Abstract
Three small nucleolar RNAs (snoRNAs), E1, E2 and E3, have been described that have unique sequences and interact directly with unique segments of pre-rRNA in vivo. In this report, injection of antisense oligodeoxynucleotides into Xenopus laevis oocytes was used to target the specific degradation of these snoRNAs. Specific disruptions of pre-rRNA processing were then observed, which were reversed by injection of the corresponding in vitro-synthesized snoRNA. Degradation of each of these three snoRNAs produced a unique rRNA maturation phenotype. E1 RNA depletion shut down 18 rRNA formation, without overaccumulation of 20S pre-rRNA. After E2 RNA degradation, production of 18S rRNA and 36S pre-rRNA stopped, and 38S pre-rRNA accumulated, without overaccumulation of 20S pre-rRNA. E3 RNA depletion induced the accumulation of 36S pre-rRNA. This suggests that each of these snoRNAs plays a different role in pre-rRNA processing and indicates that E1 and E2 RNAs are essential for 18S rRNA formation. The available data support the proposal that these snoRNAs are at least involved in pre-rRNA processing at the following pre-rRNA cleavage sites: E1 at the 5′ end and E2 at the 3′ end of 18S rRNA, and E3 at or near the 5′ end of 5.8S rRNA.
Eukaryotic mature rRNA is generated by a complex series of processing steps (for review, see refs. 1–4). rRNA processing requires several small nucleolar RNA (snoRNA) species (for review, see refs. 5–7). Processing at some pre-rRNA cleavage sites requires more than one snoRNA species, and some snoRNAs function at more than one pre-rRNA processing site. E1/U17 (8–13) and E2 and E3 (8–11) snoRNAs do not belong to the main class of snoRNAs, since they lack the conserved C and D boxes that are present in most snoRNAs, do not show obvious sequence homology with any other snoRNAs, and do not associate with the nucleolar protein fibrillarin. Several snoRNAs, including E1, E2, and E3, have the triplet ACA near the 3′ end (14). These three snoRNAs are housekeeping RNAs since they are present in all tissues tested (8). The genes for E1, E2, and E3 RNAs, among other snoRNAs, reside in introns of protein-encoding genes (5). The coding sequences for E1 (10, 12, 13), E2 (15), and E3 (10, 16) RNAs lie in introns of genes for the cell cycle regulatory protein named RCC1, a laminin-binding protein, and protein synthesis initiation factor 4AII, respectively. The nucleolar localization of E1, E2, and E3 RNAs and their direct contact with pre-rRNA in vivo (9) suggest that they are involved in ribosome biogenesis. They may play new roles in ribosome formation, since they interact directly in vivo with unique segments of pre-rRNA. They psoralen-crosslink to four different sites of pre-rRNA in vivo: in the 5′ external transcribed spacer (5′ETS) and 18S rRNA for E1, 28S rRNA for E2, and 18S rRNA for E3 (9). Unlike the snoRNAs that determine the location of ribose methylations in pre-rRNA (17–19), E1, E2, and E3 RNAs do not show extensive complementarity (>12 bases) to sites of ribose methylation in pre-rRNA and lack sequence boxes D and D′. These three snoRNAs, in cell extracts, have accessible sites that make possible their specific degradation by antisense oligonucleotide-targeted digestion with RNase H (9).
Yeast is a very valuable organism to study the functions of various snoRNAs in vivo (5, 6), but yeast homologs of E1, E2, or E3 RNAs have not been detected. Oligodeoxynucleotide-targeted RNA degradation after microinjection into Xenopus laevis oocytes is an important system to study functions of vertebrate small nuclear RNAs (20, 21). It made possible the analysis of the role of three vertebrate snoRNAs, U3, U8, and U22, in pre-rRNA processing (22–25). We used this approach to test whether E1, E2, and E3 RNAs are involved in rRNA processing. The results indicate that they are involved and suggest that each of these snoRNAs plays a different role in rRNA processing.
MATERIALS AND METHODS
Oligodeoxynucleotides.
