of identity < 0.001) between all pairs of sites.
Phasing Analysis.
Probes for phasing analysis were prepared by PCR amplification of fragments between 349 and 366 bp in length containing the AP-1 site and the intrinsic bend at the center. Intrinsic DNA bend standards containing different numbers of phased A tracts at the middle or at the end of probes were prepared by NheI and BamHI cleavage of PCR products generated by using plasmids pJT170-2 through -11 (25) as templates.
Heterodimers were prepared by incubation of a 2-fold molar excess of Fos with Jun to ensure quantitative heterodimerization. Homo- and heterodimers were incubated with the probes under the conditions described previously (13). The complexes were analyzed by electrophoresis in 8% polyacrylamide (29 acrylamide/1 bisacrylamide) gels in 25 mM Tris, 195 mM glycine buffer at a field strength of 10 V/cm at 4°C for 22 h. To examine the effects of multivalent cations on DNA bending, 1 mM MgCl2, 1 mM spermidine trihydrochloride, or 0.1 mM Co(NH3)6Cl3 was added to both the gel mix and the running buffer.
Data Analysis.
The mobilities of the complexes and the free probes were measured either manually or by automatic band recognition of PhosphorImager data (Molecular Dynamics). The complex mobilities were normalized for differences in probe mobilities to an average relative mobility of 1. This normalization is based on the assumption that any pre-existing DNA structures outside of the reference DNA bend have the same effect on the mobilities of the DNA fragments alone and on the protein–DNA complexes. The relatively small mobility differences of most of the DNA fragments alone and the reciprocal effects of the exchange of flanking sequences on protein induced DNA bending regardless of its effects on DNA structure alone (i.e. compare M vs. M-6T and X vs. X-6G) indicate that the variation in probe mobilities does not affect interpretation of the data. The relative complex mobilities were highly reproducible with an average relative standard deviation of <1%. To calculate the DNA bend angles and directions, the best fit of the phasing function (14) to the relative mobilities was determined. The relative mobilities of all of the complexes yielded an excellent fit to this function (average r2 = 0.98).
The direction of protein-induced DNA bending was determined by comparison of the phase of the mobility variation for each complex with that of DNA fragments containing two intrinsic bends (16). A tracts were assumed to bend DNA toward the minor groove at a position 0.5 bp toward the 3′ end of the center of the A run (29). The DNA bend direction was defined at the center of the AP-1 site such that bending away from the leucine zipper (toward the minor groove) was assigned the value 0°, and positive bend angles correspond to clockwise rotation of the torsion angle when viewed from the side of the AP-1 site opposite to the reference bend (the left side in Fig. 2A). Thus, the DNA bend direction corresponds to the projection of the DNA helix axis on one side of the AP-1 site onto a plane perpendicular to the DNA helix axis on the other side of the AP-1 site (27).
The DNA bend angle was calculated from the amplitude of the phasing function. Because the intrinsic and protein-induced DNA bends were closely apposed, the overall shapes of probes containing in-phase and out-of-phase DNA bends were similar to those of probes containing single DNA bends with magnitudes that represent the sum and difference of the two bends (16). Thus, the protein-induced DNA bend angle was calculated by finding the angle which, when added to and subtracted from the angle of the reference DNA bend (three phased A tracts), resulted in DNA bend angles with a mobility difference predicted by the intrinsic bend calibration curve that was equivalent to the observed amplitude of the phasing function (28). The DNA bend angle was expressed in degrees based on the consensus estimate of 18° per A tract (29). This purely empirical approach is different from and, based on the better fit of the calibration function, more accurate than the approach used previously (14), which was based on the presumed dependence of electrophoretic mobility on the end-to-end distance of a DNA fragment (25).
The absolute DNA bend angles and directions were calculated relative to the estimated magnitude and direction of DNA bending by phased A tracts and are therefore subject to revision of these parameters. However, the relative DNA bend angles and directions induced by different complexes at the various binding sites are independent of this arbitrary standard. Protein-induced DNA bending is dynamic, and the DNA bend angles were calculated by comparison with intrinsic DNA standards that may differ in flexibility. The calculated DNA bend angles are therefore primarily intended for comparison of bending among related protein–DNA complexes.
RESULTS
Residues Adjacent to the Basic Region Bend DNA Through Charge Interactions.
