Abstract
An RNA transcribed from the antisense strand of the FGF-2 gene has been implicated in the regulation of FGF-2 mRNA stability in amphibian oocytes. We have now cloned and characterized a novel 1.1-kb mRNA (fgf-as) from neonatal rat liver. In non-central nervous system (CNS) tissues the fgf-as RNA is abundantly expressed in a developmentally regulated manner. The FGF-AS cDNA contains a consensus polyadenylylation signal and a long open reading frame (ORF) whose deduced amino acid sequence predicts a 35-kDa protein with homology to the MutT family of nucleotide hydrolases. Western blot analysis with antibodies against the deduced peptide sequence demonstrates that the FGF-AS protein is expressed in a broad range of non-CNS tissue in the postnatal period. In the developing brain, the abundance of sense and antisense transcripts are inversely related, suggesting a role for the antisense RNA in posttranscriptional regulation of FGF-2 expression in this tissue.The FGF-AS is complementary to two widely separated regions in the long 3′ untranslated region of the FGF-2 mRNA, in the vicinity of the proximal and distal polyadenylylation sites. These findings demonstrate that the FGF-2 and fgf-as RNAs are coordinately transcribed on a tissue-specific and developmentally regulated basis and suggest that interaction of the sense and antisense RNAs may result in posttranscriptional regulation of FGF-2 in some tissues.
Basic fibroblast growth factor (bFGF, FGF-2), a member of the heparin-binding growth factor family of mitogens, has been implicated in a range of normal physiological processes from embryonic mesoderm induction (1) and pattern formation (2) to angiogenesis and wound repair (3, 4). Inappropriate expression of FGF-2 mRNA is a hallmark of many types of solid tumors (5), and has also been implicated in a variety of vascular proliferative disorders including diabetic microangiopathies (6). Expression of FGF-2 is believed to be tightly regulated (7), and FGF-2 mRNA is present at extremely low levels postnatally in most normal tissues outside the central nervous system (CNS) (8, 9). However, the transcriptional and posttranscriptional controls governing the expression of FGF-2 are still largely unknown.
In Xenopus laevis oocytes, a 1.5-kb FGF-2 antisense RNA complementary to the third exon and 3′ untranslated region (3′ UTR) of the FGF-2 mRNA, has been implicated in the regulation of FGF-2 mRNA editing and stability (10). The antisense RNA also contains a long open reading frame (ORF) encoding a hypothetical 24-kDa protein of unknown function. Strand-specific cRNA probes also detect FGF-2 antisense expression in chicken (11, 12), rat (13), and human (14) tissues, and circumstantial evidence supports the notion that the antisense RNA may regulate FGF-2 expression in these species (reviewed in ref. 15). In the present study, to elucidate the relationship between the FGF-2 sense and antisense RNAs in higher vertebrates, we have cloned and characterized a cDNA from neonatal rat liver corresponding to the full-length rat FGF-2 antisense mRNA, and identified its translation product in Western blots of rat tissue extracts.
MATERIALS AND METHODS
cDNA Cloning and Sequencing.
Liver tissue for cDNA library construction was obtained from neonatal Sprague-Dawley rats and poly(A)+ RNA isolated by oligo(dT) cellulose chromatography (16). Approximately 5 μg of poly(A)+ RNA was Moloney murine leukemia virus (MMLV) reversed transcribed, size fractionated, and directionally cloned into the λZAP vector (Stratagene). Approximately 1 × 106 recombinants were screened by conventional filter hybridization with a human FGF-2 antisense cDNA fragment (17). The resulting positive clones were sequenced using sequenase 2.0 (United States Biochemical). Homology searches of the nucleotide database (Human Genome Center, Baylor College of Medicine, Houston, TX) were performed using the blast program (18). The complete sequence for the FGF-2 antisense cDNA clone was submitted to GenBank and has been assigned the accession number U58289U58289.
Northern and Reverse Transcription–PCR (RT-PCR) Analysis.
Brain, heart, kidney, and liver tissue were obtained from day 17 embryonic, day 11 neonate, and adult Sprague-Dawley rats. Poly(A)+ RNA was electrophoresed on formaldehyde-containing agarose gels and transferred to Hybond-N nylon membranes (Amersham) as previously described (13). Antisense strand-specific 32P-labeled cRNA probes were generated by in vitro transcription as previously described (13). Hybridization was carried out overnight at 60°C in 50% formamide, followed by sequential washes at increasing stringency (2 × 15 min at 60°C in 2 × SSC/0.1% SDS, followed by 2 × 15 min at 65°C, and finally 2 × 15 min in 0.1 × SSC at 65°C). Blots were exposed to x-ray film (DuPont/NEN reflection) for 5 days at −70°C.
