Abstract
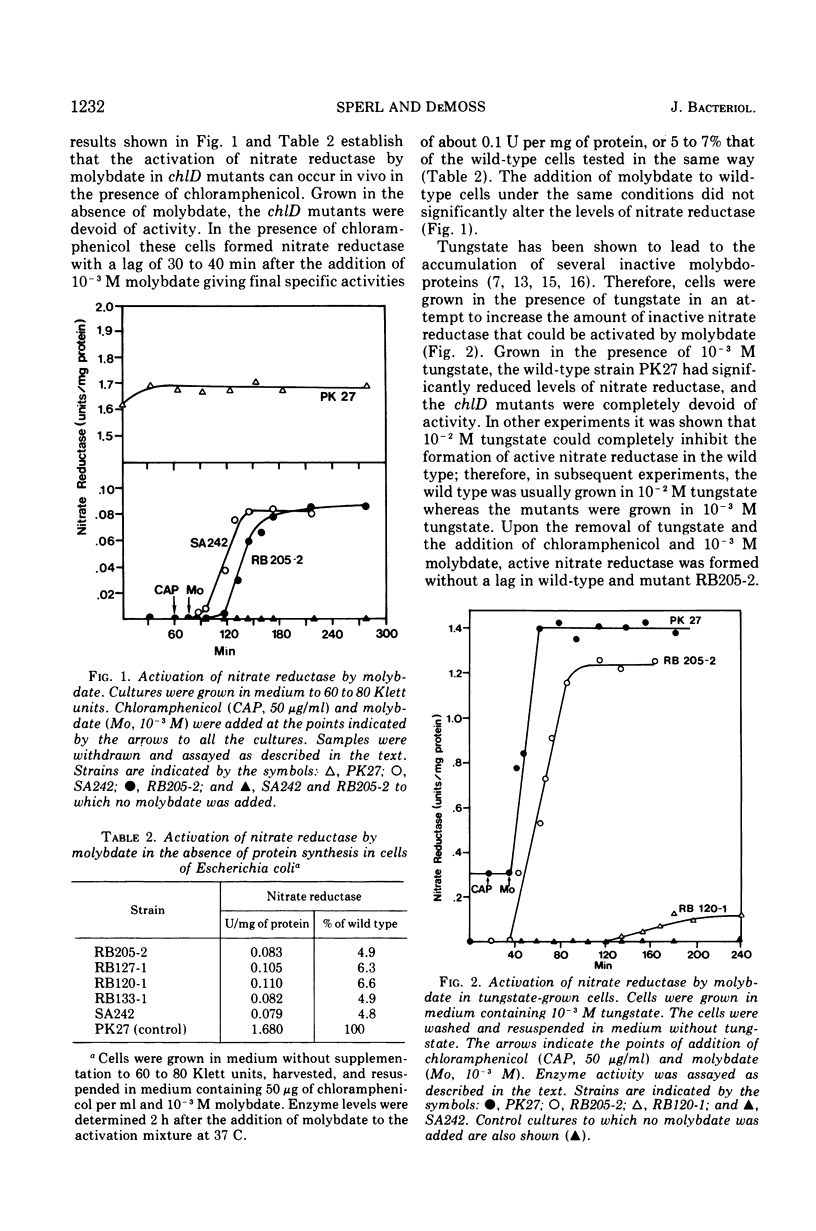
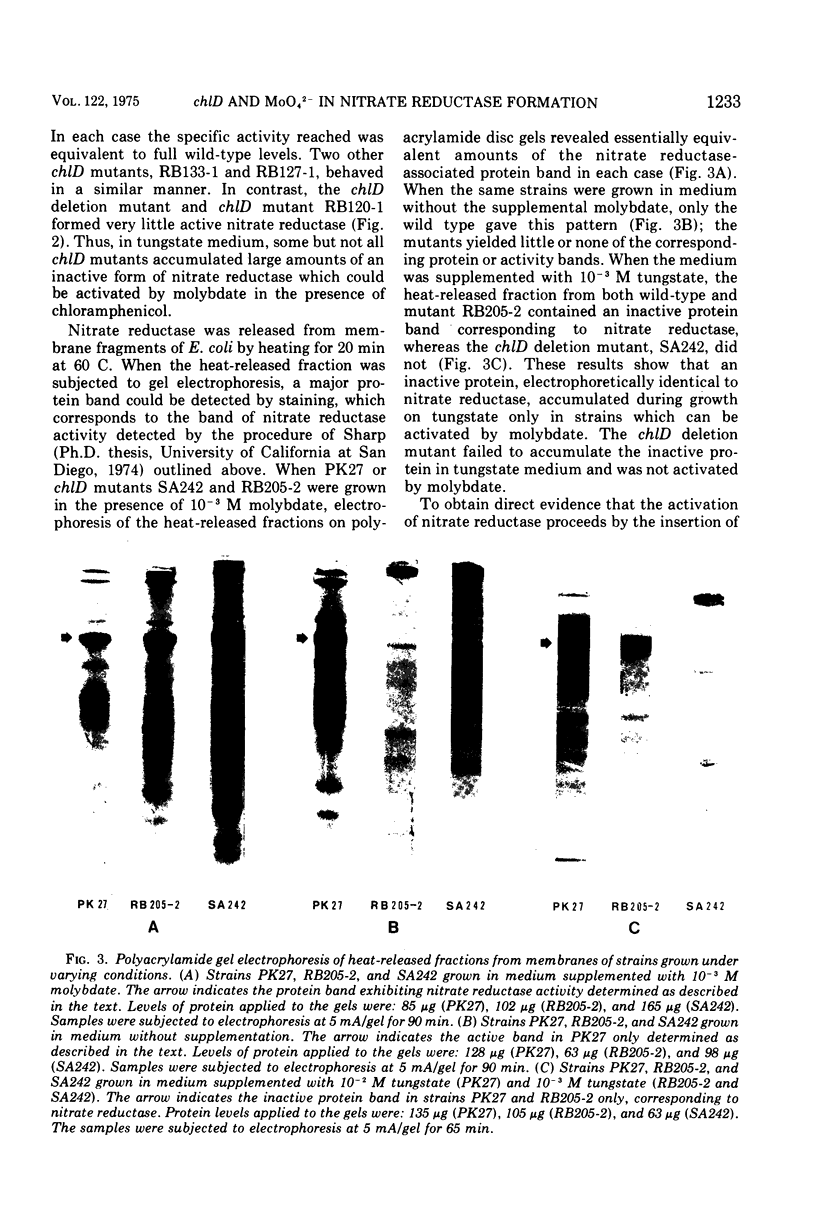
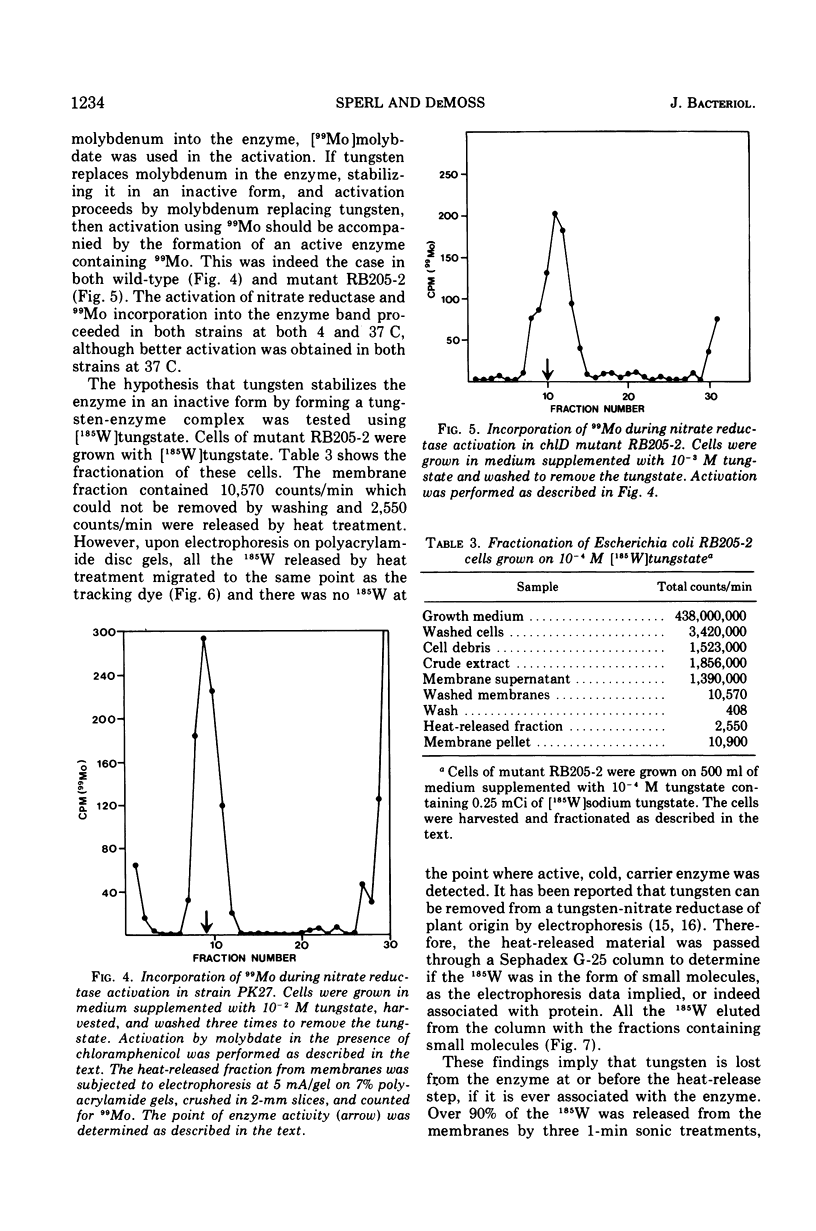
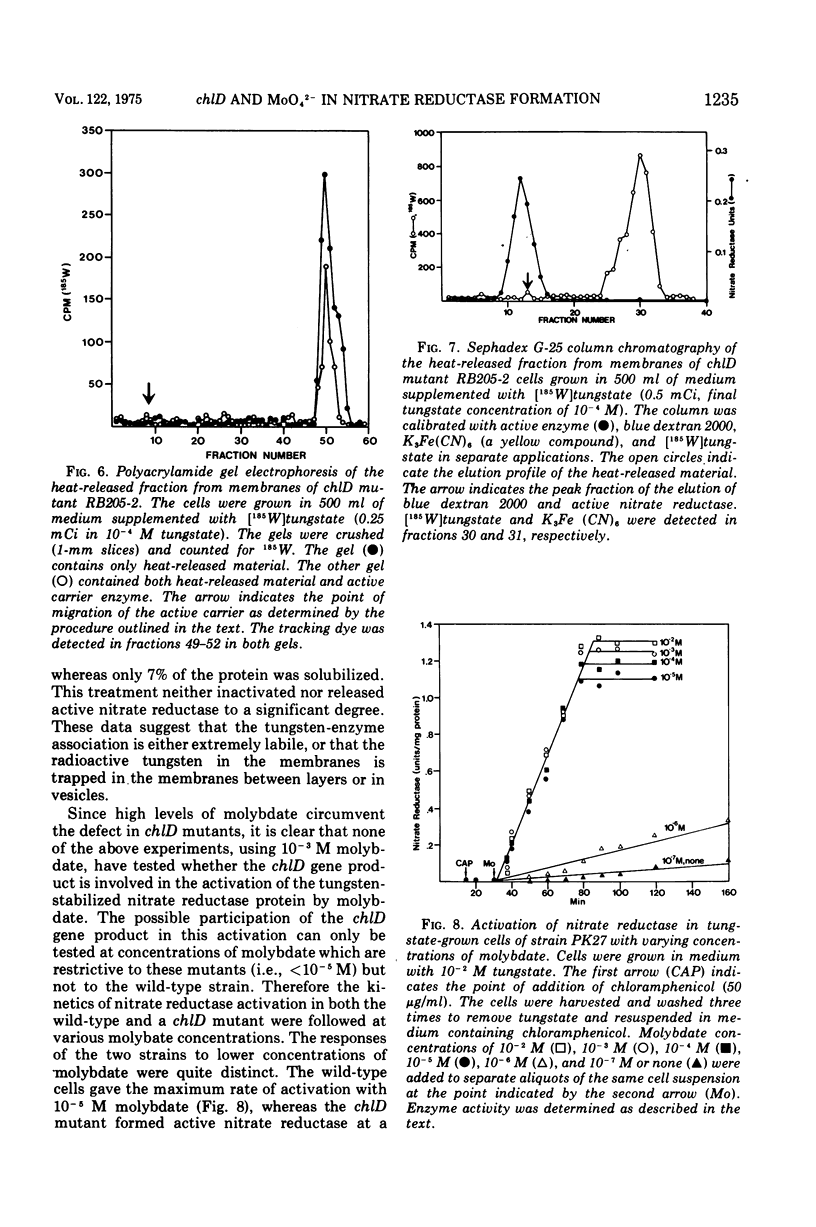
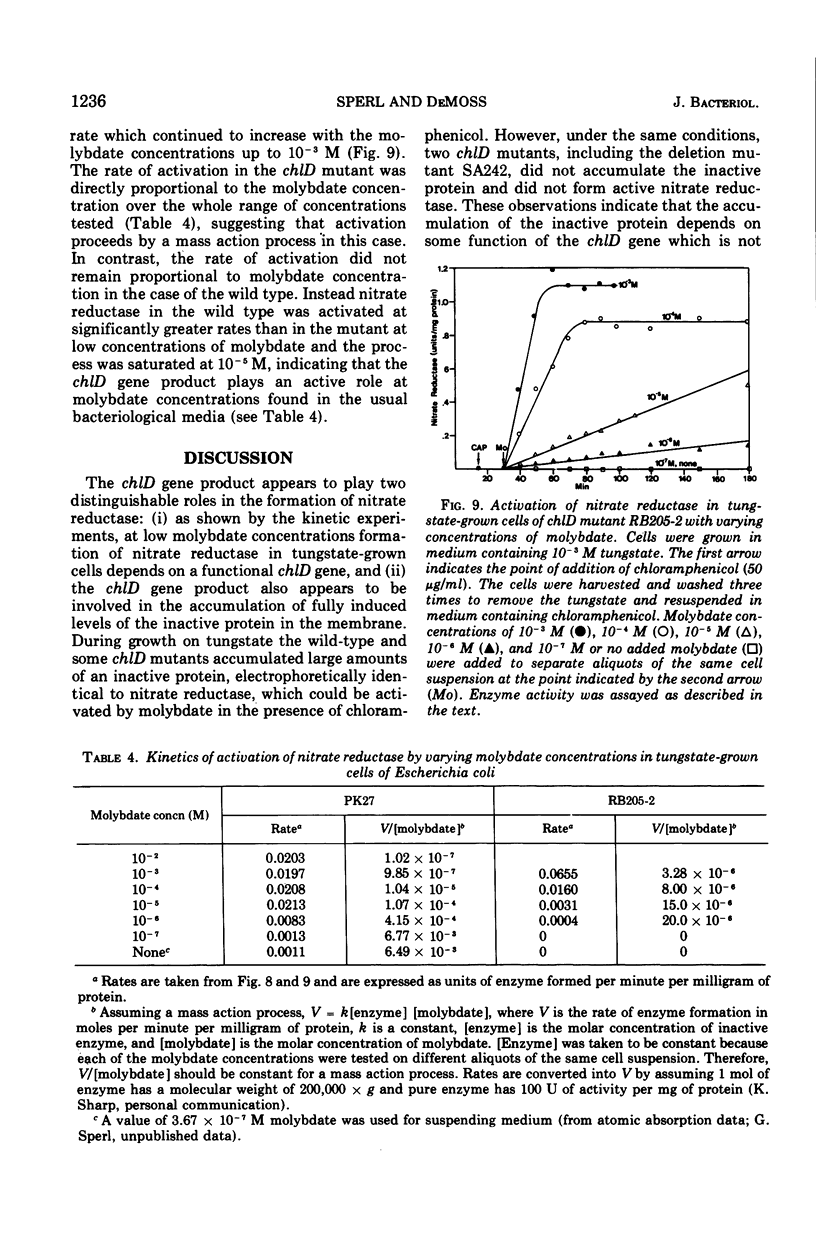
chlD mutants of Escherichia coli lack active nitrate reductase but form normal levels of this enzyme when the medium is supplemented with 10-3 M molybdate. When chlD mutants were grown in unsupplemented medium and then incubated with molybdate in the presence of chloramphenicol, they formed about 5% the normal level of nitrate reductase. Some chlD mutants or the wild type grown in medium supplemented with tungstate accumulated an inactive protein which was electrophoretically identical to active nitrate reductase. Addition of molybdate to those cells in the presence of chloramphenicol resulted in the formation of fully induced levels of nitrate reductase. Two chlD mutants, including a deletion mutant, failed to accumulate the inactive protein and to form active enzyme under the same conditions. Insertion of 99-Mo into the enzyme protein paralleled activation; 185-W could not be demonstrated to be associated with the accumulated inactive protein. The rates of activation of nitrate