To investigate the structural basis of DNA bending by the Fos and Jun bZIP domains, we have examined the effects of amino acid substitutions adjacent to the basic regions and changes in the base pairs flanking the AP-1 site. Our previous studies of DNA bending by different bZIP family proteins indicate that each protein induces a distinct DNA bend and that bending is correlated with the charge of amino acid residues on the amino terminal side of the basic region (30, 31). To test the hypothesis that the charge of these residues influences DNA bending, we analyzed DNA bending by heterodimers composed of Fos and Jun proteins containing single and double amino acid substitutions at these positions (Fig. 1). Substitution of two basic residues (RR) adjacent to the bZIP domain of Fos by neutral and acidic (AE) residues from Jun resulted in a 2-fold reduction in DNA bending. Conversely, substitution of the residues adjacent to the bZIP domain of Jun by residues from Fos resulted in an increase in DNA bending. Reciprocal exchange of these residues between Fos and Jun yielded a heterodimer that induced a DNA bend angle identical to that induced by the wild-type proteins. Single amino acid substitutions caused independent changes in DNA bending whose sum was equal to the effect of the double substitution. Substitution of a charged residue by a neutral residue had half of the effect of substitution of a charged residue by a residue of opposite charge. Therefore, alterations of the charge of this region caused proportionate changes in DNA bending.
Figure 1.
Charged residues on the amino-terminal side of the basic regions of Fos and Jun induce DNA bending. (A) Alignment of the basic regions of Fos and Jun. Residues substituted in the mutant proteins are indicated in boldface above and below the alignment. (B) Phasing analysis of selected combinations of wild-type and mutant proteins. Proteins containing the amino acid residues indicated above the lanes at positions adjacent to their basic regions were incubated with phasing analysis probes containing the M site described in Fig. 2, and the complexes were analyzed by PAGE. Each set of lanes contained probes in which the separations between the centers of the AP-1 site and the intrinsic DNA bend were 26, 28, 30, 32, 34, and 36 bp, respectively. All of the complexes were run on the same gel, but the lanes were rearranged for a more concise figure. The average mobilities of complexes from multiple experiments were normalized for differences in probe mobilities and plotted with standard deviations as a function of the separation between the centers of the AP-1 site and the intrinsic bend. (C) The relative DNA bend angles induced by all combinations between the mutant and wild-type proteins were plotted as a function of the net charge of amino acid residues adjacent to the basic regions. The charged amino acids were assumed to be fully ionized with formal charges of +1 and −1. In the vicinity of the DNA helix, these formal charges are likely to be modified by the local microenvironment (32). It is therefore possible that the absolute charges are shifted to more positive values. However, the relative charges of the various complexes are likely to remain the same. The standard bend induced by the wild-type proteins is encircled. Vertical bars indicate standard deviations or ranges (two cases) of the bend angles calculated based on independent experiments. A heterodimer containing four arginine residues at the positions examined did not bind to the AP-1 site, possibly because of the high local charge predicted for such a complex.
Analysis of the DNA bends induced by all combinations between the wild-type and mutant proteins revealed a striking correlation between the DNA bend angle and the net charge of these residues (Fig. 1C). Therefore, the effects of the charged residues on DNA bending were virtually identical at each of the positions examined in both Fos and Jun. Consistent with the role of charged residues in DNA bending by Fos and Jun, electrophoresis in the presence of multivalent cations reduced the mobility variation of all Fos and Jun complexes examined, but had no effect on the relative mobilities of intrinsic DNA bend standards (27). These results suggest that charge interactions between residues adjacent to the basic region and the phosphodiester backbone are a major determinant of DNA bending by the Fos and Jun bZIP domains.
The residues examined to date do not account for the entire DNA bend induced by these proteins. Heterodimers in which the net formal charge of these residues was 0 induced a significant bend, and heterodimers in which the net charge of these residues was negative induced a small bend in the same direction as heterodimers with a net positive charge. Thus, additional residues either within or adjacent to the bZIP domains are likely to influence DNA bending. A negatively charged region located further from the bZIP domain of Jun contributes to the opposite direction of DNA bending induced by homodimers formed by larger Jun proteins (e.g., J241–334) and counteracts bending by Fos in the context of heterodimers (Fig. 2). In addition to these regions, the transcription activation domains of Fos and Jun also induce DNA bending (13, 14, 27). These domains contain a high density of charged amino acid residues. However, other charged regions in the proteins have no effect on DNA bending. Thus, DNA bending by charged amino acid residues depends on the structural context of those residues. In the case of the residues adjacent to the bZIP domain, this structural context is most likely provided by their proximity to the α-helical DNA contact interface.
