Abstract
Sensory rhodopsin II (SRII) is a repellent phototaxis receptor in the archaeon Halobacterium salinarum, similar to visual pigments in its seven-helix structure and linkage of retinal to the protein by a protonated Schiff base in helix G. Asp-73 in helix C is shown by spectroscopic analysis to be a counterion to the protonated Schiff base in the unphotolyzed SRII and to be the proton acceptor from the Schiff base during photoconversion to the receptor signaling state. Coexpression of the genes encoding mutated SRII with Asn substituted for Asp-73 (D73N) and the SRII transducer HtrII in H. salinarum cells results in a 3-fold higher swimming reversal frequency accompanied by demethylation of HtrII in the dark, showing that D73N SRII produces repellent signals in its unphotostimulated state. Analogous constitutive signaling has been shown to be produced by the similar neutral residue substitution of the Schiff base counterion and proton acceptor Glu-113 in human rod rhodopsin. The interpretation for both seven-helix receptors is that light activation of the wild-type protein is caused primarily by photoisomerization-induced transfer of the Schiff base proton on helix G to its primary carboxylate counterion on helix C. Therefore receptor activation by helix C–G salt-bridge disruption in the photoactive site is a general mechanism in retinylidene proteins spanning the vast evolutionary distance between archaea and humans.
Keywords: halobacteria, photosensory receptor, retinal proteins, activating mutation
Sensory rhodopsin II (SRII, also called phoborhodopsin) is a repellent phototaxis receptor in the archaeon Halobacterium salinarum (previously salinarium and halobium) (1). Blue-green light photoconverts SRII (λmax 487 nm) in < 1 ms to a UV-absorbing species, SII360 (subscript denotes the absorption maximum in nm) that decays in ≈100 ms to another intermediate, SII540, which returns to the initial state SRII487 in ≈500 ms (2). The formation of these two photointermediates causes swimming reversals by the cells (3), resulting in a repellent effect of the light.
The absorption shifts during the photocycle (2) and Fourier-transform infrared (FTIR) evidence from the similar SRII of Natronobacterium pharaonis (pSRII) (4), indicate that deprotonation and reprotonation of the retinylidene Schiff base occur in the formation and decay, respectively, of SII360. Light-induced difference FTIR spectroscopy of pSRII indicates the protonation of a carboxylate during the formation of the corresponding UV-absorbing intermediate, pSII390, in the photocycle (4). The FTIR band caused by this event in pSRII is very similar in spectral shape and position to that caused by Asp-85 protonation in bacteriorhodopsin (BR; ref. 5), and this aspartyl residue is conserved in all known sensory rhodopsins (Asp-73 in SRII; for review see ref. 6). Participation of Asp-73 in proton transfers would be especially of interest because the homologous Asp-76 in sensory rhodopsin I (SRI; λmax 587 nm) is known to be protonated in the unphotolyzed state and to remain uncharged during attractant phototaxis signaling by the receptor (7). Asp-76 becomes a proton acceptor in SRI when its tightly bound transducer protein HtrI is removed and in alkaline conditions, where a purple (λmax 552 nm) proton-pumping form of SRI is produced (8–10).
The lack of a transformation procedure and selectable plasmids for N. pharaonis precluded the efficient use of site-specific mutagenesis to confirm the identity of the proton acceptor from the Schiff base in pSRII. The recent cloning of the SRII apoprotein gene sopII (sensory opsin II) and its transducer gene htrII from H. salinarum (11), a species for which molecular genetic procedures are well developed, enables recombinant DNA approaches to be taken. The sopII gene was identified as immediately adjacent to the 3′ end of the htrII gene, in an arrangement similar to that observed for the htrI–sopI gene pair (12).
The work described here identifies Asp-73 as the Schiff base proton acceptor in SRII. Absorption spectroscopy of membranes and of purified wild-type and D73N SRII preparations also indicates Asp-73 is a counterion to the protonated Schiff base in the unphotolyzed state SRII487. Constitutive signaling by the D73N receptor is observed in remarkable similarity to the analogous mutant E113Q in human rhodopsin (13). The results reported here and the great evolutionary distance of the archaeon H. salinarum from humans strongly argue for a general mechanism of seven-helix retinylidene receptor activation based on active site salt-bridge disruption, as proposed for human rhodopsin (14).
