Abstract
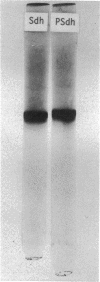
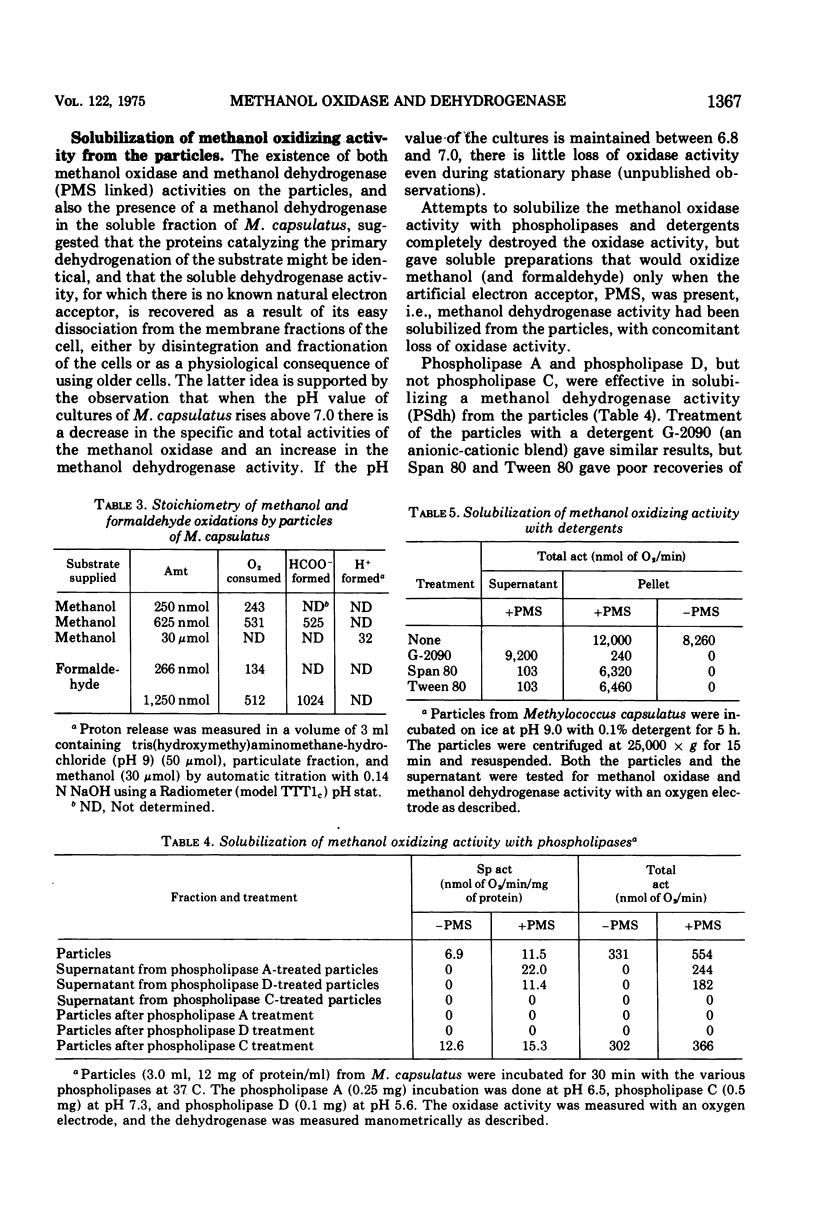
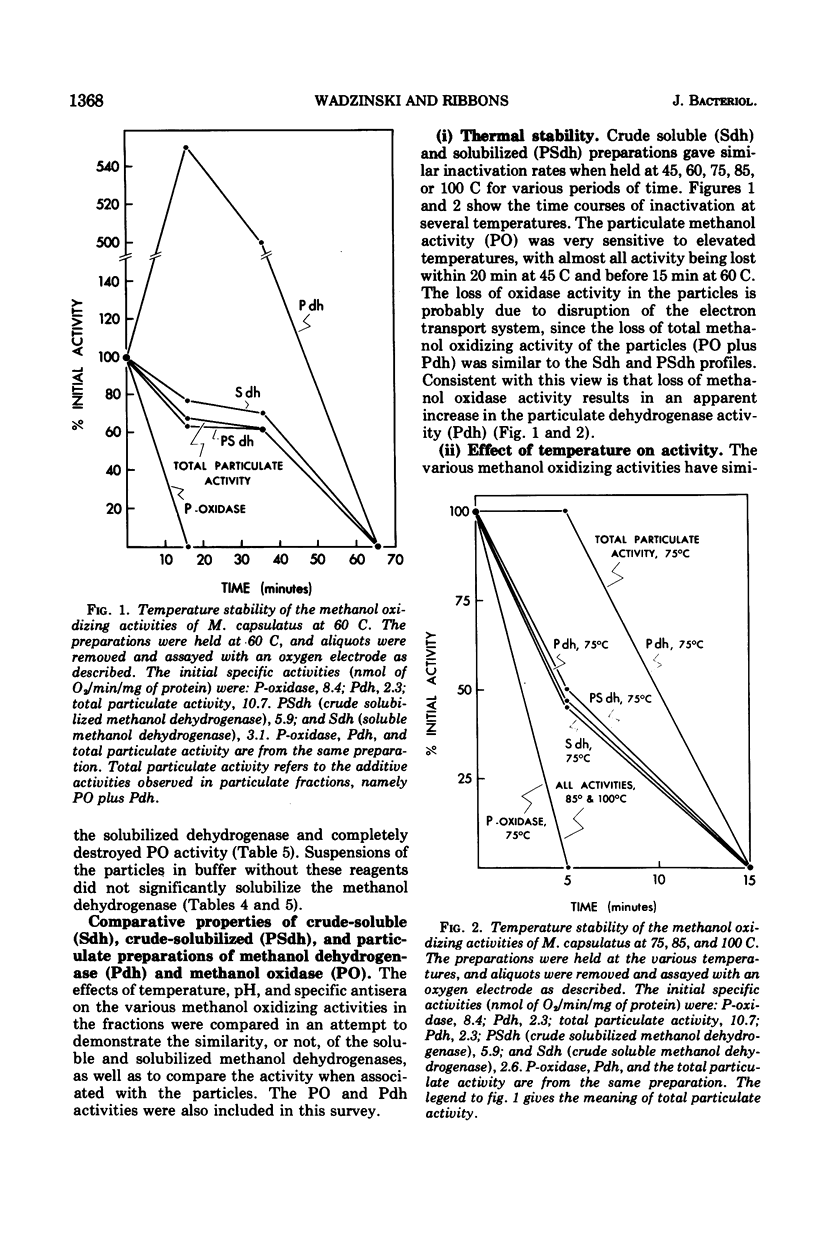
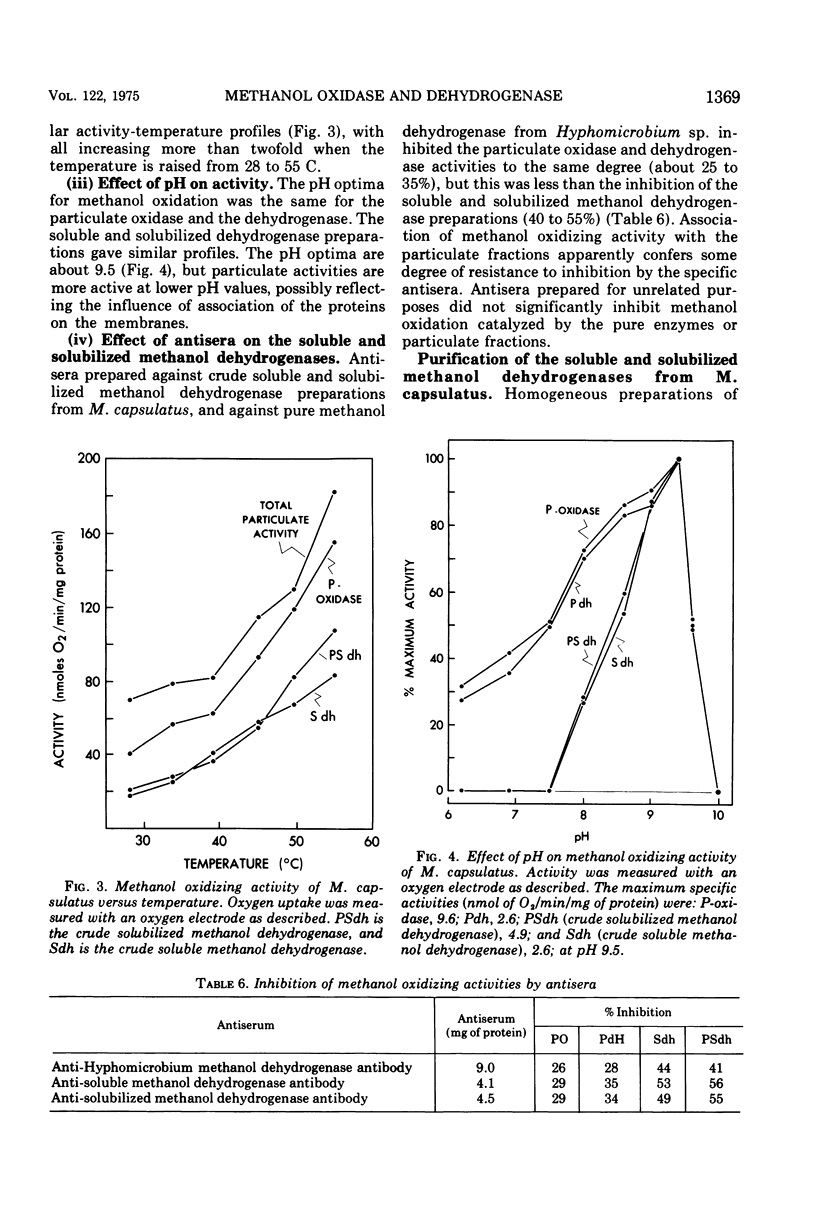
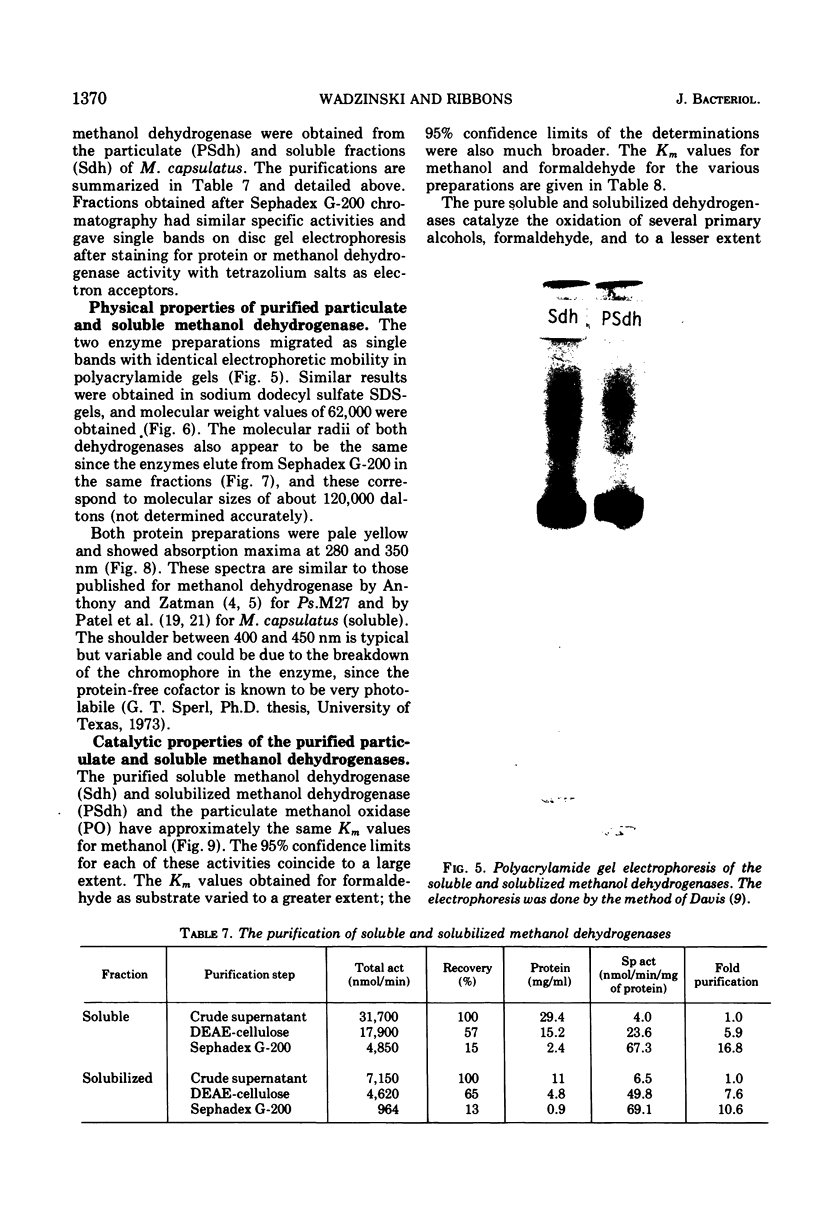
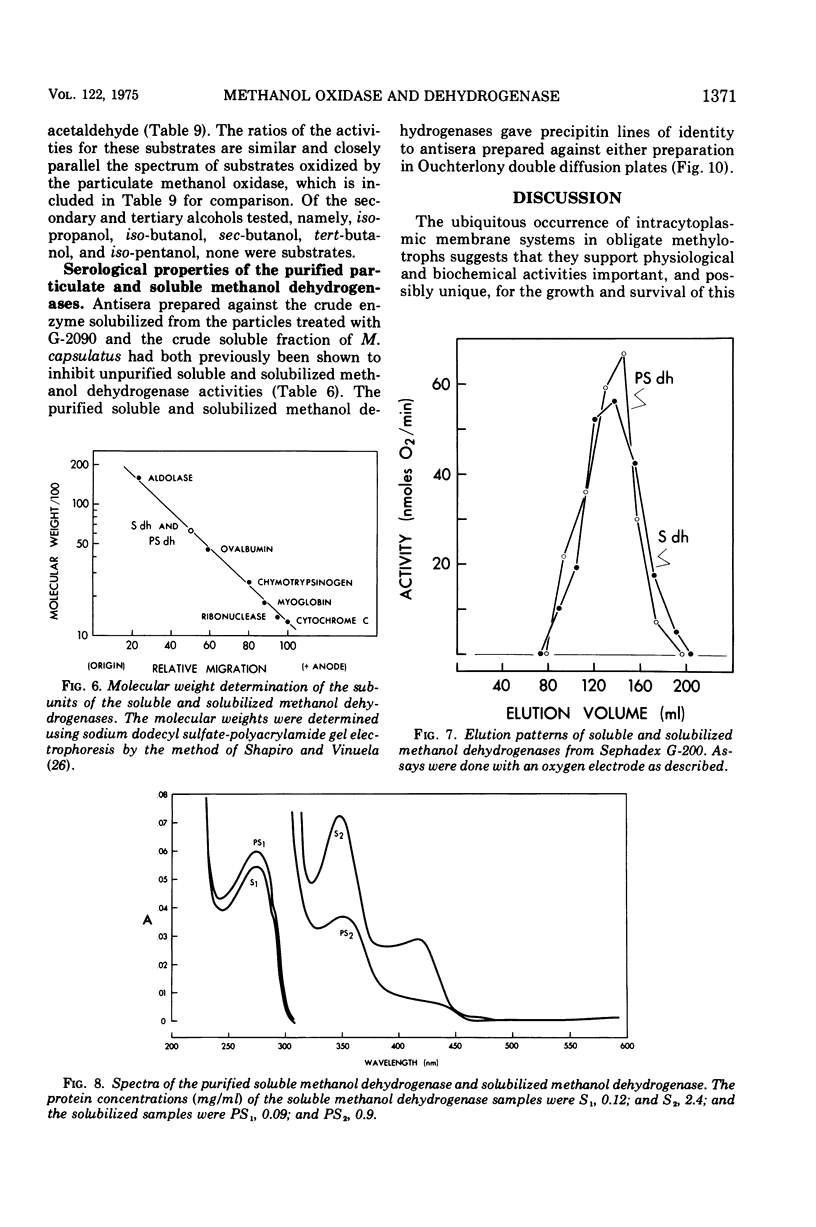
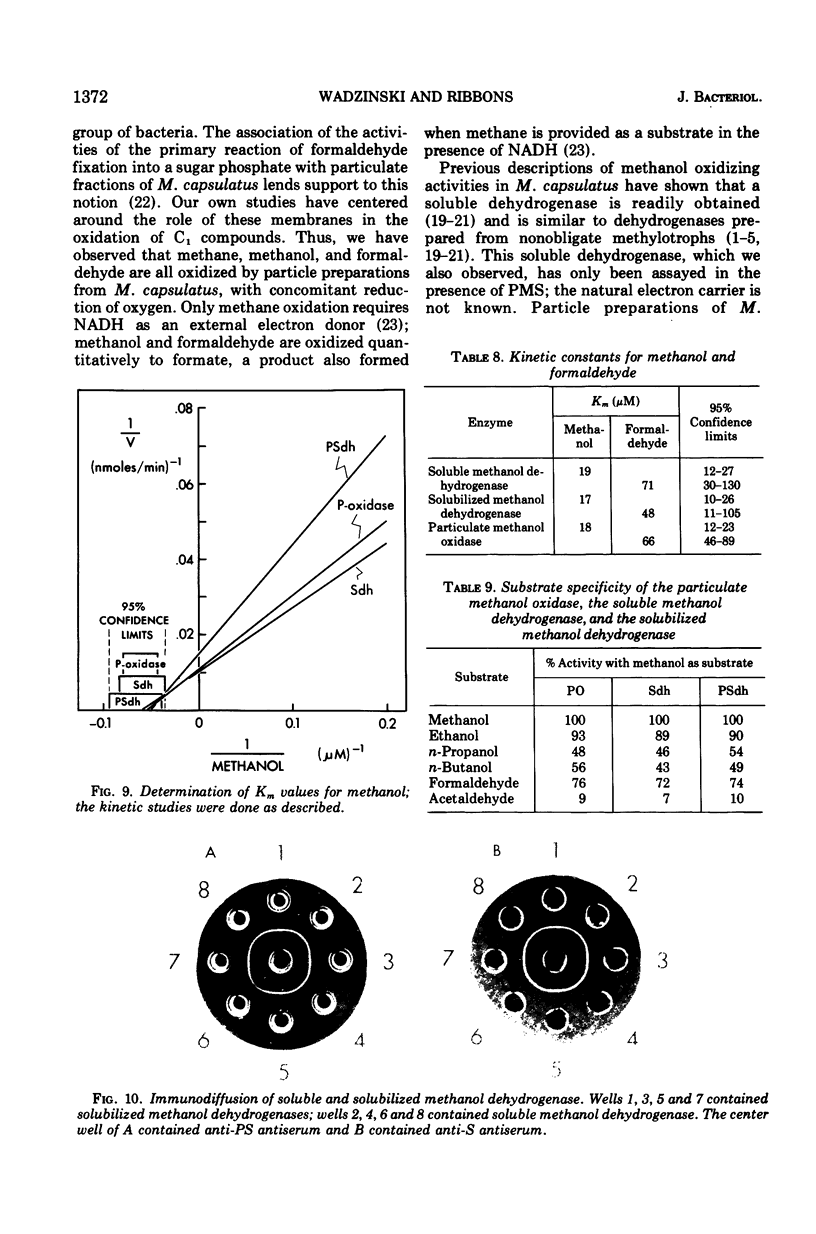
Methanol (and formaldehyde) oxidizing activities in crude extracts of Methylococcus capsulatus are associated mainly with particulate fractions sedimenting between 3,000 and 40,000 X g. Most of the phenazine methosulfate (PMS)-dependent methanol (and formaldehyde) dehydrogenase activity observed resides in the soluble fraction but represents only 40% of the total (PMS dependent plus