Sequences Flanking the AP-1 Site Have Converse Effects on DNA Bending in Opposite Directions.
Fos and Jun can bind to AP-1 sites in many different genes and mediate responses to distinct extracellular signals at these sites. To investigate possible differences in the structure of the complex at these sites, we examined DNA bending by the Fos and Jun bZIP domains at sites that differ in the sequences flanking the consensus AP-1 element (Fig. 2). We initially compared bending at an AP-1 site containing symmetrical flanking sequences (M) with bending at the site used for x-ray crystallographic analysis of the complex formed by the Fos and Jun bZIP domains (X). Truncated Fos–Jun heterodimers induced up to 2-fold more bending at the X site than at the M site. In contrast, truncated Jun homodimers induced a larger bend at the M site than at the X site. Complexes formed by other Fos and Jun proteins also induced distinct bends at these sites (28). Complexes that bent DNA in the same direction as heterodimers induced a larger bend at the X site whereas complexes that bent DNA in the same direction as homodimers induced a larger bend at the M site. Thus, sequences flanking the AP-1 site influence DNA bending by Fos and Jun, and complexes that bend DNA in opposite directions display different sequence preferences for DNA bending.
The sequences of the M and X sites differ at multiple positions outside of the conserved heptanucleotide core. To determine if base pairs outside of the region contacted in the x-ray crystal structure affected DNA bending, we exchanged a single base pair at position −6 between the M and X sites. Surprisingly, these substitutions resulted in DNA bending properties that were even more divergent than those observed at the M and X sites. DNA bends induced by truncated Fos–Jun heterodimers were up to 4-fold larger at the X-6G site than at the M-6T site. Conversely, truncated Jun homodimers induced a larger bend at the M-6T site than at the X-6G site. The difference between the DNA bends induced at the M-6T and X-6G sites was larger than the difference between the bends induced at the M and X sites for all Fos and Jun complexes examined. Thus, the exchange of a single base pair outside of the region contacted in the x-ray crystal structure affects DNA bending by Fos and Jun.
The single base pair substitution at the M-6T site resulted in an intrinsic DNA bend adjacent to the AP-1 site. The contribution of this intrinsic bend to the mobility variation of the protein–DNA complexes was taken into account by normalization to the mobilities of the free probes. Thus, the differences in the relative mobilities of Fos and Jun complexes at various binding sites reflect distinct protein induced DNA bends rather than pre-existing differences in DNA structure. Intriguingly, the difference in DNA bending between the M and M-6T sites was similar in magnitude, but opposite in direction to the difference in DNA bending between the X and X-6G sites for all complexes examined. Thus, the substitution of a T:A for a G:C base pair at the −6 position has similar effects on protein-induced DNA bending in the context of the flanking sequences at both the M and X-6G sites.
Fos–Jun Heterodimers Bind the AP-1 Site in a Preferred Orientation.
The directions of DNA bending induced by the Fos and Jun bZIP domains at AP-1 sites containing the core heptanucleotide binding motif in the same orientation were relatively similar. However, inversion of the asymmetric core recognition element by substitution of the central C:G base pair in the M site by a G:C base pair in the W site resulted in a shift in the direction of DNA bending induced by heterodimers (Fig. 2). To investigate if this shift in DNA bend direction was due to a switch in the preferred orientation of heterodimer binding to the inverted site, we examined DNA bending by heterodimers in which the arginine that contacts the central guanine (12) was substituted by an isoleucine in either Fos or Jun (33) (Fig. 3). Heterodimers in which the arginine in Jun was mutated induced bending in the same direction as the wild-type proteins whereas heterodimers in which the arginine in Fos was mutated induced bending in the opposite direction. Significantly, heterodimers containing a mutation in Fos bend the M site in the same direction as heterodimers containing a mutation in Jun bend the W site and vice versa. The reciprocal effects of inverting the central asymmetric base pair and mutating an amino acid residue in one subunit of the asymmetric dimer strongly support the hypothesis that the heterodimer binds to the site in a preferred orientation. The effect of reversing the orientation of heterodimer binding on the direction of DNA bending by the bZIP domains is also consistent with the distinct directions of DNA bending induced by transcription activation domains fused to the amino-terminal ends of the Fos and Jun bZIP domains (28).
Figure 3.