MATERIALS AND METHODS
Expression and Purification.
H. salinarum strain Pho81Wr− (15), which lacks HtrII and SRII protein, and a derivative in which the htrII–sopII region was deleted (strain Δ35), were used as recipients in plasmid transformations. The expression vector pPR5 was assembled as follows: First, a SpeI site was introduced by PCR 4 bases upstream of the initiator methionine codon, at the beginning of HtrII, to place the gene under control of the HtrI promoter. The 3.15-kbp SpeI–StuI fragment containing the htrII–sopII genes was joined with the 4.1-kbp SpeI–HindIII fragment and with the 1.9-kbp HindIII-EcoRV fragment from the plasmid pKJ301. pKJ301 is an H. salinarum expression and Escherichia coli/H. salinarum shuttle vector with the htrI–sopI gene pair under control of their native promoter. It derives from pVJY1 (15) by removal of a 2.8-kbp Eco47III–NotI fragment in the mevinolin resistance (MevR) region, which was found to be not required for the MevR phenotype (K.-H. Jung and J.L.S., unpublished results). The SRII D73N mutation was made following a two-step PCR method (16), and the plasmid pPR73N encoding mutated SRII apoprotein was constructed similarly as pPR5. The expression vectors pPR5 and pPR73N encode both the HtrII and receptor proteins.
For high-level expression and purification, 660-bp fragments containing sopII or sopII with the D73N mutation were amplified by PCR to contain NcoI–NsiI ends and were substituted in place of the sopI-polyhistidine fusion protein gene under control of the efficient bop promoter in the H. salinarum expression vector pSO12 (17) to yield pHT5 and pHT73N. The encoded proteins terminate with MHSESHHHHHH, like His-tagged SRI, after Ala-221 in wild-type SRII. His-tagged SRII and His-tagged D73N were purified by Ni2+-affinity chromatography after solubilization with lauryl maltoside following the procedure described for His-tagged SRI (17).
Immunoblotting and Autofluorography.
Cells were labeled in vivo with l-[methyl-3H]methionine in the presence of puromycin, membranes were isolated after sonication, and autofluorography of membrane proteins separated by SDS/polyacrylamide gel electrophoresis was performed as described (18). Immunoblotting with polyclonal antibody to the signaling domain of halobacterial transducers (HC23 antibody) was as described (19).
Spectroscopy.
Absorption spectra of wild-type and D73N SRII in membranes were measured after hydroxylamine bleaching and all-trans-retinal reconstitution as described (20). Purified receptors were measured directly against the final solution buffer (4 M NaCl/0.025% lauryl maltoside/25 mM Tris⋅HCl, pH 6). Spectra were recorded on an SLM-Aminco DW2000 spectrophotometer (path length 1 cm). Flash-induced absorbance changes were measured with a laboratory-constructed cross-beam kinetic spectrophotometer (20) except the actinic flash was a 6-ns, 532-nm, 40-mJ pulse from a Nd:YAG laser (Surelite I; Continuum, Santa Clara, CA), and curve fitting used the algorithm from Sigma Plot (Jandel Scientific, Corte Madera, CA). Flashes were delivered at 0.08 Hz, and 15 transients at 500 μs per point (3968 points per transient) were averaged for each measurement.
Cell Tracking and Motion Analysis.
Swimming reversal frequency measurements of cell populations used the ExpertVision system from Motion Analysis Systems (Santa Rosa, CA). Photostimuli were 10-ms pulses of 500-nm light selected from a Nikon 100W Hg–Xe lamp by a wide-band (480–520 nm) filter (Ditric Optics, Hudson, MA).
RESULTS
Rescue of Blue-Green Phototaxis Mutants by Plasmid-Mediated Expression of the htrII–sopII Gene Pair.
The htrII–sopII gene pair has been identified on a 6.5-kbp PstI fragment (11). Southern hybridization with an htrII-specific and a sopI -specific probe reveal a shift to a 7-kbp PstI fragment in the blind mutant Pho81Wr− and the presence of a BglII site not present in the wild-type PstI fragment. Sequencing analysis (J. Zhu, E.N.S., M.A., and J.L.S., unpublished work) reveals that these changes are due to insertion of the 552-bp ISH2 transposable element, which is also responsible for disruption of the htrI–sopI pair in Pho81Wr− (15).