The sequences of the oligodeoxynucleotides used are as follows: E1 1–22, 5′-CCTCATGAGATATCCACGTTGG-3′; E1 23–45, 5′-CAGGACAGAGCCCATGAGAGTAA-3′; E1 50–71, 5′-CACAGGGCGACGCTCCCATACG-3′; E1 73–92, 5′-GGGATTATACGTCACTGCGCC-3′; E1 84–101, 5′-TTGTGGAAGGGGATTATAC-3′; E1 103–127, 5′-GCCGGGGACATGCTTGTTCTCCAAC-3′; E1 128–151, 5′-TGCTGCCCACACCAGCCGAATG-3′; E1 158–178, 5′-GGCTCTGGGAAGTTGTAGGAAT-3′; E1 179–198, 5′-CTCCCCAGTCACTGCCCGAG-3′; E1 199–217, 5′-TGTATCCTGCATGGTTTGT-3′; E2 1–21, 5′-CCAAGTTCTAACTGTGTGCAA-3′; E2 23–45, 5′-TCATTGGCTGAAAAGGCCTCAGC-3′; E2 39–62, 5′-CATCCTACACTCAGAGTTCATTGG-3′; E2 62–82, 5′-CTGCCATGTTAATGTAGCACAC-3′; E2 84–100, 5′-TGGCGTTAGCGAAAAGT-3′; E2 101–121, 5′-CTGGAGCTCTAAAGCTCCTTG-3′; E2 121–143, 5′-AATTACTATGAAACTCCAGTCAC-3′; E2 139–155, 5′-AGCTGTGGCAAGAATTA-3′; E3 1–14, 5′-GTAACTAATCCTGC-3′; E3 15–35, 5′-ACAGCACTGCCCAGATATATT-3′; E3 36–54, 5′-TAGCAGGGGGAACGACAAC-3′; E3 55–77, 5′-ATAGAAGGAATCAACTTACTTTC-3′; E3 78–100, 5′-TCTGCTATAGAGAACAGCCAGGA-3′; E3 103–125, 5′-AATTGTTTGAGACCAAGCGTTCC-3′; control, 5′-GCGGAATTCAAAAAAAAAAAAAAAAAA-3′.
RNA Synthesis.
Each frog snoRNA was synthesized in vitro with bacteriophage T7 RNA polymerase and was capped at the 5′ end by including the cap analog m7G(5′)ppp(5′)G in the transcription reaction. The sequence of Xenopus laevis E1 RNA was reported earlier, and the f sequence was used since it was shown to be expressed (13). The sequences of frog E2 and E3 RNAs were from cDNA and genomic DNA (26).
Oocyte Injection and RNA Analysis.
Frog oocytes were injected twice, 2, 6, or 16 h apart, with the indicated oligodeoxynucleotide (45 ng per oocyte). Then, they either were incubated for 6 h and injected with [α-32P]GTP or were incubated for 2 h, injected with the indicated in vitro-transcribed frog snoRNA (0.8 ng per oocyte) or water, then incubated for 8 or 16 h, and injected with [α-32P]GTP. Radiolabeling was for 8 or 16 h. Injections were 20 nl per oocyte, into the cytoplasm of Xenopus laevis stage 5 and 6 oocytes (23, 24, 27), and oocyte incubations were at 19°C. Whole oocyte RNA was extracted with phenol and chloroform in the presence of 1% SDS. Small RNAs were fractionated by 10% polyacrylamide/7 M urea gel electrophoresis and electroblotted to ZetaProbe GT (Bio-Rad) membranes. Agarose gel electrophoresis in the presence of formaldehyde was as described (28).
RESULTS
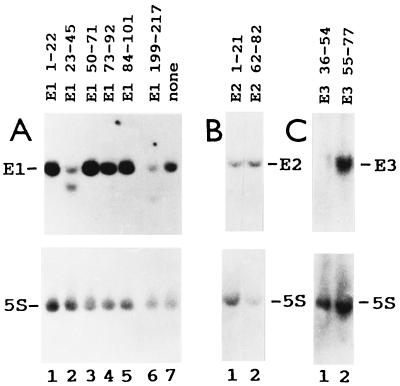
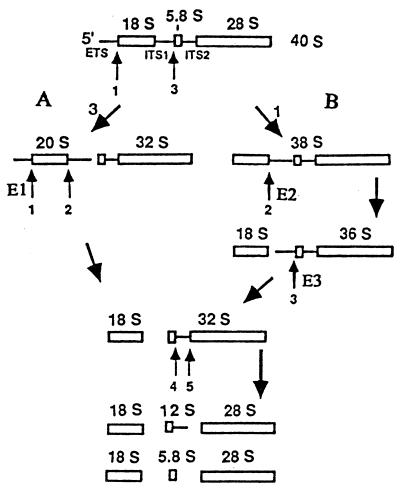
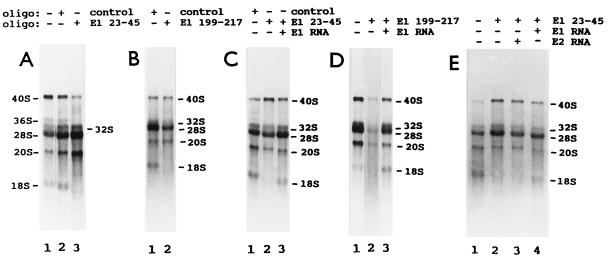
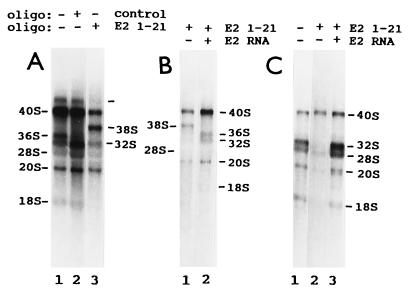
Antisense oligodeoxynucleotides specifically target the efficient degradation by RNase H of human E1, E2, and E3 RNAs in cell extracts (9). We tested first whether this occurs with frog E1, E2, and E3 RNAs in vivo, injecting into the cytoplasm of oocytes three series of complementary oligonucleotides that span the entire lengths of these snoRNAs (Fig. 1). RNA breakdown was monitored by Northern blot analysis, hybridizing first with the corresponding snoRNA probe and then with a 5S rRNA probe, to control for the number of oocyte equivalents per gel blot lane. Oligonucleotides complementary to frog E1 RNA nucleotide positions 23–45 and 199–217 (E1 23–45 and E1 199–217) targeted the specific degradation of E1 RNA in oocytes (Fig. 2A). (The Xenopus E1, E2, and E3 RNA nucleotide position numbers differ from those of their human counterparts because the snoRNAs have different lengths). Oligonucleotide E2 1–21 targeted the specific breakdown of E2 RNA in oocytes, as indicated by the decreased labeled