Semi-quantitative RT-PCR amplification of target mRNAs was performed essentially as described previously (13, 17, 19) with minor modifications. One microgram of total RNA from embryonic, neonatal, and adult rat tissues was reverse transcribed at 42°C for 60 min with MMLV reverse transcriptase (Promega) in a 25-μl reaction as previously described (19). All primer pairs were chosen to span at least one intron–exon splice boundary to eliminate the possibility of amplification of genomic DNA. Primers for PCR amplification of FGF-2 (19) and glyceraldehyde 3-phosphate dehydrogenase (13) were as previously reported, and generated RT-PCR products of 352 and 242 bp, respectively. Primers for amplification of the FGF-2 antisense product were RG-1 (ACTGCCTCGAGCGGCCTGGAGATCA) corresponding to nucleotides 115–139 of the rat FGF-AS cDNA sequence, and RG-2 (CTGGTGCTAACATCAAATACGGCA), which is complementary to nucleotides 468–491. The predicted FGF-AS amplification product with these primers is 379 bp. One microliter of the RT reaction was added to the standard PCR mixture containing 10 × buffer, 100 μM dNTPs, 1.5 mM MgCl2, 50 pmol of each primer, and 0.5 units of Taq DNA polymerase (Promega) in a final volume of 25 μl. PCR thermal cycling was performed exactly as previously described (17), except that the final 7-min elongation reaction was eliminated. Preliminary experiments for all products confirmed that under these conditions all reactions remained within the exponential phase (data not shown).
Preparation of Antisera.
Antisera were raised against synthetic peptides based on the deduced amino acid sequence of the rat FGF-AS cDNA. All peptides were synthesized by Research Genetics (Huntsville, AL). The COOH-terminal antigen was a KLH-coupled peptide containing the sequence KLYHRGLPERYKAEMGTD, corresponding to amino acids 296–313 of the FGF-AS protein. The NH2-terminal antigen was a multiple antigen peptide containing eight copies of the sequence RARRRTASSGLEITGS (amino acids 23–38) on a branched lysine core. Antisera were prepared as described previously (20).
In Vitro Translation, Immunoprecipitation, and Western Blot Analysis.
A coupled transcription/translation system (TnT, Promega) was used for in vitro translation of the rat FGF-AS cDNA clone. The full-length rat FGF-AS (1.1 kb) insert including the ATG initiation codon was cloned into the EcoRI/XhoI site of the phagemid pBK-CMV (4.5 kb, Stratagene) in each orientation and expressed using the T3 promoter. Circular plasmid DNA (1 μg) was added to 25 μl reactions containing 12.5 μl of rabbit reticulocyte lysate, [35S]methionine (1,000 Ci/mmol, 40 μCi; 1 Ci = 37 GBq), 1 × TnT buffer, and the appropriate RNA polymerase. After incubation at 30°C for 120 min, the reactions were snap frozen and stored at −80°C until further analysis. One-half of each reaction (12.5 μl) was diluted in ice-cold NET buffer (150 mM NaCl/1 mM EDTA/50 mM Tris, pH 7.4, containing 0.1% Nonidet P-40 and 0.25% gelatin), and immunoprecipitation was performed using the anti-MutT domain IgG as previously described (20). Radioactive bands were visualized by autoradiography after SDS/PAGE separation on linear 4–20% gradient minigels (Bio-Rad). Western blots of rat tissue extracts were prepared exactly as previously described (20), and immunoreactive bands were visualized by chemiluminescent detection using a commercially available kit (ECL, Amersham).
RESULTS
Isolation and Characterization of the Rat FGF-2 Antisense cDNA.