SRII production from its native promoter is at low levels and is maximal during exponential-phase growth (2). To increase expression level in stationary-phase cultures, the htrII–sopII pair was placed under control of the htrI–sopI promoter on the expression vector pPR5 and introduced by transformation into Pho81Wr− cells as well as into mutant strain Δ35, a derivative of Pho81 in which the htrII–sopII region has been excised. Southern hybridization of the transformants with a sopII-specific probe shows the inactivated copy of sopII on the 7-kbp PstI chromosomal fragment, as in untransformed Pho81Wr−, and the plasmid copy on a 6.5-kbp fragment (data not shown). No evidence of recombination, which would produce additional bands, was observed.
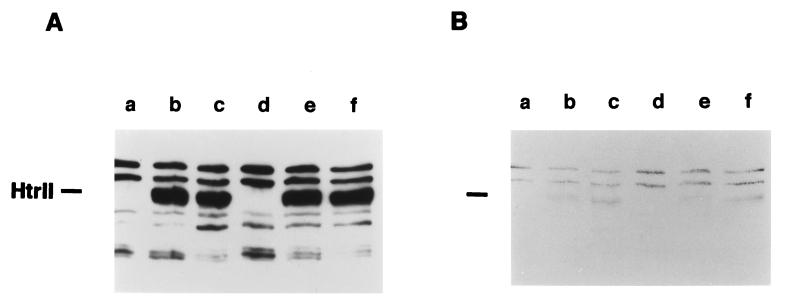
Expression of both htrII and sopII genes was evident by the restoration of (i) the absorption spectrum (see Fig. 1a) and flash-induced absorption changes characteristic of SRII (see Figs. 2 and 3), (ii) SRII-mediated phototaxis responses (see Fig. 4), and (iii) the HtrII methylation and immuno band (see Fig. 5). These data confirm that the putative htrII–sopII gene pair, assigned on the basis of sequence homologies, encode the phototaxis receptor SRII and its transducer HtrII.
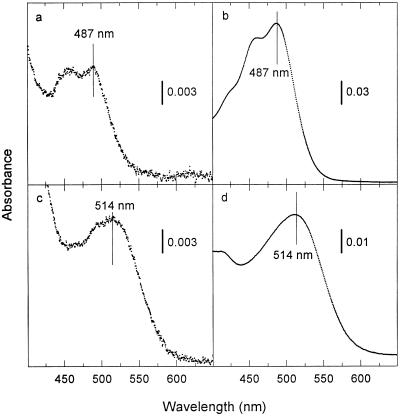
Figure 1.
Absorption spectra of wild-type and D73N SRII in membranes and purified in lauryl maltoside. (a and c) Spectra generated by all-trans-retinal addition to hydroxylamine-bleached membranes from Pho81Wr− containing wild-type and D73N SRII, respectively, in the presence of HtrII. (b and d) Spectra of purified His-tagged wild-type and D73N SRII, respectively, in lauryl maltoside.
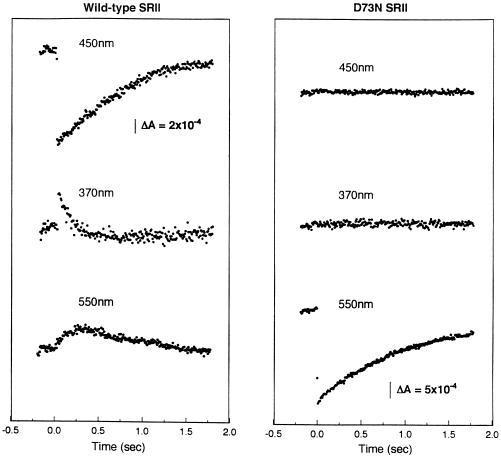
Figure 2.
Laser flash-induced absorption transients of wild-type and D73N SRII in membranes. Membranes as in Fig. 1 containing wild-type (Left) and D73N (Right) SRII were subjected to a 532-nm laser flash at time 0, and absorption changes were monitored at 500 μs per point at the indicated wavelengths.
