Abstract
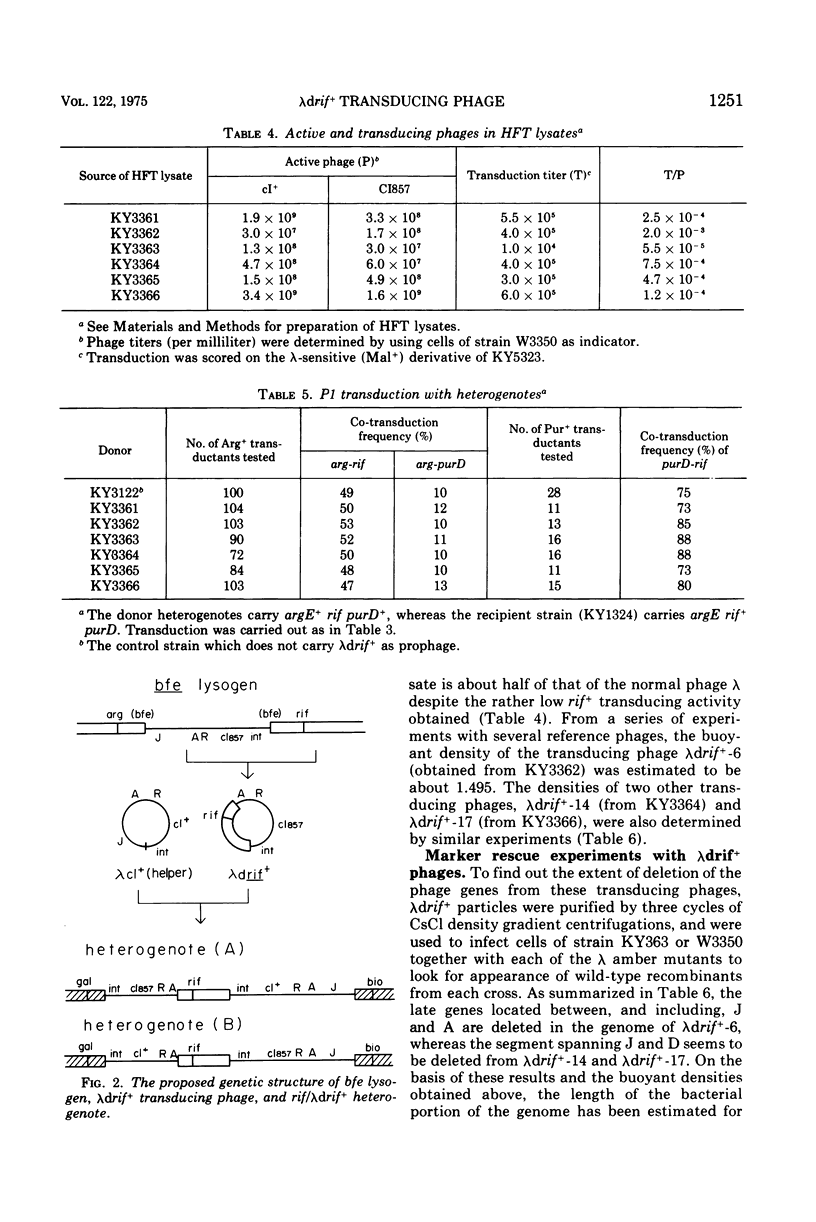
The prophage lambdac1857 was inserted into the bfe gene located near rif (the structural gene for the beta subunit of deoxyribonucleic acid [DNA]-dependent ribonucleic acid polymerase) on the Escherichia coli chromosome. Induced lysates (low-frequency transducing lysates) of such a lysogen contained defective lambda phage particles (lambdadrif+) that can specifically transduce the wild-type rif+ gene. Upon transduction into a recipient strain carrying recA, heterogenotes harboring both the wild-type and the mutant rif genes were isolated. Rec+ derivatives of these heterogenotes produce high-frequency transducing lysates that contain lambdadrif+ and normal active phages at a ratio of 1 to 2. The results of marker rescue experiments and of density determination with several transducing phages indicate that most of the late genes are deleted and replaced by a segment of the chromosomal DNA carrying the bfe-rif region. The length of the chromosomal segment seems to vary between approximately 0.5 and 0.6% of the total bacterial DNA among the three independently isolated lambdadrif+ phages. Electron microscopy of heteroduplex DNA consisting of one strand from lambdadrif+-6 and the other from lambdaimm-21 phages directly confirmed that most of the phage DNA of the "left arm" was replaced by the bacterial DNA. The heteroduplex study also demonstrated that the integration of prophage lambda into the bfe region occurred at the normal cross-over point within the phage attachment site.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burgess R. R. RNA polymerase. Annu Rev Biochem. 1971;40:711–740. doi: 10.1146/annurev.bi.40.070171.003431. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Davidson N. Electron microscope study of the structures of lambdadv DNAs. J Mol Biol. 1974 Jun 15;86(1):69–89. doi: 10.1016/s0022-2836(74)80008-2. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Parkinson J. S. Deletion mutants of bacteriophage lambda. 3. Physical structure of att-phi. J Mol Biol. 1971 Mar 14;56(2):403–423. doi: 10.1016/0022-2836(71)90473-6. [DOI] [PubMed] [Google Scholar]
- Emmons S. W. Bacteriophage lambda derivatives carrying two copies of the cohesive end site. J Mol Biol. 1974 Mar 15;83(4):511–525. doi: 10.1016/0022-2836(74)90511-7. [DOI] [PubMed] [Google Scholar]
- Feiss M., Campbell A. Duplication of the bacteriophage lambda cohesive end site: genetic studies. J Mol Biol. 1974 Mar 15;83(4):527–540. doi: 10.1016/0022-2836(74)90512-9. [DOI] [PubMed] [Google Scholar]
- Freifelder D., Chud L., Levine E. E. Requirement for maturation of Escherichia coli bacteriophage lambda. J Mol Biol. 1974 Mar 15;83(4):503–509. doi: 10.1016/0022-2836(74)90510-5. [DOI] [PubMed] [Google Scholar]
- Gottesman M. E., Yarmolinsky M. B. The integration and excision of the bacteriophage lambda genome. Cold Spring Harb Symp Quant Biol. 1968;33:735–747. doi: 10.1101/sqb.1968.033.01.084. [DOI] [PubMed] [Google Scholar]