Fos–Jun heterodimers bind the AP-1 site in a preferred orientation. (A) Diagram of the effect of the orientation of Fos (black)–Jun (gray) heterodimer binding on the direction of DNA bending. The central C:G base pair is shown in cross-section along with the conserved arginine of Fos, which contacts the guanine. (B) The heterodimers indicated on the left consisting of the wild-type (WT) Fos118–211 and Jun225–334 proteins or mutant proteins containing a substitution of the conserved arginine in Jun (R273I) or Fos (R155I) were incubated with the probes indicated above the lanes and described in Fig. 2A containing a central C:G (M) or G:C (W) base pair. The complexes were analyzed on an 8% polyacrylamide gel. The relative mobilities of the complexes were plotted as a function of the separation between the centers of the AP-1 site and the intrinsic bend.
To confirm that the difference between the directions of DNA bending induced by heterodimers containing a mutation in the conserved arginine of Fos versus Jun was due to opposite orientations of heterodimer binding to the AP-1 site, we examined bending at the cAMP response element (CRE) site, which contains a symmetric core recognition element. There was no detectable effect of substitution of the arginine in Fos or Jun on bending at the CRE site, indicating that these mutations have no effect on DNA bending apart from determining the orientation of heterodimer binding. Thus, there is a perfect agreement between the effects of mutation of the conserved arginine in Fos or Jun and reversal of the orientation of the core AP-1 recognition sequence on DNA bending. The wild-type proteins induce DNA bending in the same direction as the heterodimer containing a mutation in the conserved arginine of Jun, indicating that they have the same preferred orientation of binding to the AP-1 site. However, the apparent DNA bend angle induced by the wild-type proteins is smaller, suggesting that their orientation preference is not as strong as that of heterodimers containing mutant proteins. Consequently, the wild-type Fos and Jun bZIP domains bind to the AP-1 site in a preferred orientation, and this orientation preference is determined in part by a contact between a conserved arginine in Fos and the central guanine in the AP-1 site.
DISCUSSION
The concordant effects of amino acid substitutions adjacent to the basic region and changes in base pairs flanking the AP-1 site manifest a role for interactions between these regions in DNA bending by Fos and Jun (Fig. 4). Exchange of single amino acid residues adjacent to the basic regions of Fos and Jun affected DNA bending in direct proportion to the change in net charge. Correspondingly, single base pair substitutions at positions flanking the AP-1 site had opposite effects on bending by homo- and heterodimers. The contrasting effects of amino acid residues of opposite charge and the converse influence of base pair changes on bending in opposite directions indicate that these groups influence DNA bending directly. The direct relationship between net charge and DNA bend angle together with the influence of nearby base pairs on DNA bending suggest that Fos and Jun induce DNA bending through electrostatic interactions between residues adjacent to the basic regions and base pairs flanking the AP-1 site.
Figure 4.
Amino acid residues and base pairs that influence DNA bending by Fos and Jun. The positions of the amino acid residues that induce DNA bending by Fos (red) and Jun (blue) are shown as dot surfaces based on the x-ray crystal structure. The α-helix of Jun was extended by three residues to indicate the approximate positions of residues for which no density was detected in the x-ray crystallographic analysis. Sequences flanking the AP-1 site and the single base pair at position −6 that influence the DNA bend angle are shown in orange and purple respectively. The charged phosphate oxygens are shown as green dot surfaces. The charge interactions predicted to induce DNA bending are shown in white, with an arrow indicating the direction of DNA bending. The conserved arginine in Fos and the central base pair that influence the orientation of heterodimer binding are shown in red.
Truncated Fos and Jun induce distinct DNA bends at AP-1 sites with different sequences surrounding the core recognition element. Flanking sequences that promote DNA bending in one direction, impede bending in the opposite direction. It is therefore likely that these differences in DNA bending are not caused by altered contacts with nucleotide bases, but are due to sequence dependent differences in DNA bendability. Moreover, because changes in flanking sequences had converse effects on bending DNA in opposite directions, the bendability of these sequences is anisotropic. The extent of DNA bending by the catabolite gene activator protein (CAP) also depends on sequences flanking the core recognition element (34). Furthermore, dinucleotides that promote DNA bending at positions where the minor groove faces the CAP protein impede bending at positions half a helical turn away where the major groove faces CAP. Analysis of sequence periodicities in nucleosomal DNA has revealed similar differences in the preferred direction of DNA bending by different dinucleotides (35). Thus, the sequence dependence of DNA bending by electrostatic interactions is determined by the anisotropic bendability of different DNA sequences, and is independent of the structure of the DNA bending protein.