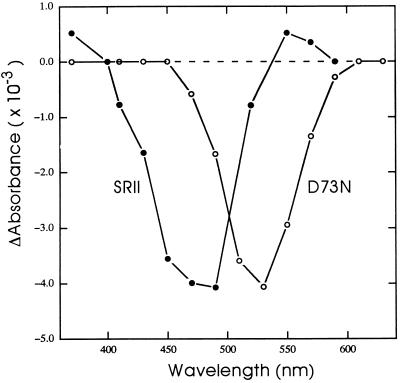
Figure 3.
Flash-induced absorption difference spectra of SRII and D73N in membranes. Maximum amplitudes of absorption changes monitored at various wavelengths as in Fig. 2 are shown for membranes containing wild-type and D73N SRII. The ordinate values are those of D73N, and SRII values were multiplied by 3 for plotting.
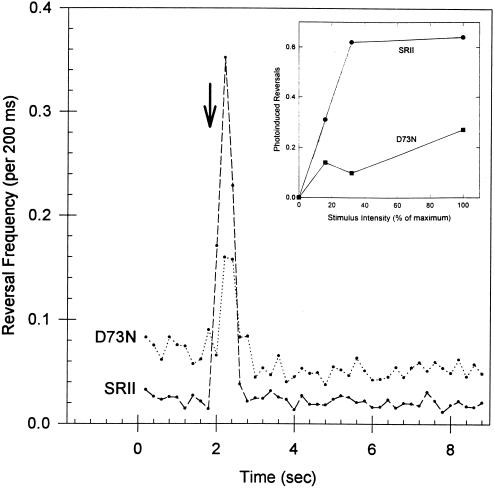
Figure 4.
Phototaxis responses of cells containing wild-type or D73N SRII. Reversal frequency transients from computerized cell-tracking and motion analysis for cells carrying SRII or D73N in the presence of HtrII are shown for a 10-ms 500-nm stimulus (arrow) at 100% stimulus intensity. (Inset) Fluence/response curves. Photoinduced reversals were assessed as the increased frequency of swimming reorientations in the 1-s period following the 10-ms stimulus. Stimuli were delivered at 30-s intervals and three sets of 16 stimuli were averaged to produce the reversal transients. Values in the fluence/response curves are each from one set of 16 stimuli.
Figure 5.
Immunoblot and methylation pattern of Pho81Wr−, Δ35, and transformant membranes. Shown are immunoblot with HC23 antibody (A) and methylation (B) fluorograms of membrane proteins from the following: a, Pho81Wr−; b, Pho81Wr− × pPR73N; c, Pho81Wr− × pPR5; d, Δ35; e, Δ35 × pPR73N; and f, Δ35 × pPR5.
Absorption Spectra of Wild-Type and D73N SRII.
Asp-73 in SRII is in the position homologous to that of the Schiff base counterions Asp-85 in BR and Asp-76 in the purple form of SRI. The known counterions have well characterized effects on the absorption spectra in BR and SRI. Accordingly, we replaced Asp-73 with Asn in SRII to test by absorption spectroscopy whether Asp-73 serves the analogous function.
Transformant membranes containing either wild-type or D73N SRII in the presence of HtrII were bleached with hydroxylamine as described (20) and reconstituted with all-trans-retinal to determine their visible absorption spectra. Wild-type SRII exhibits structured absorption with maxima at 487 nm and 460 nm (Fig. 1a), similar to the spectrum reported previously for hydroxylamine bleaching/reconstitution of Flx3b membranes (21). Purified His-tagged SRII in lauryl maltoside shows a nearly identical, but better resolved, spectrum (λmax 487 nm), with an evident vibrational fine structure, as previously reported for native SRII (21).