independent) activity. Both PMS-dependent methanol dehydrogenase activity and PMS-independent methanol oxidase activity are found in particulate fractions, and the PMS-dependent dehydrogenase is easily solubilized by treatment with certain phospholipases or detergents. The properties of the PMS-dependent dehydrogenase activities in the soluble fraction and that solubilized from the particles suggested that they may be identical proteins. Their pH optima, temperature dependence, thermolabilities, and sensitivities to the presence of specific antisera were indistinguishable. Homogeneous preparations of the enzyme proteins obtained from the soluble fractions of extracts and the particulate fractions solubilized by detergents had similar: (i) electrophoretic mobilities in native and denatured states (subunit size in sodium dodecyl sulfate 62,000 daltons); (ii) molecular radii under native conditions, (iii) visible absorption spectra, lambdamax 350 nm, (iv) kinetic constants for methanol and formaldehyde; (v) substrate specificity; and (vi) immunological characteristics--antisera to each enzyme preparation showed precipitin lines of identity to either of the enzymes. It is suggested that the major site of methanol and formaldehyde oxidation in M. capsulatus occurs on the intracytoplasmic membranes in vivo and is coupled to oxygen reduction.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AZOULAY E., HEYDEMAN M. T. Extraction and properties of alcohol dehydrogenase from Pseudomonas aeruginosa. Biochim Biophys Acta. 1963 May 7;73:1–6. doi: 10.1016/0006-3002(63)90353-6. [DOI] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. 1. Isolation and properties of Pseudomonas sp. M27. Biochem J. 1964 Sep;92(3):609–614. doi: 10.1042/bj0920609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. 2. The methanol-oxidizing enzyme of Pseudomonas sp. M 27. Biochem J. 1964 Sep;92(3):614–621. doi: 10.1042/bj0920614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. Purification and properties of the alcohol dehydrogenase of Pseudomonas sp. M27. Biochem J. 