Preliminary Northern blot analysis of developing rat tissues, using a human cRNA probe (gfg-1) specific for the FGF-2 antisense strand, detected abundant expression of FGF-2 antisense RNA in early postnatal liver (13). We constructed a cDNA library from day 12 rat liver RNA and screened it with the gfg-1 human cDNA probe. Following tertiary screening, four positives were selected, each of which yielded an approximately 1.1-kb insert when subjected to restriction enzyme digestion (data not shown). Dideoxy sequencing revealed that the rat FGF-2 antisense cDNA comprises 1,099 bp and contains a consensus polyadenylylation signal 13 nt upstream of the 18-nt poly(A)-stretch (Fig. 1). Using 5′ rapid amplification of cDNA ends (RACE), we also isolated and sequenced partial cDNA clones of the rat FGF-2 antisense RNA, which confirmed that the library-derived cDNA contained a complete 5′ end sequence (data not shown). The rat cDNA has >90% homology to the human gfg-1 FGF antisense clone and 67% overall homology to the Xenopus FGF antisense cDNA. The rat cDNA also contains a long ORF whose deduced amino acid sequence has 70% identity with the Xenopus hypothetical 24-kDa protein (see below). With the exception of its homology to the Xenopus antisense cDNA, and its complementarity with FGF-2 sequences, the rat FGF-AS sequence has no striking homology with any other nucleotide sequences in GenBank.
Figure 1.
Nucleotide sequence of the rat FGF-2 antisense cDNA. The translation start codon (ATG) and the stop codon (TGA) are denoted by the thick underline. The consensus polyadenylylation signal sequence (AATAAA) is double-underlined. The region of complementarity with the FGF-2 mRNA is shaded. This cDNA sequence has been deposited in GenBank and assigned the accession number U58289U58289.
Complementarity of the Rat FGF-2 Sense and Antisense RNAs.
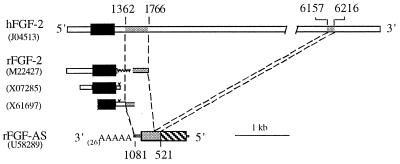
It has been presumed that the antisense RNA regulates FGF-2 mRNA by the formation of double-stranded RNA (dsRNA) complexes for subsequent RNA editing by dsRNA-specific adenosine deaminase (DRADA) (10). The Xenopus FGF-2 sense and antisense transcripts share 900 bp of contiguous complementary sequence at their 3′ ends, and the region of overlap extends into the 3′ coding region of the FGF-2 RNA. The full-length rat FGF-2 mRNA sequence has never been reported and the majority of the 3′ UTR is unknown. Comparison of the FGF-AS sequence with three partial FGF-2 cDNA sequences in the GenBank database revealed no overlap with the FGF-2 coding sequence (Fig. 2). However, comparison of the 1.1-kb rat FGF-AS cDNA with the full-length (6.7 kb) human FGF-2 cDNA (21) demonstrates that the FGF-AS transcript contains sequences complementary to two widely separated regions of the FGF-2 sense RNA; one 60-bp region (corresponding to bases 6157–6216 of the FGF-2 cDNA) includes the most distal polyadenylylation signal motif of the 6.7-kb FGF-2 mRNA, while the other (corresponding to bases 1362–1766 of the FGF-2 cDNA) lies just downstream of the proximal FGF-2 polyadenylylation signal motif (Fig. 2). The long intervening sequence, which is part of the 3′ UTR of the FGF-2 mRNA, represents an intron in the antisense gene that is removed by processing of the antisense pre-mRNA (17).
Figure 2.
The rat FGF-AS transcript is complementary to discrete regions of the 3′ UTR of the FGF-2 mRNA. The relationship of the antisense cDNA to the human (top line) and rat FGF-2 mRNA sequences is shown. GenBank accession numbers are indicated in parentheses. Regions of complementarity between the sense and antisense cDNAs are indicated by the shaded areas. Complementary sequence in the distal 3′ UTR of the 6.7-kb human FGF-2 mRNA (top line) predicts a similar region in the as yet unsequenced 3′ UTR of the 6-kb rat FGF-2 RNA.
We used 3′ RACE in an attempt to identify longer antisense transcripts that might be complementary to the coding region of the FGF-2 sense RNA. However, 3′ RACE of rat liver RNA generated a single major cDNA species whose sequence was identical to the FGF-AS cDNA isolated by conventional library screening (data not shown). The mammalian FGF-2 sense and antisense transcripts thus share sequences in their 3′ regions that are antisense and complementary. However, these sequences do not include the FGF-2 coding region, and the extent of sequence complementarity (425 bp) is considerably less than the 900 bp reported for the Xenopus cDNAs. Although we cannot rule out the possibility of tissue- or stage-specific expression of longer antisense transcripts, the present findings indicate that the major FGF antisense RNA expressed in neonatal rat liver is complementary to a portion of the 3′ UTR of the FGF-2 mRNA.