reductase at varying molybdate concentrations indicated that the chlD gene product facilitates the activation of nitrate reductase at concentrations of molybdate found in normal growth media. At high concentrations, molybdate circumvented this function in chlD mutants and appeared to activate nitrate reductase by a mass action process. We conclude that the chlD gene plays two distinguishable roles in the formation of nitrate reductase in E. coli. It is involved in the accumulation of fully induced levels of the nitrate reductase protein in the cell membrane and it facilitates the insertion of molybdenum to form the active enzyme.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Cleary P., Campbell A. A deletion analysis of prophage lambda and adjacent genetic regions. Proc Natl Acad Sci U S A. 1968 Nov;61(3):956–962. doi: 10.1073/pnas.61.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Comparison of nitrate reductase mutants of Escherichia coli selected by alternative procedures. Mol Gen Genet. 1972;116(1):1–10. doi: 10.1007/BF00334254. [DOI] [PubMed] [Google Scholar]
- Heimer Y. M., Filner P. Regulation of the nitrate assimilation pathway in cultured tobacco cells. 3. The nitrate uptake system. Biochim Biophys Acta. 1971 Feb 23;230(2):362–372. doi: 10.1016/0304-4165(71)90223-6. [DOI] [PubMed] [Google Scholar]
- Johnson J. L., Rajagopalan K. V., Cohen H. J. Molecular basis of the biological function of molybdenum. Effect of tungsten on xanthine oxidase and sulfite oxidase in the rat. J Biol Chem. 1974 Feb 10;249(3):859–866. [PubMed] [Google Scholar]
- Kahn P. L. Isolation of high-frequency recombining strains from Escherichia coli containing the V colicinogenic factor. J Bacteriol. 1968 Jul;96(1):205–214. doi: 10.1128/jb.96.1.205-214.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lester R. L., DeMoss J. A. Effects of molybdate and selenite on formate and nitrate metabolism in Escherichia coli. J Bacteriol. 1971 Mar;105(3):1006–1014. doi: 10.1128/jb.105.3.1006-1014.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor C. H., Schnaitman C. A. Reconstitution of nitrate reductase activity and formation of membrane particles from cytoplasmic extracts of chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1973 Jun;114(3):1164–1176. doi: 10.1128/jb.114.3.1164-1176.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani H. H., Shah V. K., Brill W. J. Activation of inactive nitrogenase by acid-treated component I. J Bacteriol. 