- Heil A., Zillig W. Reconstitution of bacterial DNA-dependent RNA-polymerase from isolated subunits as a tool for the elucidation of the role of the subunits in transcription. FEBS Lett. 1970 Dec;11(3):165–168. doi: 10.1016/0014-5793(70)80519-1. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Tomizawa J. I. Transducing fragments in generalized transduction by phage P1. I. Molecular origin of the fragments. J Mol Biol. 1965 Nov;14(1):85–109. doi: 10.1016/s0022-2836(65)80232-7. [DOI] [PubMed] [Google Scholar]
- Inman R. B., Schnös M. Partial denaturation of thymine- and 5-bromouracil-containing lambda DNA in alkali. J Mol Biol. 1970 Apr 14;49(1):93–98. doi: 10.1016/0022-2836(70)90378-5. [DOI] [PubMed] [Google Scholar]
- Jasper P., Whitney E., Silver S. Genetic locus determining resistance to phage BF23 and colicins E 1 , E 2 and E 3 in Escherichia coli. Genet Res. 1972 Jun;19(3):305–312. doi: 10.1017/s0016672300014555. [DOI] [PubMed] [Google Scholar]
- Kirschbaum J. B., Konrad E. B. Isolation of a specialized lambda transducing bacteriophage carrying the beta subunit gene for Escherichia coli ribonucleic acid polymerase. J Bacteriol. 1973 Nov;116(2):517–526. doi: 10.1128/jb.116.2.517-526.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschbaum J. B., Scaife J. Evidence for a lambda transducing phage carrying the genes for the beta and beta' subunits of Escherichia coli RNA polymerase. Mol Gen Genet. 1974;132(3):193–201. doi: 10.1007/BF00269392. [DOI] [PubMed] [Google Scholar]
- Konrad B., Kirschbaum J., Austin S. Isolation and characterization of phi80 transducing bacteriophage for a ribonucleic acid polymerase gene. J Bacteriol. 1973 Nov;116(2):511–516. doi: 10.1128/jb.116.2.511-516.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Low B. Formation of merodiploids in matings with a class of Rec- recipient strains of Escherichia coli K12. Proc Natl Acad Sci U S A. 1968 May;60(1):160–167. doi: 10.1073/pnas.60.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzura H., Molin S., Maaloe O. Sequential biosynthesis of the and ' subunits of the DNA-dependent RNA polymerase from Escherichia coli. J Mol Biol. 1971 Jul 14;59(1):17–25. doi: 10.1016/0022-2836(71)90410-4. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Yura T. Localization of the structural gene for the ' subunit of RNA polymerase in Escherichia coli. Biochem Biophys Res Commun. 1973 Jul 17;53(2):645–652. doi: 10.1016/0006-291x(73)90710-9. [DOI] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. I. Location of the secondary attachment sites and the properties of the lysogens. J Mol Biol. 1972 Feb 14;63(3):483–503. doi: 10.1016/0022-2836(72)90443-3. [DOI] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. II. Mutations induced by bacteriophage lambda in Escherichia coli K12. J Mol Biol. 1973 Oct 25;80(2):297–314. doi: 10.1016/0022-2836(73)90174-5. [DOI] [PubMed] [Google Scholar]
- Taylor A. L., Trotter C. D. Linkage map of Escherichia coli strain K-12. Bacteriol Rev. 1972 Dec;36(4):504–524. doi: 10.1128/br.36.4.504-524.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]