Antisense Expression Is Tissue-Specific and Developmentally Regulated.
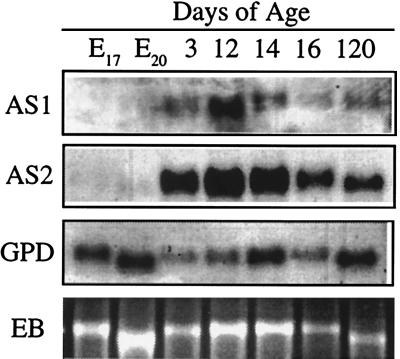
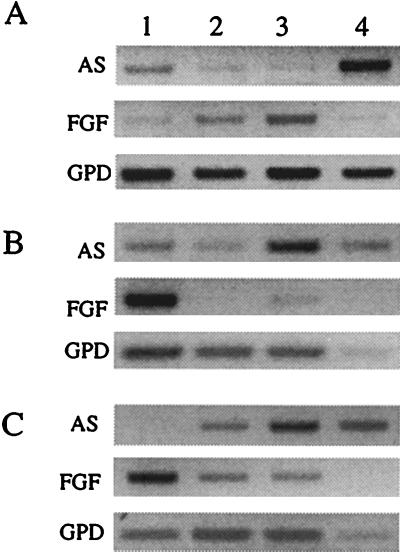
32P-labeled cRNA riboprobes (AS1 and AS2) confirmed the developmentally regulated pattern of expression of the antisense RNA in Northern blots of rat liver. The 1.5-kb transcript was detectable in liver RNA from all stages of development but was most abundant in early postnatal liver (Fig. 3). Comparison of FGF-AS expression in developing rat tissues by Northern blot and RT-PCR analysis confirmed that liver contained the highest steady-state level of FGF-AS RNA (data not shown). Amplification of FGF-2 sense and antisense RNA by RT-PCR demonstrates the tissue-specific distribution and developmental pattern of expression of the sense and antisense transcripts (Fig. 4). In heart and kidney, FGF-2 mRNA transcripts were more abundant than FGF-AS transcripts in embryonic samples, but in postnatal samples the situation reversed due to a marked increase in abundance of FGF-AS transcripts. In sharp contrast, in the developing brain, FGF-2 mRNA expression increased throughout development, while FGF-AS decreased in a reciprocal fashion. This confirms our earlier observation by Northern hybridization using the human 301-bp cRNA probe (13).
Figure 3.
Northern blot analysis of rat liver poly(A)+ RNA (5 μg per lane) isolated at various stages of embryonic and postnatal development. The Northern blot was probed sequentially with two cRNA probes (AS1, AS2) derived from the FGF-AS cDNA, and with a cDNA probe for glyceraldehyde phosphate dehydrogenase (GPD). The bottom panel shows the residual 28S ribosomal RNA band stained with ethidium bromide (EB).
Figure 4.
RT-PCR analysis of FGF-2 and FGF-AS mRNA expression in tissues isolated from embryonic (day 17 postfertilization) (A); neonatal (postnatal day 12) (B); and adult (day 120) (C). Glyceraldehyde phosphate dehydrogenase (GPD) mRNA levels were determined as a loading control. Control reactions (no RT), included for each reaction, produced no product (not shown). Lanes: 1, brain; 2, heart; 3, kidney; 4, liver.
The FGF Antisense RNA Encodes a 35-kDa Protein.
The rat FGF-2 antisense RNA contains a long ORF (nucleotides 34–975) that predicts a 35-kDa translation product containing 313 aa. The deduced amino acid sequence of the rat FGF-2 antisense clone has 70% identity to the predicted 217-aa (24-kDa) Xenopus protein (Fig. 5 Upper). However, the ORF in the rat cDNA extends upstream of the putative initiator ATG in the Xenopus cDNA, which in the rat sequence is replaced by a CTG (leucine) codon. The rat ORF predicts an NH2-terminal extension of 96 aa. This ORF is conserved in the Xenopus 5′ UTR sequence, suggesting that the original Xenopus cDNA sequence may be incomplete. Topology analysis (22, 23) predicts an intracytoplasmic NH2 terminus segment of 88 residues followed by a short transmembrane region (18 residues denoted by the heavy overline in Fig. 5 Upper) and a large extracytoplasmic COOH terminus of 207 residues.
Figure 5.