The D73N mutation shifts the absorption maximum to longer wavelengths by ≈30 nm both in membranes and in purified pigment in lauryl maltoside (Fig. 1 c and d). A loss of vibrational fine structure is especially evident in the purified pigment spectra, which have higher signal-to-noise ratios. Using the λmax values, we calculate a shift of 1078 cm−1 caused by substitution of Asn for Asp-73 (Table 1). This value is identical to that observed from the replacement of the homologous residues Asp-85 in BR (22) and Asp-76 in the purple form of SRI (9). In BR and in HtrI-free SRI the Asn-induced red shift is known to be due to the neutralization of the anionic form of the aspartyl residues Asp-85 and Asp-76, respectively, which function as counterions to the protonated Schiff base in each of the two pigments. The homologous position of Asp-73 and the identical red shift strongly indicate that Asp-73 is anionic in SRII487, as depicted in Table l, and functions similarly as an important counterion component in SRII. Further support for this interpretation is that acidic pH also induces a transition (pKa 3.0) of the wild-type SRII absorption maximum to 514 nm, analogous to formation of the 605-nm form of BR due to Asp-85 protonation, that D73E exhibits a similar transition shifted to pKa 5.1, and D73N does not exhibit a shift in absorption maximum in the range pH 1 to pH 6 (J. Zhu, E.N.S., M.A., and J.L.S., unpublished results).
Table 1.
Effect of asparagine substitution of Asp-85 in BR and of homologs in sensory rhodopsins
| Rhodopsin | Asp position | λmax, nm
|
Shift, cm−1 | |
|---|---|---|---|---|
| Asp− | Asn | |||
| BR | 85 | 568 | 605 | 1076 |
| SRI | 76 | 552 | 587 | 1080 |
| SRII | 73 | 487 | 514 | 1078 |
Flash-Induced Absorption Changes in Membranes Containing Wild-Type and D73N SRII.
The main Schiff base counterion of BR, of purple SRI, and of human rod rhodopsin serves as the primary proton acceptor from the Schiff base after photoisomerization. Accordingly, we assessed whether photoinduced deprotonation of the Schiff base is blocked in D73N SRII.
Transformant membranes containing either wild-type or D73N SRII in the presence of HtrII were analyzed by laser flash kinetic UV–visible spectroscopy. Absorption transients were monitored at 450 nm, at which in wild-type SRII the depletion and return of SRII487 are evident. The formation and decay of the two photointermediates SII360 and SII540 after a 532-nm laser flash were monitored at 370 nm and 550 nm, respectively. Transformant membranes containing wild-type SRII exhibited transients that fit the expected intermediates with formation of SII360 in <1 ms, decay of this intermediate into SII540 with a first-order half-time of 90 ms, and return of the initial state with a first-order half-time of 500 ms (Fig. 2). These values and the flash-induced absorption difference spectrum shown in Fig. 3 are similar to those reported previously (2).
D73N SRII shows no detected formation of a deprotonated intermediate as evidenced by the lack of a signal at 370 nm (Fig. 2). The flash-induced absorption difference spectrum shows a depletion with no evident increases in absorption. A similar difference spectrum is observed for the BR D85N mutant, in which Schiff base deprotonation is almost completely blocked (23), and for the light-driven chloride pump halorhodopsin, which does not deprotonate the Schiff base during its photocycle (24). The lack of positive bands indicates the photoproduct(s) of the D73N pigment mostly overlaps the absorption spectrum of the initial species. A blue-shifted photointermediate is indicated, since the depletion maximum of the difference spectrum at 530 nm is red-shifted from the 514-nm λmax of D73N. The photocycle of D73N is slower than that of wild type (Fig. 2).
Phototaxis Responses of Cells Containing Wild-Type and D73N SRII.
To assess the importance of Schiff base deprotonation to phototaxis signaling, fluence/response curves of cells containing wild-type or D73N SRII were produced by motility analysis using 10-ms stimuli with 500 ± 20 nm light for which the 487-nm SRII and 514-nm D73N receptors have comparable absorption (Fig. 1 a and c). At a stimulus strength sufficient to saturate the repellent response in wild-type SRII, cells carrying the D73N receptor exhibit 16% of the response (Fig. 4 Inset). The D73N mutant receptor and wild-type SRII are present at similar concentrations in the cells as assessed by absorption measurement (Fig. 1 a and c), and therefore the efficiency of signal generation by SRII is reduced, but not eliminated, by the mutation.