E2/5S RNA ratio (Fig. 2B). Oligonucleotide E3 36–54 induced the specific hydrolysis of E3 RNA in vivo (Fig. 2C). Several oligonucleotides were tested; only the four identified above resulted in E1, E2, or E3 RNA breakdown. Full degradation of the snoRNA molecules that were first cleaved by RNase H activity (Fig. 2 B and C) was observed earlier with various small nuclear RNA species (9, 21, 25, 27). Next we tested the effect of E1, E2, and E3 RNA degradation on frog oocyte pre-rRNA processing. Oocytes of some frogs cleave 40S pre-rRNA at the first internal transcribed spacer–5.8S rRNA junction (site 3), producing 20S and 32S rRNA intermediates (pre-rRNA processing pathway A; Fig. 3). Oocytes from other frogs have two rRNA maturation pathways. In the second pathway (pathway B), 40S pre-rRNA is cut at the 5′ETS–18S rRNA boundary (site 1), generating 38S pre-rRNA (refs. 22 and 29; Fig. 3). This intermediate is short lived, requires brief labeling for detection, and is cleaved at the 18S rRNA–first internal transcribed spacer junction (site 2) to generate 36S pre-rRNA (ref. 22; Fig. 3). Oligonucleotides that target the degradation of E1, E2, and E3 RNAs, as well as oligonucleotides that do not, were injected into frog oocytes, and rRNA processing was monitored by denaturing gel electrophoresis of newly made rRNA labeled in vivo. All the effects observed with E1, E2, and E3 antisense oligonucleotides were oligonucleotide sequence specific, since they did not occur with any other oligonucleotide tested. Formation of 18S rRNA stopped only after injection of frog oligonucleotides E1 23–45 and E1 199–217 in oocytes that have pathway A or pathways A and B (Fig. 4 A and B). This inhibition was reversed by cytoplasmic injection of in vitro-synthesized frog E1 RNA (Fig. 4 C and D). This “rescue” was snoRNA species specific, since it did not occur when E2 RNA was injected (Fig. 4E). Whole oocyte RNA was usually analyzed, because analysis of nuclear RNA did not reveal additional rRNA bands or other information. After degradation of E1 RNA, 20S pre-rRNA was made but 18S rRNA did not accumulate (Fig. 3). The three lanes of Fig. 4A have different numbers of radiolabeled cell equivalents, as can be seen by the levels of radioactive 28S rRNA. Differences in the number of radiolabeled cell equivalents per gel lane may reflect variations in (i) the amount of damage caused by four injections per oocyte in rescue experiments, (ii) the level of rRNA synthesis in oocytes of similar appearance, and (iii) the number of nanoliters delivered into each oocyte in each injection. Some RNAs in their ribonucleoprotein particles are more accessible than others to antisense oligodeoxynucleotide-targeted degradation. For example, depletion of U8 and U22 snoRNAs requires a single injection of oligonucleotide (24, 25), but two injections are needed for E1, E2, and E3 RNAs. The radioactive 20S/28S rRNA ratio did not increase substantially after injection of oligonucleotide E1 23–45 (Fig. 4A, lanes 2 and 3). Fig. 4A, lanes 2 and 3, shows a faint smear whose size falls between 40S and 36S pre-rRNAs. The level of this RNA relative to 28S rRNA did not increase appreciably after injection of oligonucleotide E1 23–45 (Fig. 4A, lanes 2 and 3). This smear differs from the sharp band of 38S pre-rRNA (Fig. 5 A, lane 3, and B, lane 1). We do not know whether the decrease in 36S pre-rRNA after injection of oligonucleotide E1 23–45 (Fig. 4A) is E1 RNA specific, since there is no evidence that it is restored by E1 RNA.
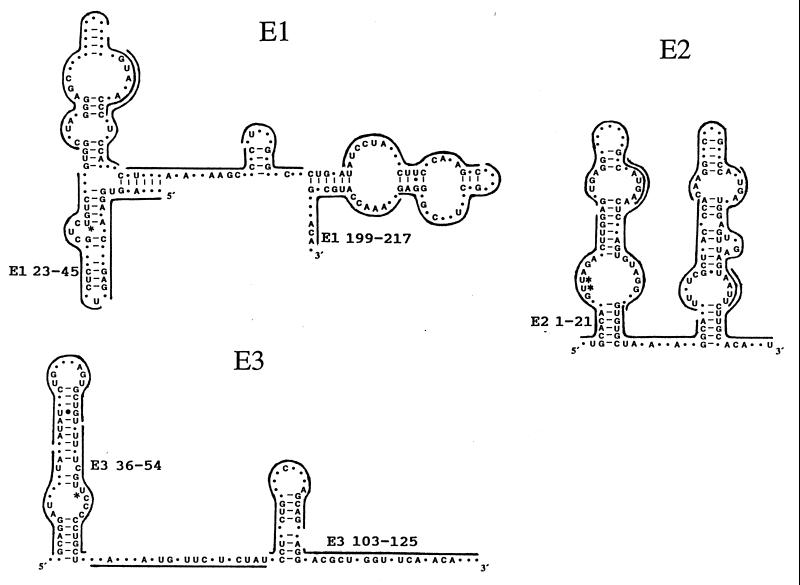
Figure 1.
Antisense oligodeoxynucleotides that were tested in vivo. Vertebrate consensus sequences and secondary structures of E1, E2 and E3 RNAs we determined earlier (26). Lines span the lengths of the complementary oligonucleotides that were tested. Oligonucleotides that affected pre-rRNA processing are labeled. Asterisks indicate the location of psoralen adducts that may be crosslinked to pre-rRNA in psoralen crosslinking experiments in vivo (9).
Figure 2.
The degradation of E1, E2, and E3 RNAs is specifically targeted by antisense oligodeoxynucleotides in frog oocytes. Xenopus laevis oocytes were injected twice with the oligodeoxynucleotides indicated and then were incubated for 16 h. Some oocytes were not injected with any oligonucleotides (A, lane 7). Whole oocyte RNA was extracted, fractionated by polyacrylamide gel electrophoresis, and blotted. The blot was hybridized first with a probe for E1, E2, and E3 RNAs (A, B, and C, respectively, Upper) and then with a probe for 5S rRNA (Lower).
Figure 3.
Pre-rRNA processing sites in which E1, E2, and E3 RNAs are proposed to be involved. Pre-rRNA processing pathways A and B of Xenopus laevis oocytes differ in the temporal order of cleavages; the final products of both pathways are the mature 18S, 5.8S, and 28S rRNAs. Numbers 1–5 indicate processing cleavage sites.
Figure 4.
Specific disruption of frog oocyte pre-rRNA processing induced by E1 RNA degradation and reversed by E1 RNA. Xenopus laevis oocytes were injected twice with the indicated antisense oligodeoxynucleotides or the control oligonucleotide. Some oocytes were incubated and then injected with [α-32P]GTP (A and B). Other oocytes were incubated, injected with the indicated snoRNA or water, incubated, and injected with [α-32P]GTP (C–E). After injection of [α-32P]GTP, all oocytes were incubated for 16 h. Some oocytes were not injected with any oligonucleotides (A, D, and E, lanes 1). Whole oocyte RNA was extracted and analyzed by formaldehyde/agarose gel electrophoresis. RNA from a single oocyte was loaded on each gel lane. A–E are five electrophoreses. The oocytes in A show both pre-rRNA processing pathways A and B. The oocytes in B–E have only pre-rRNA processing pathway A.
Figure 5.
Specific disruption of frog oocyte pre-rRNA processing induced by E2 RNA degradation and reversed by E2 RNA. The experiments were done as indicated in Fig. 4. After injections of oligonucleotides and [α-32P]GTP, oocytes were incubated for 8 (A and B) or 16 (C) h. The oocytes in C have only pre-rRNA processing pathway A. The oocytes in A and B show both pre-rRNA processing pathways A and B.
E2 RNA degradation shut down production of two rRNA species: (i) 18S rRNA in oocytes that have pathway B or only A and (ii) 36S pre-rRNA only in oocytes that have pathway B (Fig. 5). The 38S pre-rRNA is not seen after long-labeling times (22–24). In long-labeling experiments, a 38S rRNA appeared only after destruction of E2 RNA (Fig. 5). It has the properties of the known 38S pre-rRNA. (i) It has the size of 38S pre-rRNA. (ii) It accumulated only in oocytes that have pathway B (Fig. 5), and it is known that 38S pre-rRNA is generated only in pathway B (22). (iii) It accumulated only when 36S pre-rRNA formation was shut down, and its accumulation stopped when 36S pre-rRNA production was restored (Fig. 5). This is as expected, since 38S pre-rRNA is the immediate precursor of 36S pre-rRNA (22). Finally, the only way to generate an rRNA species the size of 38S pre-rRNA, using known processing sites, is by cleaving 40S pre-rRNA at site 1, as in the formation of 38S pre-rRNA in normal oocytes (Fig. 3). Injection of synthetic E2 RNA restored the production of 18S rRNA and 36S pre-rRNA and reversed the accumulation of 38S rRNA (Fig. 5B and C). The short incubation with 32P in the experiment in Fig. 5B did not radiolabel 18S rRNA well, even in control oocytes (data not shown). The rRNA species larger than 40S rRNA, present at low levels in control oocytes (Fig. 5A, lanes 1 and 2), may be a short-lived precursor of the known frog 40S pre-rRNA that is longer at one or both termini. The blockage of 18S rRNA accumulation may be more complete than the extent of E1 or E2 RNA degradation that triggered it (Figs. 2, 4, and 5). It is possible that in frog oocytes some E1 and E2 RNA molecules are not functioning but stored in a less accessible form and that these molecules do not need to be broken down for a virtually full interruption of 18S rRNA accumulation.
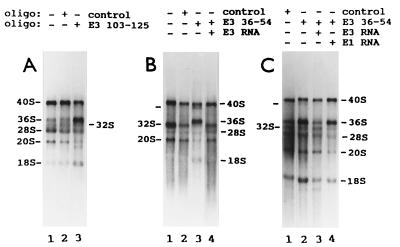
Human oligonucleotide E3 116–135 targets the specific breakdown of human E3 RNA in cell extracts (9). Frog oligonucleotide E3 103–125 and human oligonucleotide E3 116–135 span the same region of E3 RNA (refs. 8 and 26; Fig. 1). Only two of the oligonucleotides complementary to frog E3 RNA, E3 36–54 and E3 103–125, affected oocyte pre-rRNA maturation, and both had the same effects. After E3 RNA degradation, oocytes that normally make 36S pre-rRNA overaccumulated this pre-rRNA relative to its immediate product, 32S pre-rRNA (Fig. 6A). This was reversed by injection of E3 RNA but not of E1 RNA (Fig. 6C). E3 RNA depletion in oocytes that normally do not produce 36S pre-rRNA (i) induced the formation and overaccumulation of this pre-rRNA and (ii) shut down 32S rRNA production (Fig. 6B). Both effects were reversed by injection E3 RNA (Fig. 6B). [The only way to produce an rRNA species the size of 36S pre-rRNA, using known cleavage sites, is by cutting 38S or 40S pre-rRNA at site 2 (ref. 22; Fig. 3)]. A new pre-rRNA band, slightly smaller than 40S pre-rRNA, accumulated in substantial amounts in E3-depleted oocytes that have only pathway A, and this was reversed by injection of E3 RNA (Fig. 6B, marked in lane 3). E3-depleted oocytes occasionally showed an rRNA smaller than 20S pre-rRNA (Fig. 6A, lane 3) and an rRNA smaller than 18S rRNA (Fig. 6C, lanes 2 and 3). These rRNA bands were not studied further because they were not induced reproducibly by E3 RNA degradation or were not lost consistently in E3 RNA rescue experiments (Fig. 6 and other experiments not shown). The effect of E3 RNA degradation on the level of 36S pre-rRNA was reproducible, regardless of whether any of these three new rRNA bands appeared or not (Fig. 6 and data not shown). Fig. 6 may appear to show some changes in the radioactive 20S/28S and 18S/28S rRNA ratios. These shifts were not reproducible, substantial, or specifically reversed by E3 RNA in these and other experiments not shown. RNA analysis by polyacrylamide gel electrophoresis did not reveal any disturbance in processing of small pre-rRNA after degradation of E1, E2, and E3 RNAs (data not shown). This suggests that these snoRNAs are not involved in the processing at the 3′ end of the 5.8S rRNA sequence in 12S pre-rRNA that generates mature 5.8S rRNA (Fig. 3).
Figure 6.
Specific disruption of frog oocyte pre-rRNA processing induced by E3 RNA degradation and reversed by E3 RNA. The experiments were done as indicated in Fig. 4. After injections of oligonucleotides and [α-32P]GTP, oocytes were incubated for 16 (A and C) or 8 (B) h. The oocytes in A show both pre-rRNA processing pathways A and B. The oocytes in B and C have only pre-rRNA processing pathway A.
Nonspecific effects can be a potential problem in antisense oligonucleotide targeting experiments (e.g., ref. 30). Two observations indicate that the rRNA maturation phenotypes observed in this work are specific for each snoRNA species. (i) The same unique phenotype was produced for each targeted snoRNA by oligonucleotides complementary to different segments of the snoRNA, but to no other oligonucleotide. (ii) Each phenotype was specifically reversed by injection of the corresponding snoRNA species.