1974 Nov;120(2):697–701. doi: 10.1128/jb.120.2.697-701.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nason A., Lee K. Y., Pan S. S., Ketchum P. A., Lamberti A., DeVries J. Invitro formation of assimilatory reduced nicotinamide adenine dinucleotide phosphate: nitrate reductase from a Neurospora mutant and a component of molybdenum-enzymes. Proc Natl Acad Sci U S A. 1971 Dec;68(12):3242–3246. doi: 10.1073/pnas.68.12.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notton B. A., Hewitt E. J. Comparative aspects of incorporation of vanadium, tungsten or molybdenum into protein of nitrate reductase of Spinacea oleracea L. leaves. Biochim Biophys Acta. 1972 Sep 20;275(3):355–357. doi: 10.1016/0005-2728(72)90216-2. [DOI] [PubMed] [Google Scholar]
- Notton B. A., Hewitt E. J. The role of tungsten in the inhibition of nitrate reductase activity in spinach (spinacea oleracea L.) leaves. Biochem Biophys Res Commun. 1971 Aug 6;44(3):702–710. doi: 10.1016/s0006-291x(71)80140-7. [DOI] [PubMed] [Google Scholar]
- PATEMAN J. A., COVE D. J., REVER B. M., ROBERTS D. B. A COMMON CO-FACTOR FOR NITRATE REDUCTASE AND XANTHINE DEHYDROGENASE WHICH ALSO REGULATES THE SYNTHESIS OF NITRATE REDUCTASE. Nature. 1964 Jan 4;201:58–60. doi: 10.1038/201058a0. [DOI] [PubMed] [Google Scholar]
- Pateman J. A., Rever B. M., Cove D. J. Genetic and biochemical studies of nitrate reduction in Aspergillus nidulans. Biochem J. 1967 Jul;104(1):103–111. doi: 10.1042/bj1040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera J., DeMoss J. A. Nitrate reductase complex of Escherichia coli K-12: participation of specific formate dehydrogenase and cytochrome b1 components in nitrate reduction. J Bacteriol. 1969 Sep;99(3):720–729. doi: 10.1128/jb.99.3.720-729.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showe M. K., DeMoss J. A. Localization and regulation of synthesis of nitrate reductase in Escherichia coli. J Bacteriol. 1968 Apr;95(4):1305–1313. doi: 10.1128/jb.95.4.1305-1313.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TANIGUCHI S., ITAGAKI E. Nitrate reductase of nitrate respiration type from E. coli. I. Solubilization and purification from the particulate system with molecular characterization as a metalloprotein. Biochim Biophys Acta. 1960 Nov 4;44:263–279. doi: 10.1016/0006-3002(60)91562-6. [DOI] [PubMed] [Google Scholar]