(Upper) Alignment of the deduced amino acid sequence of rat and Xenopus FGF-2 antisense cDNA clones. Double and single dots represent amino acid identity and similarity, respectively. Percentages of identity and similarity between rat and Xenopus antisense amino acid sequences are 69% and 89%, respectively. The location of a putative transmembrane domain (amino acids 89–107) is indicated by a heavy overline. The conserved MutT domain is shaded. (Lower) Comparison of the MutT domain and flanking sequence of the FGF-AS protein with those of other members of the MutT/Nudix family of proteins. In the consensus sequence (top line) U represents a bulky aliphatic amino acid (I, L, or V) and x may be any amino acid. Bold letters indicate identities with the rat FGF-AS protein sequence. The indicated sequences are P13420P13420 (Xenopus FGF-AS); U30313U30313 (human Ap4Aase); D49977D49977 and P36639P36639 (rat and human 8-oxo-dGTPase); Q01976Q01976 (yeast YSA1); and P08377P08377 (Escherichia coli MutT).
The GenBank protein database was searched for protein sequences sharing homology with the rat FGF-2 antisense protein using the blast algorithm (18). The deduced amino acid sequence was found to share homology with the consensus sequence of the MutT domain of prokaryotic and eukaryotic “nudix” hydrolases. The MutT domain [first defined by Koonin (24), and recently modified by Bessman et al. (25)] contains the signature sequence G(X)5E(X)7REUXEEXXU (where X = any amino acid; U = a bulky aliphatic amino acid I, L, or V). Within the consensus sequence, the rat FGF-2 antisense amino acid sequence has 48–78% identity with the corresponding domains in proteins from a range of organisms (Fig. 5 Lower). However, further comparison of the FGF-AS with the MutT hydrolase family of proteins using protomat did not reveal any other motifs in common.
The FGF-AS RNA Is Translated in Vivo.
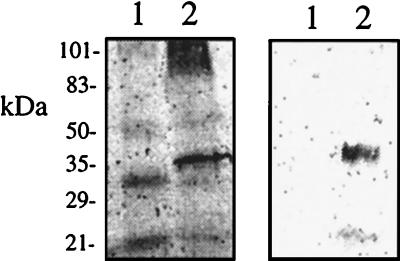
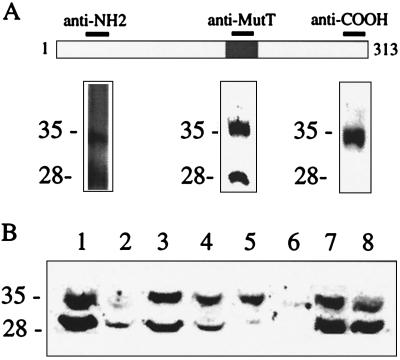
In vitro translation of the rat FGF-AS cDNA generated the predicted 35-kDa protein, which was immunoprecipitated with a polyclonal antiserum raised against the conserved MutT domain (Fig. 6). We have recently reported that the anti-MutT domain antiserum detects two immunoreactive proteins of approximately 28 kDa and 35 kDa in Western blots of rat liver homogenates (20). To confirm the identity of the immunoreactive bands as products of the FGF-AS gene, we raised two additional antisera against the deduced amino acid sequence of the FGF-AS protein. As shown in Fig. 7A, all three antisera detected an abundant 35-kDa protein in rat liver homogenates. The antisera against the NH2 terminus or the MutT domain also recognized the 28-kDa band, whereas the COOH-selective antiserum detected the 35-kDa product exclusively. These results indicate that the 35-kDa and 28-kDa bands are both derived from the same primary transcript, and suggest that the 28-kDa isoform may be generated by proteolytic cleavage at the COOH terminus. Western blot analysis of postnatal rat tissues demonstrates that the FGF-AS protein is abundantly expressed in a variety of tissues including liver, heart, kidney, and adrenal (Fig. 7B). The varying ratio of 35-kDa to 28-kDa isoforms may reflect differences in tissue protease levels. The immunoreactive bands were only faintly detectable in extracts from postnatal brain, in agreement with the low level of FGF-AS mRNA detected in this tissue by RT-PCR.
Figure 6.
In vitro translation (Left) and immunoprecipitation (Right) of the rat FGF-AS protein product. The migration of molecular mass markers (kDa) are indicated on the left. In vitro translation was performed with the inserts in noncoding orientation (lane 1) and in the coding orientation (lane 2).
Figure 7.