Cells carrying D73N exhibit higher reversal frequencies in the dark than wild type, as is evident in the prestimulus computerized motion analysis data (Fig. 4). Visual single cell tracking confirms that the D73N receptor increases the spontaneous reversal frequency of the cells observed with nonactinic infrared background illumination. The mean ± SEM for four independent measurements was 3.5 ± 0.2 min−1 and 10.8 ± 1.8 min−1 for transformants carrying wild-type SRII or D73N, respectively. This finding implies that the excitation signal that induces reversals is constitutively generated by the D73N receptor and is incompletely compensated by the adaptation (methylation) system. However the D73N receptor is not fully activated, since photostimuli still elicit a partial response from cells carrying the mutant SRII.
D73N Causes HtrII Demethylation in the Dark.
According to the E. coli chemotaxis paradigm, repellent signals result in transducer demethylation in the adaptation process. Light-induced methyl group turnover measurements (25, 26) and effects of mutations on swimming behavior (27, 28) strongly support that this paradigm applies, although light-induced changes of methylation extent of the Htr proteins have not been directly demonstrated. In analogy to E. coli transducers, if the D73N receptor is in a constitutively active signaling state, the HtrII protein should demethylate as an adaptation response (6). In vivo l-[methyl-3H]methionine labeling confirms this expectation. The amounts of HtrII protein expressed with SRII or D73N are identical according to immunoblotting (Fig. 5A) but methylation of HtrII is greatly reduced in the dark in the presence of the D73N receptor (Fig. 5B). Moreover, HtrII migrates slightly slower in the D73N background, consistent with demethylation, which slows the migration of E. coli transducers (29).
DISCUSSION
We conclude from the spectroscopic results that Asp-73 in SRII is (i) a counterion to the protonated Schiff base in the unphotolyzed repellent receptor SRII487 and (ii) the proton acceptor from the Schiff base during photoconversion to the SII360 intermediate. This role of Asp-73 is analogous to that of Glu-113 in human rod rhodopsin. Glu-113 is the Schiff base counterion and proton acceptor during photoconversion to the G-protein-activating state metarhodopsin-II380 (14). Disruption of the Glu-113-protonated Schiff base salt bridge by mutagenic replacement of Glu-113 with Gln constitutively activates rhodopsin (13). This is a key observation supporting a model proposed for rhodopsin that the counterion–Schiff base ion pair constrains the protein in an inactive conformation and the constraint is released by its light-induced disruption (14). The demethylated state of the HtrII protein when complexed with D73N SRII demonstrates analogous constitutive repellent signaling by the mutant SRII receptor and provides direct evidence that the salt-bridge locked-switch mechanism applies to both. It has been noted that this mechanism may also play an important role in the alternating access conformations in ion transport by BR (30). The higher spontaneous reversal frequency of D73N transformants further confirms the active repellent signaling state of D73N and indicates the constitutive excitation signal is not completely compensated by the demethylation, which is expected to provide a counterbalancing adaptation signal.
The role of Asp-73 as counterion and proton acceptor is in contrast to that obtained with the attractant receptor SRI, in which the corresponding Asp-76 is protonated in the unphotolyzed pigment SRI587, and apparently does not participate in proton transfers during the orange light-driven photocycle that produces attractant signals (7). If Asp-76 is deprotonated in SRI by removal of the bound HtrI transducer at alkaline pH, the Asp-76 carboxylate then functions as a Schiff base counterion and proton acceptor during the photocycle of the purple form SRI552 (10). Therefore the position of this residue near the Schiff base must be similar in both SRI and SRII; however, in the former, interaction with transducer raises its pKa and the proton therefore is transferred elsewhere. The distinct proton release pathways may have significance for the opposite signals produced by Schiff base deprotonation in the two receptors. SRI has been proposed to exist in the dark in an equilibrium mixture capable of being shifted to either conformation to explain its dual attractant and repellent signaling functions (31). Weakening of the counterion by neutralization of Asp-76 may provide the mechanism for generating the equilibrium mixture, not needed in SRII.