DISCUSSION
To our knowledge, the present results are the first to show that E2 RNA has a function and that it involves pre-rRNA processing. This is the first evidence that E1 and E3 RNAs have functions in rRNA maturation, which was obtained in vivo, after degradation of the snoRNA in ribonucleoprotein particle form, and with phenotypes that are specifically reversed by the corresponding snoRNA. We report also the first use of antisense oligonucleotides to specifically degrade these three snoRNAs in vivo and demonstrate that destruction of anyone of these three snoRNAs generates a unique alteration in pre-rRNA processing that is specifically reversed by the appropriate snoRNA species.
When rRNA processing at both sites 1 and 2 is blocked (Fig. 3), 18S rRNA formation stops and its immediate precursor, 20S pre-rRNA, overaccumulates relative to other pre-rRNA and rRNA species in U22 RNA-depleted oocytes (25). (Processing blocked at both ends of 20S pre-rRNA might produce a more stable RNA.) Both after E2 RNA and E1 RNA degradation, 18S rRNA production ceased but 20S pre-rRNA did not overaccumulate, indicating that either site 1 or site 2 was affected in each case. Pre-rRNA processing at both sites 1 and 2 requires U22 snoRNA (25). If both sites required E1 or E2 RNA for processing, degradation of E1 or E2 RNA would be expected to generate a pre-rRNA maturation phenotype identical to that of U22 RNA depletion, but this did not occur. Cleavage of 38S pre-rRNA at site 2 makes 36S pre-rRNA (22). After E2 RNA destruction, 36S pre-rRNA formation shut down and 38S pre-rRNA accumulated, each event indicating that site 2 was blocked. Collectively, these results indicate that E2 RNA is at least involved in rRNA processing at the 3′ end of 18S rRNA (site 2). As mentioned above, rRNA processing in E1-depleted oocytes is interrupted at either site 1 or site 2. Apparently it is not site 2 since 38S pre-rRNA did not accumulate. These observations strongly suggest that E1 RNA is at least involved in rRNA processing at the 5′ end of 18S rRNA (site 1). The present results support the idea that the cleavages at the ends of frog oocyte 18S rRNA are not tightly coupled. Inhibition of rRNA processing at site 1 might have resulted in overaccumulation of 40S pre-rRNA in E1-depleted oocytes that have pathway B, but this was not detected. Perhaps 40S pre-rRNA that is not cleaved at site 1 is either very unstable or is processed at site 3. The 36S pre-rRNA intermediate generates 32S pre-rRNA by cleavage at site 3 (ref. 22; Fig. 3) that is located at or near the 5′ terminus of 5.8S rRNA (25, 31, 32). During specific E3 RNA depletion, oocytes that normally make 36S pre-rRNA overaccumulated this pre-rRNA relative to 32S pre-rRNA, indicating that site 3 was affected. Oocytes that have only pathway A do not produce 36S pre-rRNA and all of their 32S pre-rRNA is generated by cleavage of 40S pre-rRNA at site 3 (Fig. 3). The specific depletion of E3 RNA in oocytes that have only pathway A induced the formation and overaccumulation of 36S pre-rRNA and shut down 32S rRNA production. Each of these two observations indicates that site 3 was blocked. These results indicate that E3 RNA is involved in rRNA processing at site 3 (at or near the 5′ end of 5.8S rRNA). The continued formation of some mature 28S rRNA after E3 RNA degradation is compatible with several interpretations. First, E3 RNA might be essential for rRNA processing but 28S rRNA might still be made by cleavage at site 5 of intermediates blocked at site 3, or a full phenotype might not be seen unless virtually all the E3 RNA molecules in the oocyte are destroyed, and this may not have happened in these experiments. Alternatively, E3 RNA might facilitate rRNA processing. Verification of the termini of the pre-rRNA and rRNA molecules made after degradation of E1, E2, and E3 RNAs is not expected to reveal a novel cleavage site located far from the known processing sites or to alter substantially the present conclusions, for the following reasons. (i) Depletion of various snoRNA species in yeast and frog oocytes has shown many blockages of known pre-rRNA processing sites, but no novel cleavage of pre-rRNA (5, 6). (ii) The rRNA molecules generated after depletion of these three snoRNAs are unlikely to have grossly unexpected sequence spans because the present rRNA maturation phenotypes are internally consistent.
Degradation of E1, E2, or E3 RNA each produces a unique pre-rRNA maturation phenotype, different from those observed after depletion of other snoRNA species that function at rRNA processing sites 1, 2, or 3. Production of mature 18S rRNA stops after degradation of U22, E1 and E2 snoRNAs, but the level of 20S pre-rRNA rises relative to the other pre-rRNA and rRNA species after U22 RNA depletion (25) and not after degradation of E1 or E2 RNAs. E2 is the only known snoRNA species whose degradation induces the accumulation of 38S pre-rRNA. Degradation of E3 RNA, but not of U3 snoRNA (22), induces the appearance of 36S pre-rRNA in frog oocytes that normally do not make this pre-rRNA species. U8 snoRNA depletion blocks formation of mature 28S and 5.8S rRNAs (23). Yeast U14 snoRNA depletion leads to impaired processing of the initial 35S pre-rRNA to 20S pre-rRNA, the precursor to 18S rRNA (33).