The effects of single amino acid and base pair substitutions on the electrophoretic mobilities of Fos and Jun complexes validate phasing analysis and argue against alternative interpretations of the phase dependent mobility variation (15). Although changes in charged amino acid residues could alter protein conformation, there is no significant effect of these substitutions on the average complex mobilities whereas there is an up to 2-fold difference in the phase-dependent mobility variation. Likewise, the sequence of the binding site could induce different conformations on proteins that bind to different recognition elements. However, there is no evidence for such conformational differences in the average complex mobilities. Nor is there any reason why such conformational changes would have converse effects on complexes with opposite phase-dependent electrophoretic mobility variations. Both amino acid substitutions adjacent to the basic region as well as changes in the sequences flanking the AP-1 site had moderate effects on the binding affinities of the complexes. However, there was no overall correlation between DNA binding affinity and DNA bending. Furthermore, although the effects on DNA bending were observed regardless of dimerization partner or binding site, the effects on binding affinity were observed only for particular dimer combinations at selected binding sites. Thus, differences in binding affinity within the range observed in these experiments do not influence quantitation of DNA bending by phasing analysis. Consequently, when appropriate probes are used (16), phasing analysis is a reliable method for studies of DNA bend direction and magnitude.
Although neither the residues adjacent to the basic region nor the base pairs flanking the AP-1 site were reported to participate in intermolecular contacts based on the x-ray crystal structure, they are in close proximity and can be brought into direct contact by DNA bending (Fig. 4). The lack of DNA bending in the x-ray crystal structure may be due to shielding of the charge interactions by the high concentrations of salts used during crystallization. This possibility is consistent with the reduction in DNA bending by Fos and Jun in phasing analysis performed in the presence of multivalent cations (27). In addition, stacking of the oligonucleotides in the crystal is likely to restrict the conformational flexibility of the binding sites. The converse effects of base substitutions on DNA bending in opposite directions suggest that DNA bending by Fos and Jun depends on the bendability of sequences flanking the AP-1 site. Thus, constraints imposed on the flexibility of the flanking sequences are likely to reduce or prevent DNA bending.
The effects of charged amino acid residues and sequences flanking the AP-1 site on DNA bending by Fos and Jun are consistent with both phosphate neutralization (36, 37) as well as electrostatic force (14, 27) models for DNA bending. These models represent alternative molecular mechanisms for DNA bending caused by asymmetric charge distribution as originally proposed by Mirzabekov and Rich (38). The mechanisms are not mutually exclusive and may enhance each other as electrostatic attraction brings the phosphate backbone to a distance where neutralization can occur. We favor a model in which the charged amino acid residues impose an electrostatic force on the phosphodiester backbone that causes DNA bending. This model is consistent with the linear relationship between overall charge and DNA bend angle since the bending force is predicted to be approximately proportional to net charge and the extent of DNA bending is predicted to be a linear function of the bending force for small DNA bend angles. The phosphate neutralization model is more difficult to reconcile with the effects of negatively charged residues, which would presumably have to affect bending through an indirect mechanism.
The role of electrostatic interactions in DNA bending by Fos and Jun raises the question whether such interactions may be involved in bending by other DNA binding proteins. The close correlation between the direction of DNA bending and the charge of residues adjacent to the basic region among a wide variety of bZIP family proteins (30) suggests that this mechanism is general among members of this family. Shielding of electrostatic interactions under conditions of crystallization may also explain in part the differences between results from studies of DNA bending by other proteins using x-ray crystallography and phasing analysis (39–44). However, such shielding is not likely to eliminate DNA bending under physiological conditions since electrophoresis in the presence of multivalent cations causes only a modest reduction in DNA bending by Fos and Jun (27). Electrostatic interactions may also contribute to bending by DNA binding proteins that do bend DNA in crystals. Several basic side chains contact the phosphodiester backbone at positions flanking the core binding sites in both the CAP–DNA and serum response factor–DNA crystal structures (45, 46) and may contribute to bending DNA toward the convex protein surface by these proteins. Conversely, the negatively charged transcription activation domains of Fos and Jun induce DNA bending in a direction diametrically opposite from the positions of the domains in phasing analysis assays (27). Therefore, electrostatic interactions may be a general mechanism of DNA bending by many structural classes of DNA binding proteins.