Immunodetection of the FGF-AS protein in rat tissue extracts. (A) Antisera raised against the indicated regions of the deduced FGF-AS protein were used to probe Western blots containing 50 μg of crude rat liver homogenate. (B) Tissue distribution of FGF-AS immunoreactivity. Lanes: 1 and 8, liver; 2, brain; 3, heart; 4, kidney; 5, thymus; 6, lung; 7, adrenal.
DISCUSSION
Antisense transcription of the FGF-2 gene was originally reported in Xenopus laevis oocytes (26), where it was implicated in the regulation of FGF-2 mRNA stability (10). In unfertilized Xenopus oocytes the antisense transcript is present in 20-fold excess relative to the FGF sense transcript, and the complementary RNAs form stable dsRNA complexes in the cytoplasm. During oocyte maturation all of the FGF-2 mRNAs are extensively edited in the overlap region by DRADA (27) and subsequently degraded (10). DRADA has been implicated in the physiologically significant RNA editing of glutamate-gated ion channels in brain (28). Although DRADA-mediated editing may target the FGF-2 mRNA for rapid degradation, the antisense RNA could conceivably inhibit FGF-2 expression by a variety of other posttranscriptional mechanisms including inhibition of transcript processing, transport, or translation (29, 30).
The present findings demonstrate that the rat FGF-AS RNA is not complementary to the FGF-2 coding region, indicating that any DRADA-directed RNA editing is unlikely to result in modified FGF-2 proteins. The rat FGF-AS is complementary to 464 nucleotides present in two widely separated regions in the 3′ UTR of the 6.7-kb human FGF-2 mRNA. One 60-bp region includes the most distal polyadenylylation signal motif of the FGF-2 mRNA, while the other lies just downstream of the proximal FGF-2 polyadenylylation site. This raises the possibility that physical interaction of the FGF-2 and FGF-AS mRNAs may play a role in the regulation of FGF-2 polyadenylylation site usage. Multiple size classes of FGF-2 mRNA (1.2–7.0 kb in length), differing exclusively in the length of their 3′ UTR, are expressed in a tissue- and developmental-stage-specific fashion. Differential 3′ UTR expression may well have functional consequences; the 6.7-kb human FGF-2 mRNA contains 15 copies of an AU-rich motif associated with regulation of mRNA stability and perinuclear localization, whereas the 3.7-kb mRNA contains only 2 such sequences, and the 1.2-kb mRNA contains none. Alternatively, the 3′ UTR may be an important target for other mechanisms of AS-mediated regulation including pre-mRNA splicing, mRNA trafficking, and stability.
The longest FGF-2 mRNA detected in rat tissues is approximately 6 kb in length (9). However, the rat FGF-2 cDNAs characterized to date contain only short 3′ UTR sequences (8, 31, 32). The FGF-2 cDNA with the shortest 3′ UTR (31) has no sequence complementarity with the FGF-AS cDNA. A 722-bp FGF-2 cDNA from rat brain (32), with a longer 3′ UTR sequence, is 100% complementary to the 1.1-kb FGF-AS cDNA in a region corresponding to the site of overlap with the human FGF cDNA (Fig. 2). The longest rat FGF-2 cDNA, from pregnant mare serum gonadotropin-primed rat ovary (8), lacks 307 bp of 3′ UTR sequence present in the rat brain cDNA and may represent a minor splice variant or a cloning artifact (32). Beyond the site of this apparent deletion, the 3′ UTR contains an additional 256 nucleotides that are 100% complementary to the antisense cDNA. By homology with the human FGF cDNA, an additional 60-bp region of homology is predicted in the distal 3′ UTR of the 6-kb rat FGF transcript.
We have previously reported that the increase in expression of the 6-kb FGF-2 RNA transcript in the developing rat brain coincides with a decrease in the expression of the antisense transcript (13). The present RT-PCR data demonstrate that the overall level of FGF-2 mRNA in brain (regardless of transcript size) is inversely related to the level of FGF-AS mRNA in that tissue. This confirms a recent in situ hybridization study that reported an inverse relationship between the level of FGF sense and antisense RNA abundance in the developing rat CNS (33). Because the antisense transcript is complementary to regions of the 3′ untranslated region of the FGF-2 mRNA, it is conceivable that formation of dsRNA helices in these regions may in some way regulate polyadenylylation site usage and/or mRNA half-life.