The identical shift of 1080 cm−1 caused by the elimination of the negative charge of Asp-85 in BR, Asp-76 in SRI, and Asp-73 in SRII, is due presumably to weakening of the protonated Schiff base/counterion interaction. Resonance Raman measurements comparing the blue form SRI587 (protonated Asp-76) and the purple form BR568 (deprotonated Asp-85) show a lower value of the Schiff base C=N stretching frequency in the former and its smaller shift upon deuteration, providing a direct indication for a weaker counterion (32). In a study of the mechanism of color regulation in SRII using chromophore analogs, it was found that ring/chain coplanarization in the 6-s-trans conformation is sufficient to explain nearly all of the opsin shift of 2200 cm−1 in SRII487 (21). Much of the additional shift in BR568, HR578, and SRI587 has been attributed to a weakened counterion interaction (33). Thus, the small opsin shift in SRII compared with that in BR and SRI implies a strong counterion interaction with protein residues, although the strength of the interaction with Asp-73 is evidently comparable, since its neutralization causes the same wavenumber shift as that observed in BR and SRI. It has been proposed that the stronger counterion in SRII leads to a reduction in the inhomogeneous broadening of the absorbance bands seen in the other pigments, revealing the underlying vibrational fine structure only in SRII (21). Supporting this explanation, when the counterion is weakened by the substitution of Asn for Asp-73, broadening is introduced, diffusing the fine structure (Fig. 1 c and d).
Cells carrying the D73N mutation exhibit a strongly reduced but still detectable taxis response. We conclude that proton transfer is important for forming the signaling state, but some signal is generated without the transfer of the Schiff base proton to Asp-73. This result is analogous to that obtained with visual pigments, in which proton transfer from the Schiff base to Glu-113 is an important factor in stabilizing the G-protein-activating state (14), and other determinants also contribute significantly (34).
Acknowledgments
We thank Walther Stoeckenius and Wouter Hoff for critical reading of the manuscript. This research was supported by National Institutes of Health Grant R01 GM27750 to J.L.S. and Grant R55 GM53149–01A1 to M.A.
ABBREVIATIONS
- SRI
sensory rhodopsin I
- SRII
sensory rhodopsin II
- BR
bacteriorhodopsin
References
- 1.Takahashi T, Tomioka H, Kamo N, Kobatake Y. FEMS Microbiol Lett. 1985;28:161–164. [Google Scholar]
- 2.Tomioka H, Takahashi T, Kamo N, Kobatake Y. Biochem Biophys Res Commun. 1986;139:389–395. doi: 10.1016/s0006-291x(86)80003-1. [DOI] [PubMed] [Google Scholar]
- 3.Yan B, Takahashi T, Spudich J L. Biochemistry. 1991;30:10686–10692. doi: 10.1021/bi00108a012. [DOI] [PubMed] [Google Scholar]
- 4.Engelhard M, Scharf B, Siebert F. FEBS Lett. 1996;395:195–198. doi: 10.1016/0014-5793(96)01041-1. [DOI] [PubMed] [Google Scholar]
- 5.Braiman M S, Mogi T, Marti T, Stern L J, Khorana H G, Rothschild K J. Biochemistry. 1988;27:8516–8520. doi: 10.1021/bi00423a002. [DOI] [PubMed] [Google Scholar]
- 6.Hoff W D, Jung K-H, Spudich J L. Annu Rev Biophys Biomolec Struct. 1997;26:221–256. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- 7.Rath P, Olson K D, Spudich J L, Rothschild K J. Biochemistry. 1994;33:5600–5606. doi: 10.1021/bi00184a032. [DOI] [PubMed] [Google Scholar]
- 8.Bogomolni R A, Stoeckenius W, Szundi I, Perozo E, Olson K D, Spudich J L. Proc Natl Acad Sci USA. 1994;91:10188–10192. doi: 10.1073/pnas.91.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spudich J L. Cell. 1994;79:747–750. doi: 10.1016/0092-8674(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 10.Rath P, Spudich E N, Neal D D, Spudich J L, Rothschild K J. Biochemistry. 1996;35:6690–6696. doi: 10.1021/bi9600355. [DOI] [PubMed] [Google Scholar]