The reciprocal effects of mutation of the conserved arginine and inversion of the core AP-1 site on DNA bending indicate a direct relationship between the orientation of heterodimer binding and the direction of DNA bending. Thus, the shift in the direction of DNA bending induced by wild-type proteins upon inversion of the binding site indicates that Fos-Jun heterodimers bind to the AP-1 site in a preferred orientation. This orientation preference is also consistent with the distinct directions of DNA bending induced by transcription activation domains fused to the amino terminal ends of the basic regions of Fos and Jun (28). The absence of a detectable orientation preference in the x-ray crystallography and peptide affinity cleavage studies (12, 47) may be due to differences in the proteins or the sequences of the AP-1 sites used in the various studies. Alternatively, crystal packing forces or contacts with the DNA cleavage reagent may influence the binding orientation. At sites with symmetric flanking sequences (M and W), the orientation preference is principally due to contacts with the central base pair. Interactions with asymmetric flanking sequences may modify the orientation preference. Consequently, the sequence of the AP-1 site has a profound impact on the structure of the Fos–Jun–AP-1 complex, which may contribute to the functional versatility of the heterodimer in the context of different transcription factor complexes.
Acknowledgments
We thank Lily Ng and Tom Curran for expression vectors encoding the Fos118–211 R155I and Jun225–318 R273I proteins (33), David Paolella and Alanna Schepartz for phasing analysis plasmids containing the cAMP response element site (31), and Ronald Diebold, Jack Dixon, David Engelke, Neelam Taneja, and Dennis Thiele for critical comments on the manuscript.
ABBREVIATION
- bZIP
basic leucine zipper
References
- 1.Robertson L M, Kerppola T K, Vendrell M, Luk D, Smeyne R J, Bocchiaro C, Morgan J I, Curran T. Neuron. 1995;14:241–252. doi: 10.1016/0896-6273(95)90282-1. [DOI] [PubMed] [Google Scholar]
- 2.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 3.Thanos D, Maniatis T. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 4.O’Neil K T, Hoess R H, DeGrado W F. Science. 1990;249:774–778. doi: 10.1126/science.2389143. [DOI] [PubMed] [Google Scholar]
- 5.Talanian R V, McKnight C J, Kim P S. Science. 1990;249:769–771. doi: 10.1126/science.2389142. [DOI] [PubMed] [Google Scholar]
- 6.Patel L, Abate C, Curran T. Nature (London) 1990;347:572–574. doi: 10.1038/347572a0. [DOI] [PubMed] [Google Scholar]
- 7.Weiss M A, Ellenberger T E, Wobbe C R, Lee J P, Harrison S C, Struhl K. Nature (London) 1990;347:575–578. doi: 10.1038/347575a0. [DOI] [PubMed] [Google Scholar]
- 8.Oakley M G, Dervan P B. Science. 1990;248:847–850. doi: 10.1126/science.2111578. [DOI] [PubMed] [Google Scholar]
- 9.O’Shea E K, Klemm J D, Kim P S, Alber T. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 10.Ellenberger T E, Brandl C J, Struhl K, Harrison S C. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 11.Konig P, Richmond T J. J Mol Biol. 1993;233:139–154. doi: 10.1006/jmbi.1993.1490. [DOI] [PubMed] [Google Scholar]
- 12.Glover J N, Harrison S C. Nature (London) 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 13.Kerppola T K, Curran T. Cell. 1991;66:317–326. doi: 10.1016/0092-8674(91)90621-5. [DOI] [PubMed] [Google Scholar]
- 14.Kerppola T K, Curran T. Science. 1991;254:1210–1214. doi: 10.1126/science.1957173. [DOI] [PubMed] [Google Scholar]
- 15.Sitlani A, Crothers D M. Proc Natl Acad of Sci USA. 1996;93:3248–3252. doi: 10.1073/pnas.93.8.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerppola T K. Proc Natl Acad Sci USA. 1996;93:10117–10122. doi: 10.1073/pnas.93.19.10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker J C, Nikroo A, Brabletz T, Reisfeld R A. Proc Natl Acad Sci USA. 1995;92:9727–31. doi: 10.1073/pnas.92.21.9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crothers D M, Haran T E, Nadeau J G. J Biol Chem. 1990;265:7093–7096. [PubMed] [Google Scholar]