1967 Sep;104(3):953–959. doi: 10.1042/bj1040953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. The alcohol dehydrogenase of Pseudomonas sp. M27. Biochem J. 1965 Sep;96(3):808–812. doi: 10.1042/bj0960808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. The prosthetic group of the alcohol dehydrogenase of Pseudomonas sp. M27: a new oxidoreductase prosthetic group. Biochem J. 1967 Sep;104(3):960–969. doi: 10.1042/bj1040960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss C. I., James A. T. Fitting the rectangular hyperbola. Biometrics. 1966 Sep;22(3):573–602. [PubMed] [Google Scholar]
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- Davies S. L., Whittenbury R. Fine structure of methane and other hydrocarbon-utilizing bacteria. J Gen Microbiol. 1970 May;61(2):227–232. doi: 10.1099/00221287-61-2-227. [DOI] [PubMed] [Google Scholar]
- Foster J. W., Davis R. H. A methane-dependent coccus, with notes on classification and nomenclature of obligate, methane-utilizing bacteria. J Bacteriol. 1966 May;91(5):1924–1931. doi: 10.1128/jb.91.5.1924-1931.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARRINGTON A. A., KALLIO R. E. Oxidation of methanol and formaldehyde by pseudomonas methanica. Can J Microbiol. 1960 Feb;6:1–7. doi: 10.1139/m60-001. [DOI] [PubMed] [Google Scholar]
- Johnson P. A., Quayle J. R. Microbial growth on C-1 compounds. 6. