Abstract
By using mRNA polymerase chain reaction differential display technique (DDPCR), we have identified one early responsive cDNA fragment, TDD5, from a 5α-reductase-deficient T cell hybridoma. The DDPCR profiles of TDD5 suggest that its expression can be repressed by testosterone (T) within 2 hr. More importantly, both DDPCR and Northern blot analysis further demonstrated that the expression of TDD5 was differentially repressed by T and dihydrotestosterone (DHT) at the mRNA level. To our knowledge, this is the first androgen target gene to show a preference in response to T over DHT in cell culture. TDD5 is expressed in several tissues with particular abundance in kidney. Full-length TDD5 cDNA (2,916 bp) encodes a protein with a calculated molecular weight of 42,000. Finally, our animal studies further confirm that TDD5 mRNA levels can be repressed to the basal level 8 hr after DHT administration. The isolation and characterization of the early-responsive androgen target gene TDD5 and the fact that TDD5 mRNA level can be differentially regulated by T and DHT may provide a useful tool to study the molecular mechanism of androgen preference on target gene regulation.
Keywords: early responsive gene, T cell, 5α-reductase
Androgen plays an essential role in the determination of sexual differentiation, development of both internal and external sex accessory organs, and some general but distinct characteristics between sexes, e.g., muscle development and hair growth patterns in certain areas (1, 2). Androgens are synthesized and secreted into the blood stream largely in the form of testosterone (T). After entering its target cells, T will either be metabolized by aromatase to estradiol in hypothalamus where mental/social sex determination occurs (3, 4) or be metabolized by 5α-reductase to 5α-dihydrotestosterone (DHT) in most of the male reproductive organs (5). In addition to prior metabolic activation, T may directly activate the development of muscle where very limited 5α-reductase activity is detected (6). From studies of inherited 5α-reductase-deficient patients, investigators have concluded that T is able to stimulate psychosexual behavior and the development of Wolffian duct, muscle, voice deepening, spermatogenesis, and axillary as well as pubic hair growth (7, 8). DHT seems to be necessary for the development of prostate and external genitalia, the male pattern hair growth, and male-form baldness (9). From 5α-reductase insufficiency and the complete form of testicular feminization, in which no trace of androgen function is detected due to a nonfunctional androgen receptor (AR) gene mutation (10), it has been generally accepted that only one AR is responsible for both T and DHT stimulation. This one AR hypothesis was further supported by the fact that only one AR gene has been cloned by various groups (11–14). Although the biological significance of T and DHT is clear, the mechanisms that T and DHT rely on for exerting different functions in various target organs are under intensive investigation at present.
Several studies have been done on experimental animal models to target this question. For example, the development of rat prostate was severely impaired but not completely abolished when they were treated with the 5α-reductase inhibitor finasteride (15). These results suggest that the development of rat prostate is also largely DHT-dependent. However, large amounts of T may achieve the same effect via the same AR. And also, the development of Wolffian-duct-derived organs, with the exception of seminal vesicles (SVs), was not affected by finasteride treatment. In the study of 7α-methyl-19-nortestosterone, researchers have shown that this nonreducible synthetic T affected androgen-dependent muscle mass without affecting either prostate or SV development (16). In summary, these results have confirmed in animals that the development of muscle and most Wolffian-duct-derived organs is T-dependent, whereas the development of prostate and SVs is DHT-dependent.
Deslypere et al. (17) have demonstrated that DHT at a concentration that is 10-fold less than T can induce the same mouse mammary tumor virus–chloramphenicol acetyltransferase (MMTV-CAT) reporter gene expression activity in Chinese hamster ovary (CHO) cells. The limited 5α-reductase activity found in CHO cells and also the apparent Kd values for the interaction of DHT and T with AR allowed the authors to conclude that the different potencies between DHT and T are due to their affinity to the AR. Recently researchers have also proposed that the lesser stable T–AR complex versus DHT–AR complex may also contribute to why DHT is more potent than T in target gene transcriptional induction (18, 19). However, this conclusion does not explain phenomena where DHT-specific effects were observed. For example, DHT but not T reduced the production of interleukin 4 (IL-4), IL-5, and γ-interferon from mouse T cell hybridomas (20). In this case, a close interaction between T cells and local macrophages, which metabolize T to DHT for adjacent T cells, is proposed as a peripheral activation mechanism. Another potential DHT-specific gene, the FAR17a gene, has been identified from the golden Syrian hamster flank organ to be up-regulated by DHT but not by T in the presence of a 5α-reductase competitive inhibitor, progesterone (21, 22).
Since the T cell hybridoma is a unique androgen target cell without detectable 5α-reductase activity, we have looked for any differentially regulated mRNA species by the polymerase chain reaction differential display technique (DDPCR). Herein we report the identification and characterization of one early-responsive androgen target gene, TDD5, that can be differentially regulated by T versus DHT in cell culture.
EXPERIMENTAL PROCEDURES
Cell Culture and Metabolism of [3H]T.
T cell hybridoma 312.13 was obtained from R. A. Daynes’ laboratory (20). This line was maintained in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS) and cultured in a 5% CO2/95% air tissue culture incubator. The metabolism of T by 312.13 cells was examined by growing 105 cells for 48 hr in 1 ml of RPMI 1640 medium supplemented with 10% charcoal/dextran-treated FBS (CDFBS) and [1,2,6,7-3H(N)]testosterone (specific activity = 100 Ci/mmol, DuPont/NEN) at a concentration of 10 nM (23). In parallel, as a control, PC3 cells (24), a 5α-reductase-positive control, were incubated in the same medium. After a 48-hr incubation, conditioned medium (CM) and cells were collected separately for steroid extraction. The samples were extracted sequentially with 10 ml of ice-cold diethyl ether and then 1 ml of methanol. The extraction efficacy was estimated from radioactivity in 10 μl of methanol extract compared to the initial inoculation. The rest of methanol extract was dried and dissolved in 1 ml of mobile phase (50% acetonitrile/50% water) for HPLC analysis. Possible T metabolites were separated by passing 15 μl of medium extract through a C18 reverse-phase micropore column (Beckman). Radioactivities were detected by using a Radioisotope flow detector (Beckman). Prior to and after the experimental sample, the whole HPLC setup was calibrated with a battery of [3H]steroid standards manufactured by DuPont/NEN. Any possible metabolite with more than 1% abundance was expected to be visualized as a distinct peak in HPLC profile.
Experimental Design and Preparation of Total RNA from T Cell Hybridoma Cells.
The 312.13 cells were treated with RPMI 1640 medium containing 10% CDFBS alone or further supplemented with either T or DHT at a concentration of 10 nM. Cell pellets were quickly frozen in a dry ice/ethanol bath and stored at −70°C for total RNA preparation. These cell pellets were homogenized in guanidine thiocyanate solution and total RNA was purified by centrifugation. Any possible DNA contaminant was digested with RNase-free DNase.
Reverse Transcription of Total RNA.
RNAs were reverse-transcribed into cDNAs with three anchor primers, HT11G (AAGCTTTTTTTTTTTG), HT11A (AAGCTTTTTTTTTTTA), and HT11C (AAGCTTTTTTTTTTTC) according to the RNAimage kit (GenHunter, Brookline, MA). In brief, 0.2 μg of DNA-free total RNA was reverse-transcribed into cDNA in the presence of 0.2 μM anchor primer and all four dNTPs (each at 20 μM). Anchor primer and total RNA were incubated at 65°C for 5 min to break down any secondary structure and then reverse-transcribed at 37°C for 60 min followed by 75°C for 5 min to inactivate murine leukemia virus reverse transcriptase activity.
Differential Display of mRNA by PCR Amplification of cDNAs with an Arbitrary Primer.
Reverse transcription products were then subjected to PCR amplification with one arbitrary primer and the matched anchor primer. One-tenth (2 μl) of the reverse transcription product was PCR-amplified with 0.2 μM primers in the presence of dNTPs (each at 2 μM) and 10 μCi of deoxyadenosine 5′-[α-[35S]thio]triphosphate. The PCR was run for 40 cycles with each cycle consisting of 94°C for 30 sec, 40°C for 2 min, and 72°C for 30 sec.
Separation of PCR-Amplified Products on a Sequencing Gel.
Each 3.5-μl PCR-amplified sample was then mixed with 2 μl of stop solution (98% formamide/10 mM EDTA, pH 8.0) and loaded onto a 6.25% sequencing gel. The gel was electrophoresed at a constant 60 W at the maximal limits of 1,700 V and 300 mA. The electrophoresis was terminated after the xylene cyanol dye emerged from the gel. The gel was dried, traced with radioactive stickers, and exposed to x-ray film for 16–72 hr at room temperature before development. Any band of interest was excised from dried gel. The DNA was eluted and PCR-amplified with corresponding arbitrary and anchor primers. The reamplified differentially displayed DNA was then subcloned into pT7-blue vector (Novagene) for sequencing and also for the template to generate Northern blot probes.
5′ Rapid Amplification of cDNA Ends (RACE)/PCR for Full-Length TDD5 cDNA.
To obtain the full-length TDD5 cDNA clone from 312.13 total RNA, we synthesized a 26-base primer (5′-GCAAAGTTACAAATTTATTGGTCTGG-3′; the putative complementary poly(A) signal is underlined) containing the 3′ most end of the TDD5 fragment and subjected it to the RACE/PCR. A full-length TDD5 was PCR-amplified, cloned, and confirmed by Southern blot analysis.
Northern Blot Analysis.
Twenty-five micrograms of total RNA was loaded into each lane in electrophoresis and transferred onto a Hybond-N membrane (Amersham). The equal RNA loading, integrity of RNA, and transfer rate were confirmed by showing consistent intensities of 28S and 18S rRNA bands. A TDD5 probe was generated by PCR amplification as described with minor modifications (25). TDD5 3′ most end segment (288 bp) was excised from pT7-blue vector and purified as a template. The PCR amplification was performed with H-T11G and H-AP12 (AAGCTTGAGTGCT) primers. The membranes were hybridized with a radiolabeled probe for 24–48 hr at 42°C and washed twice at room temperature with 1× standard saline citrate (SSC)/0.1% SDS followed by once with 0.25× SSC/0.1% SDS at 55°C for 15 min. Dam wet membrane was wrapped in Saran wrap and exposed to a PhosphorImager or x-ray films. The signal intensities were quantified with imagequant software (Molecular Dynamics) and normalized according to the β-actin hybridization results.
Animal Studies.
Two-month-old male C57BL6 mice were obtained from Harlan Sprage Dawley, Indianapolis, IN. A few days after settlement, the animals were either sacrificed for tissue distribution studies or castrated under ether anesthesia for the studies on the effect of androgen. Four days after castration, we started treating these animals with 500 μg of DHT in 100 μl of sesame oil/ethanol, 9: 1 (vol/vol), daily through peritoneal administration. Animals were sacrificed 1, 2, 4, 8, 24, and 48 hr after initial treatment. Kidneys and thymuses were collected for Northern blot analyses.
RESULTS
The 312.13 T Cell Hybridoma Is a Unique Androgen Target Cell with a Deficiency in 5α-Reductase Activity.
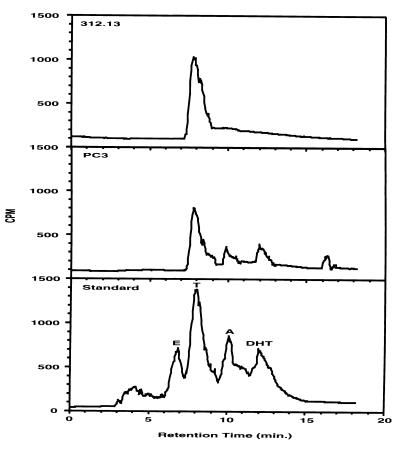
To investigate whether different androgens, T and DHT, may exert distinct regulatory effects on target gene expression, investigators have tried to block the conversion of T to DHT by applying 5α-reductase inhibitor or competitor, e.g., finasteride or progesterone (15, 21, 22). Unfortunately, the 5α-reductase system is very complicated because two isoforms have been isolated with distinct characteristics and because of the lack of pan-specific inhibitors of the isoforms, the complicated pharmacological kinetics, and the species specificity of these inhibitors. As a result, complete blockage of the conversion of T to DHT has never been accomplished in animals. For the convenience of control T- or DHT-specific stimulation, we sought an established androgen target cell line with minimal 5α-reductase activity as starting material. The 312.13 line is a T cell hybridoma generated by R. A. Daynes’ laboratory. The majority of these cell lines are shown to respond to androgen treatment by altering the secretion of various lymphokines (20). We have carried out MMTV-CAT transient transfection experiments to confirm the expression of functional AR (data not shown). To confirm its lack of 5α-reductase activities, we have incubated these cells with 10 nM [3H]testosterone to analyze how these cells metabolize T. The 3H-labeled activity was recovered from 48-hr 312.13 CM along with control medium and CM from human prostate PC3 cells (known to have 5α-reductase activity) by ether and methanol extraction. Consistently, 80–85% of total radioactivity was recovered from CM and control medium. On the other hand, less than 5% of radioactivity was recovered from both 312.13 and PC3 cell pellets. This high and consistent extraction efficacy from CM suggests that no significant glucuronidation metabolism had occurred to form water-soluble steroids. To further analyze the contents of these extracts, we performed HPLC analysis. Results show that a single radioactive population found in 312.13 CM (retention time = 7.2 min) comigrated with the T in the standard (retention time = 7.5 min; Fig. 1). In addition, like T, this peak showed a strong 246-nm absorption in a parallel UV absorption profile (data not shown). This result was also reconfirmed by using a second solvent system, 65% methanol/35% water (data not shown). It is possible that this single peak radioactivity found in 312.13 CM represents the T supplied earlier. These results provide direct evidence to confirm that these 312.13 cells are deficient in converting T to DHT (20). This HPLC profile may also suggest a deficiency of conversion to estradiol, which will show up as a distinct peak in the profile at approximately 7.5 min if it is more than 1% of radioactivity in the extract. This characteristic makes this cell line valuable for studying the differential effect of T and DHT on target gene regulation. To verify the working condition of HPLC analysis, we also examined an extract from PC3 CM. The results indicate that in addition to the T supplied, two T metabolites, DHT and androstenedione, were visualized along with a much more polar compound (retention time = 16.0 min) with an unknown identity. The 312.13 and PC3 cell pellet extracts gave similar profiles with much lower intensity than their CM counterparts (data not shown).
Figure 1.
Characterization of a T cell hybridoma lacking 5α-reductase activities. The 312.13 T cell hybridoma cells and PC3 human prostate cancer cells were incubated with 10 nM [3H]T for 48 hr. Steroid metabolites were extracted sequentially with ether and methanol and then dissolved in HPLC mobile-phase solvent (50% acetonitrile/50% water). Samples were passed through a C18 HPLC column for separation. HPLC profiles of 312.13- and PC3-CM lipophilic fractions are shown as a representative of two experiments. [3H]Steroid standard was run prior to and after experimental samples for calibration and identification. The standards, in order of hydrophobicity, are 17β-estradiol (E), T, androstenedione (A), and DHT.
mRNA Differential Display by PCR.
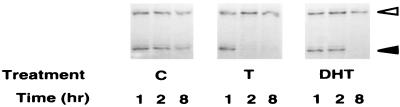
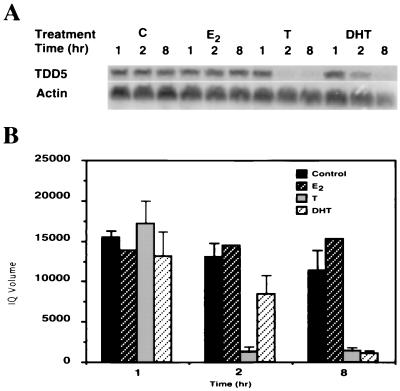
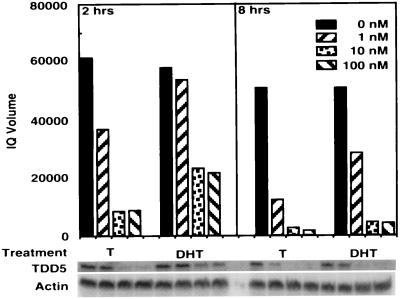
We then used the DDPCR technique to search for potential target genes that are regulated differentially by T/DHT. The 312.13 cells were treated with 10% CDFBS alone or further supplemented T or DHT at 10 nM. To identify the earlier androgen target genes and also to minimize the possibility of false-positive results, we harvested cells 1, 2, and 8 hr after treatment. TDD5, shown in Fig. 2, was identified from the PCR amplification with anchor primer H-T11G and arbitrary primer H-AP12. The intensity of TDD5 DDPCR product decreased when cells were treated with T for 2 hr or with DHT for 8 hr. It remained fairly high and consistent in the control group throughout 8 hr of treatment. To confirm the differential effect of T versus DHT on TDD5 expression and also to rule out the possibility that this T preference may have resulted from an effect of estradiol, we conducted Northern blot analysis in parallel experiments with the same 312.13 cells (Fig. 3A). The results, in Fig. 3B, show that although TDD5 mRNA decreased slightly when cells were treated with 10% CDFBS alone (controls), further supplementation with T (10 nM) or DHT (10 nM) repressed the TDD5 mRNA level much faster. On the other hand, estradiol (10 nM) did not alter significantly the quantity of TDD5 mRNA in 8 hr. More important, the repression in the level of TDD5 mRNA was more profound and faster in response to T than to DHT; only 7% and 64% of TDD5 mRNA remained in cells 2 hr after the administration of T and DHT, respectively (Fig. 3B). To investigate whether this T versus DHT preference on TDD5 is also dose-dependent, we performed 2- and 8-hr incubations with T or DHT in the concentration range from 1 to 100 nM. The results show that 1 nM T suppressed the TDD5 mRNA level to 60% in 2 hr and further to 14% in 8 hr, whereas 1 nM DHT did not have a significant effect until 8 hr (56% of TDD5 remained; Fig. 4). These results suggest that by 2 hr T is more potent than DHT even though the dose–response results of both T and DHT to TDD5 appeared to shift to the left by 8 hr. As expected, this suppression reached its maximum at 10 nM T/DHT, the same concentration used earlier in DDPCR experiments. As a result of this observation, we decided to isolate the full-length TDD5 cDNA for further characterization.
Figure 2.
Differential display profiles of 312.13 mRNA showing a potential androgen target fragment. The 312.13 cells were treated with 10% CDFBS (C, control) only or further supplemented with 10 nM T or 10 nM DHT for 1, 2, and 8 hr. Total RNA preparations were subjected to DDPCR. A partial differential display profile done with primers H-T11G and H-AP12 is shown. Open arrowhead, TDD5 DNA bands; solid arrowhead, common bands used for normalization in quantification. Profile shown is representative of three experiments.
Figure 3.
Northern blot confirmation of differential expression of TDD5 mRNA. The 312.13 cells were treated parallel to those for DDPCR. Total RNA (25 μg per sample per lane) was resolved by electrophoresis in a formaldehyde gel, transferred to a nylon membrane, and probed with a 288-bp differential display TDD5 DNA fragment. (A) The 312.13 cells were treated with 10% CDFBS only (C, control) or further supplemented with 17β-estradiol (E2), T, or DHT for 1, 2, and 8 hr. A representative Northern blot autoradiogram of three experiments with C, T, and DHT groups and only one for E2 group is shown. (B) PhosphorImage imagequant quantification of Northern blots in A. Averages of three experiments are shown; bars represent the SD. The quantification was normalized according to β-actin-hybridized band intensities (as shown in A). Four different treatments were performed as described in A: control, ▪; E2, □; T, □; DHT, □.
Figure 4.
Effect of T and DHT on the expression level of TDD5-dose-dependent and time course studies. The 312.13 cells were treated with T or DHT at 0 nM (□), 1 nM (□), 10 nM (□), or 100 nM (□) similar to those for differential display for 2 or 8 hr. Total RNA (25 μg per sample per lane) was resolved by electrophoresis in a formaldehyde gel, transferred to a nylon membrane, and hybridized with the 288-bp TDD5 differential display DNA fragment. TDD5- and β-actin-specific bands are shown and were subjected to PhosphorImage imagequant quantification. The quantification results were normalized to the intensity of the β-actin bands.
Isolation of Full-Length TDD5 cDNA Sequences.
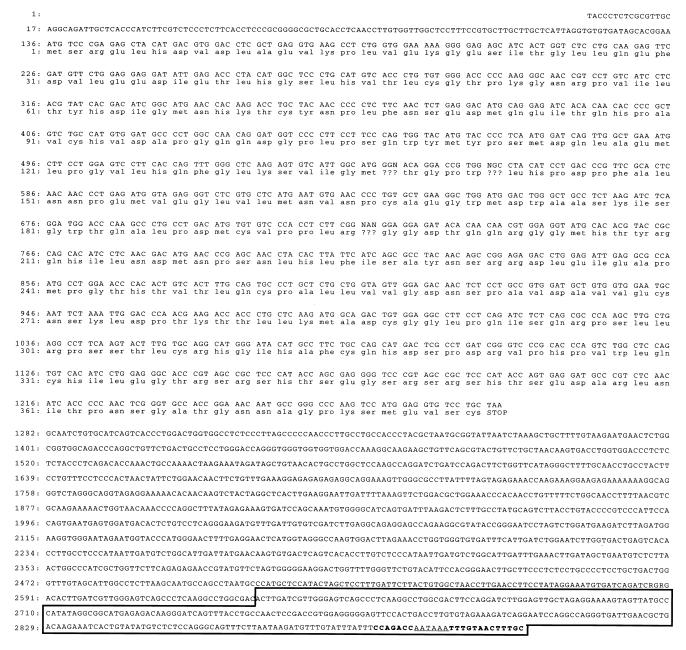
When the 3′ most 288 bp (within the box in Fig. 5) of the TDD5 sequence was isolated from our DDPCR, we had failed to identify its homologue among listed sequences with known functions in all DNA databases we examined. Consequently, we used 5′ RACE to clone the full-length cDNA. With one gene-specific primer covering the last 26 bp of TDD5 primer along with a RACE adapter primer, we succeeded in isolating one 2,916-bp cDNA fragment from a total RNA preparation from 312.13 cells. The sequence studies show that it contains the putative first ATG (136 bp) with the longest reading frame until the stop codon found at 1,279 bp (Fig. 5). The deduced protein size according to this open reading frame is 42 kDa, which is confirmed by the size of the in vitro-transcribed and -translated product (data not shown). After confirming the full-length TDD5 cDNA, we set forth to characterize its expression specificity and androgen suppression properties in mice.
Figure 5.
cDNA sequences and deduced amino acid sequences of TDD5. The nucleotide sequences and the deduced amino acid sequences are numbered on the left. The nucleotide sequence of the eukaryotic polyadenylylation signal AATAAA is underlined. The DNA sequence of isolated TDD5 fragment from DDPCR is boxed. The 26-bp complementary oligomer, located at the 3′ most end, for RACE/PCR amplification covers the region shown in boldface type.
Tissue Specificity of TDD5 Expression in Adult Male Mice.
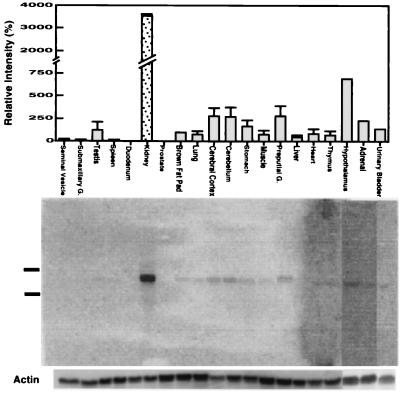
To explore the tissue distribution of TDD5 expression, we collected organs from 2-month-old C57BL/6 male mice for Northern blot analyses. As shown in Fig. 6, the data indicated that TDD5 can be expressed most abundantly in kidney followed by nervous tissues (hypothalamus, cerebellum, and cerebrum) and preputial gland. The other tested organs, except duodenum and prostate, also express TDD5 in much smaller quantities.
Figure 6.
Tissue specificity of TDD5 expression in adult male mice. C57BL/6 mice were sacrificed and several organs were collected. Northern blots were performed with the 3′ most 288 bp of TDD5 as probe. A computer-generated graph by the scanned Northern blot done with PhosphorImager is shown. (Upper) Relative intensity of TDD5-positive bands (mean ± SD) collected from three blots. The intensities of TDD5 contributed from brown fat pad (as 100) were used to normalize between blots. It should be noted that the first 17 organs, followed by hypothalamus, adrenal, and urinary bladder (Upper) correspond to the three Northern blots below (Lower). The 28S and 18S rRNAs found in total RNA preparation migrated to the positions indicated. The bands hybridized with β-actin probe for within blot normalization are shown at the bottom.
Animal Studies of the Repression of TDD5 mRNA Induced by DHT.
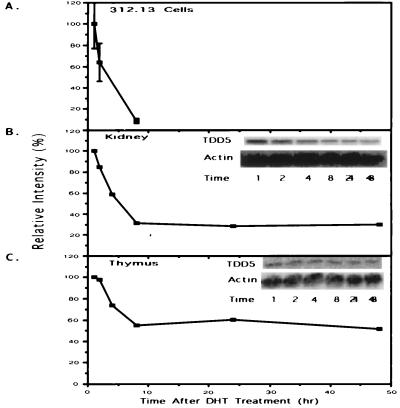
We investigated whether the androgen suppression of TDD5 expression in cell culture may also occur in animals. Because of the complexity of the 5α-reductase system in animals and the lack of effective tools to block completely the metabolism of T to DHT in mice, we decided to focus on the effect of DHT. Kidneys were collected because they express the highest quantity of TDD5 and thymuses were collected because of their production of T cells. Northern blot analysis of kidney RNA showed that TDD5 mRNA level was repressed 8 hr after the administration of DHT (Fig. 7B). This quick repression also happened in the thymus (Fig. 7C). These animal studies confirmed our previous observation done with a T cell hybridoma illustrating that TDD5 is an early androgen-responsive gene that can be repressed by androgens (Fig. 7A).
Figure 7.
Expression of TDD5 is consistently repressed by DHT administration both in cells and in animals. (A) Northern blot quantification of TDD5 shown in Fig. 3B is replotted to compare to animal studies (B and C). Results of TDD5 Northern blots from either kidney (B) or thymus (C) obtained from 4-day orchiectomized mice are shown. (B and C Insets) TDD5-specific bands (marked as TDD5) and β-actin bands (marked as Actin for normalization). The relative intensities of TDD5 changes (plotted on the y axis) are calculated against the intensity of normalized 1-hr point obtained from each group. A representative experiment of two experiments on TDD5 mRNA changes during the time course of DHT treatment (1, 2, 4, 8, 24, and 48 hr) is shown.
DISCUSSION
The AR activation mechanism is one distinct feature among the steroid receptors. Two forms of androgen, T and DHT, are available in circulation or after intracellular 5α-reduction before they interact with AR. On one hand DHT is more potent than T in stimulating the expression of MMTV-CAT reporter gene in CHO cells (17). Also DHT is approximately 10 times more potent than T in supporting yeast transfectant proliferation resulting from a specific interaction between an AR bait and a novel AR-associated activator (ARA70) (26). On the other hand, T supports the development of most Wolffian-duct-derived organs when finasteride is administrated (15). Muscle development in 5α-reductase-deficient patients is largely unaffected (16). Thus, T itself may be involved more in anabolic effects (e.g., muscle development) and DHT may be more potent in androgenic effects (e.g., growth of male reproductive tract) (27). A direct T/DHT-specific target androgen-response element should provide a potential tool for the development of new drugs that can support anabolism without any androgenic activity or vice versa.
Araneo et al. (20) have demonstrated that IL-4 secretion from T cells is negatively regulated by DHT but not by T. Expression of the hamster FAR17a gene has also been reported to be induced only by DHT (21, 22). However, due to the slow response to androgen treatment, in days, it is possible that these two potential DHT-specific target genes may not be regulated directly by androgen. Indeed, we have tried without success to identify any potential DHT-specific cis-acting elements from both human IL-4 and hamster FAR17a 5′ promoter region (28). Consequently, for the purpose of identifying potential androgen-response elements that can distinguish between T–AR and DHT–AR, we started isolating more differentially regulated androgen target genes.
The T cell hybridoma 312.13 cell was chosen because it responses differently to T/DHT stimulation by the production of several lymphokines. More important, this cell line lacks a significant ability to metabolize T to DHT. By comparing differentially displayed mRNA species among androgen-free, T, and DHT treatments, we have identified an early androgen-responsive cDNA fragment, TDD5, that responds differently to T and DHT. TDD5 responded to T (10 nM) in 2 hr and to DHT (10 nM) in 8 hr by a similar decrease in the amounts of TDD5 transcript (Fig. 3). In addition, a 2-hr treatment with 1 nM T was sufficient to cause a significant suppression but same concentration of DHT failed unless the treatment was extended to 8 hr (Fig. 4). This T preference is not due to the ability of T to be converted to estradiol, because there is no significant conversion of T to estradiol in 312.13 cells and also estradiol treatment does to alter the quantity of TDD5 mRNA in 312.13 cells. The lack of appropriate tools to block the conversion of T to DHT completely in animals has hindered our effort to confirm the T versus DHT preference in animals. Instead, we have shown that the injection of DHT into 4-day castrated mice can repress TDD5 mRNA levels in both kidney and thymus by 8 hr. This is in agreement with our T cell hybridoma studies. Thus, our data demonstrate that TDD5 is a fast-responding androgen target gene that has a preference to respond more to T than to DHT in 312.13 cells. As yet, we have not determined the physiological meaning of this kinetic change between T and DHT, nor can we ignore factors other than androgens that may be involved in this phenomenon. Understanding of the physiological cascade of TDD5 function may provide a better clue in the near future.
Full-length TDD5 cDNA was isolated as a 2,916-bp sequence that corresponds to the mRNA size on Northern blots. After we obtained the full-length cDNA, our first attempt at sequence comparison using blastn software (29) revealed that this was a novel gene. The sequence comparisons conducted recently have revealed that the full-length TDD5 sequence is highly homologous to the mouse cytoplasmic protein Ndr1 (more than 95% homology in both cDNA and amino acid sequences) and the human reducing agents and tunicamycin-responsive protein (RTP; 80–85% and 90–95% homology in cDNA and amino acid sequences, respectively). Ndr1 was identified as a gene downstream of N-myc with a GenBank citation referring to an unpublished reference. Whether Ndr1 may mediate the function of N-myc in stimulating cell proliferation or have other functions is currently under investigation. RTP was identified as a homocysteine-inducible gene isolated from human umbilical vein endothelial cells. The expression of RTP has been proposed to be important to homocysteine-induced vascular diseases, e.g., arteriosclerosis and thrombosis (30). Although the physiological function of RTP remains unclear, whether androgen may prevent the consequences of homocysteinemia by suppressing the expression of RTP in endothelial cells should be investigated.
A tissue distribution survey done by Northern blot analyses indicated that TDD5 is expressed almost exclusively in the kidney, followed by much lower expression in the nervous system and even less in almost all other tested organs. To date, several androgen-inducible AR target genes have been identified from kidney, e.g., ornithine decarboxylase (ODC), kidney androgen-regulated gene (KAP), alcohol dehydrogenase (ADH), β-glucuronidase (GUS), renal protein 2 gene (RP2), and renal cytochrome P450s (for review, see ref. 31). Almost all of these genes are expressed in the proximal tubule cells of the renal nephron. The expression of RP2, ODC, GUS, P450V, and ADH are induced by T and DHT (32). Due to their slow response, it does not surprise us that androgen-responsive elements identified cannot confer complete androgen responsiveness in reporter gene assays (31, 32). Some other factors may be involved in the regulation of these renal genes. On the contrary, the fast-responding TDD5 may be a much better candidate to search for androgen-responsive elements that can distinguish between T and DHT. One may also argue that several mechanisms may be involved in negative regulation of any gene expression. Direct interaction between trans-acting factors and specific cis-acting elements or indirect interaction, e.g., cytoplasmic sequestration of transcription factors, blocking of transcription factor response elements, direct inhibition of transcriptional factors by protein–protein interactions, and interference with trans-activation of DNA-bound transcription factors were proposed (for review, see ref. 33). As yet, we do not have evidence to support or deny any of them. However, because of the early response of TDD5 to androgens and also the transcriptional regulatory nature of AR, we propose that some cis-acting DNA elements are responsible to exert the T over DHT preference in target gene regulation. With our recent observation of ARA70, which functions as an AR-specific coactivator (26), we hypothesize that some AR coactivators may be involved in determining the T preference for TDD5 gene suppression. TDD5 promoter studies should provide us a better in vitro assay system to clarify different aspects of the specific functions of T versus DHT. Compounds that can specifically block the function of T or DHT may also be developed with the help of these studies. With the high level of kidney expression of TDD5, some kidney-specific androgen response elements for the purpose of organ-specific targeting may also be identified.
Acknowledgments
We thank Dr. R. A. Daynes for supply of 312.13 T cell hybridoma cells, Drs. H. Michael Theobald and Richard E. Peterson for usage of their HPLC equipment and supplying [3H]steroid standard, and Pei-Wen Hsiao for technical assistant on DNA sequencing. This work was supported by National Institutes of Health Grants CA55639, DK47258, and CA09471.
ABBREVIATIONS
- DDPCR
mRNA polymerase chain reaction differential display technique
- AR
androgen receptor
- T
testosterone
- DHT
5α-dihydrotestosterone
- IL
interleukin
- CM
conditioned medium
- CDFBS
charcoal-dextran treated fetal bovine serum
- RACE
5′ rapid amplification of cDNA ends
- MMTV-CAT
mouse mammary tumorvirus-chloramphenical acetyltransferase
Footnotes
References
- 1.Mooradian A D, Morley J E, Korenman S G. Endocr Rev. 1987;8:1–28. doi: 10.1210/edrv-8-1-1. [DOI] [PubMed] [Google Scholar]
- 2.Randall V A. Baillieres Clin Endocrinol Metab. 1994;8:405–431. doi: 10.1016/s0950-351x(05)80259-9. [DOI] [PubMed] [Google Scholar]
- 3.Dohler K D. Int Rev Cytol. 1991;131:1–57. doi: 10.1016/s0074-7696(08)62016-1. [DOI] [PubMed] [Google Scholar]
- 4.Kaplan M E, McGinnis M Y. Horm Behav. 1989;23:10–26. doi: 10.1016/0018-506x(89)90071-8. [DOI] [PubMed] [Google Scholar]
- 5.Bruchovsky N, Wilson J D. J Biol Chem. 1968;243:2012–2021. [PubMed] [Google Scholar]
- 6.Saartok T, Dahlberg E, Gustafsson J A. Endocrinology. 1984;114:2100–2106. doi: 10.1210/endo-114-6-2100. [DOI] [PubMed] [Google Scholar]
- 7.Imperato-McGinley J, Guerrero L, Gautier T, Peterson R E. Science. 1974;186:1213–1215. doi: 10.1126/science.186.4170.1213. [DOI] [PubMed] [Google Scholar]
- 8.Imperato-McGinley J, Peterson R E, Gautier T, Sturla E. N Engl J Med. 1979;300:1233–1237. doi: 10.1056/NEJM197905313002201. [DOI] [PubMed] [Google Scholar]
- 9.Peterson R E, Imperato-McGinley J, Gautier T, Sturla E. Am J Med. 1977;62:170–191. doi: 10.1016/0002-9343(77)90313-8. [DOI] [PubMed] [Google Scholar]
- 10.French F S, Lubahn D B, Brown T R, Simental J A, Quigley C A, et al. Recent Prog Horm Res. 1990;46:1–38. doi: 10.1016/b978-0-12-571146-3.50005-5. [DOI] [PubMed] [Google Scholar]
- 11.Chang C S, Kokontis J, Liao S T. Science. 1988;240:324–326. doi: 10.1126/science.3353726. [DOI] [PubMed] [Google Scholar]
- 12.Lubahn D B, Joseph D R, Sullivan P M, Willard H F, French F S, Wilson E M. Science. 1988;240:327–330. doi: 10.1126/science.3353727. [DOI] [PubMed] [Google Scholar]
- 13.Trapman J, Klaassen P, Kuiper G G, van der Korput J A, Faber P W, vanRooij H C, Geurts van Kessel A, Voorhorst M M, Mulder E, Brinkmann A O. Biochem Biophy Res Commun. 1988;153:241–248. doi: 10.1016/s0006-291x(88)81214-2. [DOI] [PubMed] [Google Scholar]
- 14.Lubahn D B, Joseph D R, Sar M, Tan J, Higgs H N, Larson R E, French F S, Wilson E M. Mol Endocrinol. 1988;2:1265–1275. doi: 10.1210/mend-2-12-1265. [DOI] [PubMed] [Google Scholar]
- 15.Imperato-McGinley J, Sanchez R S, Spencer J R, Yee B, Vaughan E D. Endocrinology. 1992;131:1149–1156. doi: 10.1210/endo.131.3.1324152. [DOI] [PubMed] [Google Scholar]
- 16.Kumar N, Didolkar A K, Monder C, Bardin C W, Sundaram K. Endocrinology. 1992;130:3677–3683. doi: 10.1210/endo.130.6.1597164. [DOI] [PubMed] [Google Scholar]
- 17.Deslypere J P, Young M, Wilson J D, McPhaul M J. Mol Cell Endocrinol. 1992;88:15–22. doi: 10.1016/0303-7207(92)90004-p. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Z X, Lane M V, Kemppainen J A, French F S, Wilson E M. Mol Endocrinol. 1995;9:208–218. doi: 10.1210/mend.9.2.7776971. [DOI] [PubMed] [Google Scholar]
- 19.Grino P B, Griffin J E, Wilson J D. Endocrinology. 1990;126:1165–1172. doi: 10.1210/endo-126-2-1165. [DOI] [PubMed] [Google Scholar]
- 20.Araneo B A, Dowell T, Diegel M, Daynes R A. Blood. 1991;78:688–699. [PubMed] [Google Scholar]
- 21.Hisaoka H, Ideta R, Seki T, Adachi K. Arch Dermatol Res. 1991;283:269–273. doi: 10.1007/BF01106114. [DOI] [PubMed] [Google Scholar]
- 22.Seki T, Ideta R, Shibuya M, Adachi K. J Invest Dermatol. 1991;96:926–931. doi: 10.1111/1523-1747.ep12475453. [DOI] [PubMed] [Google Scholar]
- 23.Damassa D A, Lin T M, Sonnenschein C, Soto A M. Endocrinology. 1991;129:75–84. doi: 10.1210/endo-129-1-75. [DOI] [PubMed] [Google Scholar]
- 24.Roberson K M, Padilla G M, Schmidt J M, Petrow V, Robertson C N. Prostate. 1995;26:28–34. doi: 10.1002/pros.2990260107. [DOI] [PubMed] [Google Scholar]
- 25.Mertz L M, Rashtchian A. Anal Biochem. 1994;221:160–165. doi: 10.1006/abio.1994.1392. [DOI] [PubMed] [Google Scholar]
- 26.Yeh S, Chang C. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sundaram K, Kumar N, Monder C, Bardin C W. J Steroid Biochem Mol Biol. 1995;53:253–257. doi: 10.1016/0960-0760(95)00056-6. [DOI] [PubMed] [Google Scholar]
- 28.Chang C, Lin T-M, Hsiao P-W, Su C, Riebe J, Chang C-T, Lin D-L. In: Pharmacology, Biology, and Clinical Application of Androgens. Bhasin S, Gabelnick H L, Spieler J M, Swerdloff R S, Wang C, Kelly C, editors. New York: Wiley–Liss; 1996. pp. 65–71. [Google Scholar]
- 29.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Kokame K, Kato H, Miyata T. J Biol Chem. 1996;271:29659–29665. doi: 10.1074/jbc.271.47.29659. [DOI] [PubMed] [Google Scholar]
- 31.Chang C, Saltzman A, Yeh S, Young W, Keller E, Lee H -J, Wang C, Mizokami A. Crit Rev Eukaryotic Gene Expression. 1996;5:97–125. doi: 10.1615/critreveukargeneexpr.v5.i2.10. [DOI] [PubMed] [Google Scholar]
- 32.Asadi F K, Dimaculangan D D, Berger F G. Endocrinology. 1994;134:1179–1187. doi: 10.1210/endo.134.3.8119157. [DOI] [PubMed] [Google Scholar]
- 33.Burcin M, Kohne A C, Runge D, Steiner C, Renkawitz R. Semin Cancer Biol. 1994;5:337–346. [PubMed] [Google Scholar]