The relationship of the FGF-2 sense and antisense RNAs is analogous to that of the c-erbAα/Rev-ErbAα sense/antisense pair. Rev-ErbAα is an mRNA transcribed from the opposite strand of the thyroid hormone receptor c-erbAα gene in the antisense orientation (34, 35). The c-erbAα sense and antisense transcripts overlap at their 3′ ends, and the complementary sequence in Rev-ErbAα has been shown to inhibit c-erbAα pre-mRNA splicing in HeLa cell nuclear extracts (36). In addition to its regulatory role at the RNA level, the Rev-ErbAα transcript encodes a nuclear hormone receptor-related protein (37). Our identification of a translated protein product of the FGF-AS RNA extends the analogy and raises the possibility that the FGF-AS RNA plays a dual role as a template for protein synthesis and as a regulator of FGF-2 mRNA.The rat FGF-2 antisense transcript encodes a novel member of the MutT-like family of enzymes. The E. coli MutT protein, the prototypic member of this family, is a nucleoside triphosphate pyrophosphohydrolase whose function is to remove mutagenic forms of dGTP from the nucleotide pool. The human and rat homologues of MutT are 8-oxo-dGTPases that suppress the occurrence of spontaneous oxidative mutagenesis by hydrolyzing 8-oxo-dGTP (38). The conserved MutT domain has recently been shown to be present in a larger family of nucleoside diphosphatases present in organisms ranging from viruses to humans. A common feature of this family of enzymes is their “house-cleaning” role in the hydrolysis of potentially hazardous compounds or metabolites (25). The rapid postnatal induction of FGF-AS expression in rat liver is consistent with this model, reflecting the increased metabolic activity that occurs at this time. However, identification of the physiological function of the FGF-AS product must await targeted manipulation of the FGF-AS gene in vivo.
Acknowledgments
This work was funded by grants to P.R.M. from the Medical Research Council of Canada, the Cancer Research Society, and the Atlee Endowment.
ABBREVIATIONS
- bFGF or FGF-2
basic fibroblast growth factor
- FGF-AS
FGF antisense
- ORF
open reading frame
- UTR
untranslated region
- CNS
central nervous system
- RT-PCR
reverse transcription–PCR
- dsRNA
double-stranded RNA
- DRADA
dsRNA-specific adenosine deaminase
Footnotes
References
- 1.Kimelman D, Abraham J, Haaparanta T, Palisi T, Kirschner M. Science. 1988;242:1053–1056. doi: 10.1126/science.3194757. [DOI] [PubMed] [Google Scholar]
- 2.Northrop J L, Kimelman D. Dev Biol. 1994;161:490–503. doi: 10.1006/dbio.1994.1047. [DOI] [PubMed] [Google Scholar]
- 3.Klagsbrun M. Annu Rev Physiol. 1991;53:217–239. doi: 10.1146/annurev.ph.53.030191.001245. [DOI] [PubMed] [Google Scholar]
- 4.Tsuboi R, Rifkin D B. J Exp Med. 1990;172:245–251. doi: 10.1084/jem.172.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy P R, Myal Y, Sato Y, Sato R, West M, Friesen H G. Mol Endocrinol. 1989;3:225–231. doi: 10.1210/mend-3-2-225. [DOI] [PubMed] [Google Scholar]
- 6.Karpen C W, Spanheimer R G, Randolph A L, Lowe W J. Diabetes. 1992;41:222–226. doi: 10.2337/diab.41.2.222. [DOI] [PubMed] [Google Scholar]
- 7.Moffett J, Kratz E, Florkiewicz R, Stachowiak M K. Proc Natl Acad Sci USA. 1996;93:2470–2475. doi: 10.1073/pnas.93.6.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimasaki S, Emoto N, Koba A, Mercado M, Shibata F, Cooksey K, Baird A, Ling N. Biochem Biophys Res Commun. 1988;157:256–263. doi: 10.1016/s0006-291x(88)80041-x. [DOI] [PubMed] [Google Scholar]
- 9.Powell P P, Finklestein S P, Dionne C A, Jaye M, Klagsbrun M. Mol Brain Res. 1991;11:71–77. doi: 10.1016/0169-328x(91)90023-q. [DOI] [PubMed] [Google Scholar]
- 10.Kimelman D, Kirschner M W. Cell. 1989;59:687–696. doi: 10.1016/0092-8674(89)90015-9. [DOI] [PubMed] [Google Scholar]
- 11.Borja A Z M, Meijers C, Zeller L. Dev Biol. 1993;157:110–118. doi: 10.1006/dbio.1993.1116. [DOI] [PubMed] [Google Scholar]
- 12.Savage M P, Fallon J F. Dev Dyn. 1995;202:343–353. doi: 10.1002/aja.1002020404. [DOI] [PubMed] [Google Scholar]
- 13.Li A, Seyoum G, Shiu R, Murphy P R. Mol Cell Endocrinol. 1996;118:113–123. doi: 10.1016/0303-7207(96)03772-0. [DOI] [PubMed] [Google Scholar]
- 14.Knee R S, Pitcher S E, Murphy P R. Biochem Biophys Res Commun. 1994;205:577–583. doi: 10.1006/bbrc.1994.2704. [DOI] [PubMed] [Google Scholar]
- 15.Knee, R. & Murphy, P. R. (1997) Neurochem. Int., in press. [DOI] [PubMed]
- 16.Aviv H, Leder P. Proc Natl Acad Sci USA. 1972;69:1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy P R, Knee R. Mol Endocrinol. 1994;8:852–859. doi: 10.1210/mend.8.7.7984147. [DOI] [PubMed] [Google Scholar]
- 18.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Murphy P R, Sato Y, Knee R. Mol Endocrinol. 1992;6:877–884. doi: 10.1210/mend.6.6.1323055. [DOI] [PubMed] [Google Scholar]
- 20.Li A W, Too C K L, Murphy P R. Biochem Biophys Res Commun. 1996;223:19–23. doi: 10.1006/bbrc.1996.0839. [DOI] [PubMed] [Google Scholar]
- 21.Prats H, Kaghad M, Prats A C, Klagsbrun M, Lelias J M, Liauzun P, Chalon P, Tauber J P, Amalric F, Smith J A, Caput D. Proc Natl Acad Sci USA. 1989;86:1836–1840. doi: 10.1073/pnas.86.6.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rost B, Casadio R, Fariselli P, Sander C. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rost B. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 24.Koonin E V. Nucleic Acids Res. 1993;21:4847. doi: 10.1093/nar/21.20.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bessman M J, Frick D N, O’Handley S F. J Biol Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 26.Volk R, Koster M, Poting A, Hartmann L, Knochel W. EMBO J. 1989;8:2983–2988. doi: 10.1002/j.1460-2075.1989.tb08448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bass B. Dev Biol. 1992;3:425–433. [Google Scholar]
- 28.Seeburg P. J Neurochem. 1996;66:1–5. doi: 10.1046/j.1471-4159.1996.66010001.x. [DOI] [PubMed] [Google Scholar]
- 29.Simons R W, Kleckner N. Annu Rev Genet. 1988;22:567–600. doi: 10.1146/annurev.ge.22.120188.003031. [DOI] [PubMed] [Google Scholar]
- 30.Simons R W. In: Antisense Research and Applications. Crooke S T, LeBleu B, editors. Boca Raton, FL: CRC; 1993. pp. 97–124. [Google Scholar]
- 31.Kurokawa T, Seno M, Igarashi K. Nucleic Acids Res. 1988;16:5201. doi: 10.1093/nar/16.11.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.el-Husseini A E, Paterson J A, Myal Y, Shiu R P. Biochim Biophys Acta. 1992;1131:314–316. doi: 10.1016/0167-4781(92)90031-t. [DOI] [PubMed] [Google Scholar]
- 33.Grothe C, Meisinger C. Neurosci Lett. 1995;197:175–178. doi: 10.1016/0304-3940(95)11917-l. [DOI] [PubMed] [Google Scholar]
- 34.Miyajima N, Horiuchi R, Shibuya Y, Fukushige S, Mutsubara K, Toyoshima K, Yamamoto T. Cell. 1989;57:31–39. doi: 10.1016/0092-8674(89)90169-4. [DOI] [PubMed] [Google Scholar]
- 35.Lazar M A, Hodin R A, Darling D S, Chin W W. Mol Cell Biol. 1989;9:1128–1136. doi: 10.1128/mcb.9.3.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munroe S H, Lazar M A. J Biol Chem. 1991;266:22083–22086. [PubMed] [Google Scholar]
- 37.Harding H P, Lazar M A. Mol Cell Biol. 1993;13:3113–3121. doi: 10.1128/mcb.13.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mo J-Y, Maki H, Sekiguchi M. Proc Natl Acad Sci USA. 1992;89:11021–11025. doi: 10.1073/pnas.89.22.11021. [DOI] [PMC free article] [PubMed] [Google Scholar]