- 11.Zhang W, Brooun A, Müller M M, Alam M. Proc Natl Acad Sci USA. 1996;93:8230–8235. doi: 10.1073/pnas.93.16.8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao V J, Spudich J L. Proc Natl Acad Sci USA. 1992;89:11915–11919. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson P R, Cohen G, Oprian D D. Neuron. 1992;9:719–725. doi: 10.1016/0896-6273(92)90034-b. [DOI] [PubMed] [Google Scholar]
- 14.Rao R, Oprian D D. Annu Rev Biophys Biomol Struct. 1996;25:287–314. doi: 10.1146/annurev.bb.25.060196.001443. [DOI] [PubMed] [Google Scholar]
- 15.Yao V J, Spudich E N, Spudich J L. J Bacteriol. 1994;176:6931–6935. doi: 10.1128/jb.176.22.6931-6935.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen B, Przybyla A E. Biotechniques. 1994;17:657–659. [PubMed] [Google Scholar]
- 17.Krebs M P, Spudich E N, Spudich J L. Protein Expression Purif. 1995;6:780–788. doi: 10.1006/prep.1995.0009. [DOI] [PubMed] [Google Scholar]
- 18.Spudich E N, Hasselbacher C A, Spudich J L. J Bacteriol. 1988;170:4280–4285. doi: 10.1128/jb.170.9.4280-4285.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Brooun A, McCandless J, Banda P, Alam M. Proc Natl Acad Sci USA. 1996;93:4649–4654. doi: 10.1073/pnas.93.10.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spudich E N, Sundberg S A, Manor D, Spudich J L. Proteins. 1986;1:239–246. doi: 10.1002/prot.340010306. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi T, Yan B, Mazur P, Derguini F, Nakanishi K, Spudich J L. Biochemistry. 1990;29:8467–8474. doi: 10.1021/bi00488a038. [DOI] [PubMed] [Google Scholar]
- 22.Kataoka M, Mihara K, Kamikubo H, Needleman R, Lanyi J K, Tokunaga F. FEBS Lett. 1993;333:111–113. doi: 10.1016/0014-5793(93)80385-8. [DOI] [PubMed] [Google Scholar]
- 23.Tittor J, Schweiger D, Oesterhelt D, Bamberg E. Biophys J. 1994;67:1682–1690. doi: 10.1016/S0006-3495(94)80642-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varo G, Zimanyi L, Fan X, Sun L, Needleman R, Lanyi J K. Biophys J. 1995;68:2062–2072. doi: 10.1016/S0006-3495(95)80385-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alam M, Lebert M, Oesterhelt D, Hazelbauer G L. EMBO J. 1989;8:631–639. doi: 10.1002/j.1460-2075.1989.tb03418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spudich E N, Takahashi T, Spudich J L. Proc Natl Acad Sci USA. 1989;86:7746–7750. doi: 10.1073/pnas.86.20.7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sundberg S A, Alam M, Lebert M, Spudich J L, Oesterhelt D, Hazelbauer G L. J Bacteriol. 1990;172:2328–2335. doi: 10.1128/jb.172.5.2328-2335.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudolph J, Oesterhelt D. J Mol Biol. 1996;258:548–554. doi: 10.1006/jmbi.1996.0267. [DOI] [PubMed] [Google Scholar]
- 29.Hazelbauer G L, Harayama S. Int Rev Cytol. 1983;81:33–70. doi: 10.1016/s0074-7696(08)62334-7. [DOI] [PubMed] [Google Scholar]
- 30.Lanyi J K. Nature (London) 1995;375:461–463. doi: 10.1038/375461a0. [DOI] [PubMed] [Google Scholar]
- 31.Spudich J L, Lanyi J K. Curr Opin Cell Biol. 1996;8:452–457. doi: 10.1016/s0955-0674(96)80020-2. [DOI] [PubMed] [Google Scholar]
- 32.Fodor S P, Gebhard R, Lugtenburg J, Bogomolni R A, Mathies R A. J Biol Chem. 1989;264:18280–18283. [PubMed] [Google Scholar]
- 33.Mathies R A, Lin S W, Ames J B, Pollard W T. Annu Rev Biophys Biophys Chem. 1991;20:491–518. doi: 10.1146/annurev.bb.20.060191.002423. [DOI] [PubMed] [Google Scholar]
- 34.Fahmy K, Siebert F, Sakmar T P. Biophys Chem. 1995;56:171–181. doi: 10.1016/0301-4622(95)00030-2. [DOI] [PubMed] [Google Scholar]