- 19.Goodsell D S, Kaczor G M, Dickerson R E. J Mol Biol. 1994;239:79–96. doi: 10.1006/jmbi.1994.1352. [DOI] [PubMed] [Google Scholar]
- 20.Dickerson R E, Goodsell D S, Neidle S. Proc Natl Acad Sci USA. 1994;91:3579–3583. doi: 10.1073/pnas.91.9.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haran T E, Kahn J D, Crothers D M. J Mol Biol. 1994;244:135–143. doi: 10.1006/jmbi.1994.1713. [DOI] [PubMed] [Google Scholar]
- 22.DiGabriele A D, Sanderson M R, Steitz T A. Proc Natl Acad Sci USA. 1989;86:1816–1820. doi: 10.1073/pnas.86.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DiGabriele A D, Steitz T A. J Mol Biol. 1993;231:1024–1039. doi: 10.1006/jmbi.1993.1349. [DOI] [PubMed] [Google Scholar]
- 24.Sprous D, Zacharias W, Wood Z A, Harvey S C. Nucleic Acids Res. 1995;23:1816–1821. doi: 10.1093/nar/23.10.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson J F, Landy A. Nucleic Acids Res. 1988;16:9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zinkel S S, Crothers D M. Nature (London) 1987;328:178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]
- 27.Kerppola T K, Curran T. EMBO J. 1997;16:2907–2916. doi: 10.1093/emboj/16.10.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajaram N, Kerppola T K. EMBO J. 1997;16:2917–2926. doi: 10.1093/emboj/16.10.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crothers D M, Drak J. Methods Enzymol. 1992;212:46–71. doi: 10.1016/0076-6879(92)12005-b. [DOI] [PubMed] [Google Scholar]
- 30.Kerppola T K, Curran T. Mol Cell Biol. 1993;13:5479–5489. doi: 10.1128/mcb.13.9.5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paolella D N, Palmer C R, Schepartz A. Science. 1994;264:1130–1133. doi: 10.1126/science.8178171. [DOI] [PubMed] [Google Scholar]
- 32.Lamm G, Pack G R. Proc Natl Acad Sci USA. 1990;87:9033–9036. doi: 10.1073/pnas.87.22.9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng L, Forrest D, Curran T. Nucleic Acids Res. 1993;21:5831–5837. doi: 10.1093/nar/21.25.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gartenberg M R, Crothers D M. Nature (London) 1988;333:824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- 35.Satchwell S C, Travers A A. EMBO J. 1989;8:229–238. doi: 10.1002/j.1460-2075.1989.tb03368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strauss J K, Maher L J., III Science. 1994;266:1829–1834. doi: 10.1126/science.7997878. [DOI] [PubMed] [Google Scholar]
- 37.Strauss J K, Roberts C, Nelson M G, Switzer C, Maher L J., III Proc Natl Acad Sci USA. 1996;93:9515–9520. doi: 10.1073/pnas.93.18.9515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mirzabekov A D, Rich A. Proc Natl Acad Sci USA. 1979;76:1118–1121. doi: 10.1073/pnas.76.3.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wechsler D S, Dang C V. Proc Natl Acad Sci USA. 1992;89:7635–7639. doi: 10.1073/pnas.89.16.7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fisher D E, Parent L A, Sharp P A. Proc Natl Acad Sci USA. 1992;89:11779–11783. doi: 10.1073/pnas.89.24.11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferre-D’Amare A, Prendergast G C, Ziff E B, Burley S K. Nature (London) 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 42.Ferre-D’Amare A R, Pognonec P, Roeder R G, Burley S K. EMBO J. 1994;13:180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Natesan S, Gilman M Z. Genes Dev. 1993;7:2497–2509. doi: 10.1101/gad.7.12b.2497. [DOI] [PubMed] [Google Scholar]
- 44.Houbaviy H B, Usheva A, Shenk T, Burley S K. Proc Natl Acad Sci USA. 1996;93:13577–13582. doi: 10.1073/pnas.93.24.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schultz S C, Shields G C, Steitz T A. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 46.Pellegrini L, Tan S, Richmond T J. Nature (London) 1995;376:490–498. doi: 10.1038/376490a0. [DOI] [PubMed] [Google Scholar]
- 47.Chen L, Oakley M G, Glover J N M, Jain J, Dervan P, Hogan P, Rao A, Verdiner G. Curr Biol. 1995;5:882–889. doi: 10.1016/s0960-9822(95)00178-3. [DOI] [PubMed] [Google Scholar]