E1, E2, and E3 RNAs each is apparently involved in the rRNA processing site discussed above, but the available results do not rule out the possibility that it might also function in another rRNA processing site or step in ribosome formation. However, the roles of E1, E2, and E3 RNAs are not identical since the degradation of each snoRNA leads to a different disturbance of rRNA processing. These snoRNAs psoralen-crosslink to four different pre-rRNA sites in vivo: E1 within 5′ETS positions 697-1163 and 18S rRNA positions 664-1021; E2 within 18S rRNA positions 3282–3667; and E3 within 18S rRNA positions 1021–1639 (9). These direct interactions with pre-rRNA may be part of their roles at these pre-rRNA processing sites or may reflect functions in additional steps of ribosome formation. How these snoRNAs are involved in pre-rRNA processing is unknown. They could participate in folding of pre-rRNA or in the encounter of pre-rRNA cleavage sites with nucleases. Some of the molecules of E1, E2, and E3 RNAs associate with very large complexes in vivo (9). Several snoRNAs and proteins are required for pre-rRNA processing and it has been proposed that they assemble into a large complex needed for rRNA maturation (5, 6). E1, E2, and E3 RNAs may be additional components of this complex. These three snoRNAs injected into the cytoplasm restored normal pre-rRNA processing after snoRNA degradation, suggesting that they migrate to their functional sites within the nucleolus. E1 and E3 RNAs were reported to facilitate mouse 5′ETS rRNA processing in a cell-free system (34). If this occurred in vivo, it would be difficult to detect in frog oocytes, since only ∼1% of Xenopus oocyte pre-rRNA is cleaved at the 5′ETS processing site (35, 36).
Analysis of frog small nucleolar ribonucleoprotein particles in vivo revealed four new accessible sites, targeted by oligonucleotides E1 23–45, E1 199–217, E2 1–21, and E3 36–54, that had not been detected in human small nucleolar ribonucleoprotein particles in vitro (9). It also confirmed an accessible site that was observed earlier in human cell extracts (9), that was shown by frog oligonucleotide E3 103–125, equivalent to human oligonucleotide E3 116–135. Five accessible segments were detected in these three snoRNAs in whole cells. These sites may be functional since (i) accessible segments of small RNAs are usually functional (37, 38), (ii) most of the sequence at each accessible site is evolutionarily conserved (ref. 26; Fig. 1) and conserved sequences usually have a role, and (iii) they include residues that may be psoralen-crosslinked to pre-rRNA in vivo (ref. 9; Fig. 1). Our work identified in these three snoRNAs accessible conserved sequences that may be involved in RNA–RNA intermolecular interactions and inaccessible conserved sequences that may participate in RNA–protein long-term interactions. Our results suggest that each of these three snoRNAs has unique conserved domains (Fig. 1).
Acknowledgments
We thank Francesco Amaldi for the Xenopus laevis E1 genomic DNA clone, James W. Brown for the folded RNA structures in Fig. 1, and Brenda A. Peculis and Kazimierz T. Tycowski for advice on the oocyte depletion–rescue procedures. This work was supported by a grant from the National Institutes of Health.
Footnotes
Abbrevations: snoRNA, small nucleolar RNA; 5′ETS, 5′ external transcribed spacer.
References
- 1.Eichler E D, Craig N. Prog Nucleic Acid Res Mol Biol. 1994;49:197–239. doi: 10.1016/s0079-6603(08)60051-3. [DOI] [PubMed] [Google Scholar]
- 2.van Nues R W, Venema J, Rientjes J M J, Dirks-Mulder A, Raué H A. Biochem Cell Biol. 1995;73:789–801. doi: 10.1139/o95-087. [DOI] [PubMed] [Google Scholar]
- 3.Venema J, Tollervey D. Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- 4.Sollner-Webb B, Tyc K T, Steitz J A. In: Ribosomal RNA: Structure, Evolution, Gene Expression, and Function in Protein Synthesis. Zimmermann R A, Dahlberg A E, editors. Boca Raton, FL: CRC; 1996. pp. 469–490. [Google Scholar]
- 5.Maxwell E S, Fournier M J. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 6.Lafontaine D, Tollervey D. Biochem Cell Biol. 1995;73:803–812. doi: 10.1139/o95-088. [DOI] [PubMed] [Google Scholar]
- 7.Gerbi S A. Biochem Cell Biol. 1995;73:845–858. doi: 10.1139/o95-092. [DOI] [PubMed] [Google Scholar]
- 8.Ruff E A, Rimoldi O J, Raghu B, Eliceiri G L. Proc Natl Acad Sci USA. 1993;90:635–638. doi: 10.1073/pnas.90.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rimoldi O J, Raghu B, Nag M K, Eliceiri G L. Mol Cell Biol. 1993;13:4382–4390. doi: 10.1128/mcb.13.7.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nag M K, Thai T T, Ruff E A, Selvamurugan N, Kunnimalaiyaan M, Eliceiri G L. Proc Natl Acad Sci USA. 1993;90:9001–9005. doi: 10.1073/pnas.90.19.9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selvamurugan N, Nag M K, Eliceiri G L. Biochim Biophys Acta. 1995;1260:230–234. doi: 10.1016/0167-4781(94)00222-o. [DOI] [PubMed] [Google Scholar]
- 12.Kiss T, Filipowicz W. EMBO J. 1993;12:2913–2920. doi: 10.1002/j.1460-2075.1993.tb05953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cecconi F, Mariottini P, Loreni F, Pierandrei-Amaldi P, Campioni N, Amaldi F. Nucleic Acids Res. 1994;22:732–741. doi: 10.1093/nar/22.5.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balakin A G, Smith L, Fournier M J. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 15.Selvamurugan N, Eliceiri G L. Genomics. 1995;30:400–401. [PubMed] [Google Scholar]
- 16.Séraphin B. Trends Biochem Sci. 1993;18:330–331. doi: 10.1016/0968-0004(93)90067-w. [DOI] [PubMed] [Google Scholar]
- 17.Kiss-László Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 18.Nicoloso M, Qu L-H, Michot B, Bachellerie J-P. J Mol Biol. 1996;260:178–195. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- 19.Tycowski K T, Smith C M, Shu M-D, Steitz J A. Proc Natl Acad Sci USA. 1996;93:14480–14485. doi: 10.1073/pnas.93.25.14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan S-Q, Prives C. Science. 1988;241:1328–1331. doi: 10.1126/science.2970672. [DOI] [PubMed] [Google Scholar]
- 21.Pan Z-Q, Ge H, Fu X-Y, Manley J L, Prives C. Nucleic Acids Res. 1989;17:6553–6568. doi: 10.1093/nar/17.16.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savino R, Gerbi S A. EMBO J. 1990;9:2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peculis B A, Steitz J A. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- 24.Peculis B A, Steitz J A. Genes Dev. 1994;8:2241–2255. doi: 10.1101/gad.8.18.2241. [DOI] [PubMed] [Google Scholar]
- 25.Tycowski K T, Shu M-D, Steitz J A. Science. 1994;266:1558–1561. doi: 10.1126/science.7985025. [DOI] [PubMed] [Google Scholar]
- 26.Selvamurugan N, Joost O H, Haas E S, Brown J W, Galvin N J, Eliceiri G L. Nucleic Acids Res. 1997;25:1591–1596. doi: 10.1093/nar/25.8.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prives C, Foukal D. Methods Cell Biol. 1991;36:185–210. doi: 10.1016/s0091-679x(08)60278-2. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 29.Wellauer P K, Dawid I B. J Mol Biol. 1974;89:379–395. doi: 10.1016/0022-2836(74)90526-9. [DOI] [PubMed] [Google Scholar]
- 30.Dunbar D A, Ware V C, Baserga S J. RNA. 1996;2:324–333. [PMC free article] [PubMed] [Google Scholar]
- 31.Bowman L H, Goldman W E, Goldberg G I, Hebert M B, Schlessinger D. Mol Cell Biol. 1983;3:1501–1510. doi: 10.1128/mcb.3.8.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hadjiolova K V, Georgiev O I, Nosikov V V, Hadjiolov A A. Biochim Biophys Acta. 1984;782:195–201. doi: 10.1016/0167-4781(84)90024-1. [DOI] [PubMed] [Google Scholar]
- 33.Li H V, Zagorski J, Fournier M J. Mol Cell Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enright C A, Maxwell E S, Eliceiri G L, Sollner-Webb B. RNA. 1996;2:1094–1099. [PMC free article] [PubMed] [Google Scholar]
- 35.Savino R, Gerbi S A. Biochimie. 1991;73:805–812. doi: 10.1016/0300-9084(91)90060-e. [DOI] [PubMed] [Google Scholar]
- 36.Mougey E B, O’Reilly M, Osheim Y, Miller O L, Jr, Beyer A, Sollner-Webb B. Genes Dev. 1993;7:1609–1619. doi: 10.1101/gad.7.8.1609. [DOI] [PubMed] [Google Scholar]
- 37.Steitz J A, Black D L, Gerke V, Parker K A, Krämer A, Frendeway D, Keller W. In: Structure and Function of Major and Minor snRNPs. Birnstiel M, editor. Heidelberg: Springer; 1988. pp. 115–154. [Google Scholar]
- 38.Baserga S J, Steitz J A. In: The RNA World. Gesteland R F, Atkins J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 359–381. [Google Scholar]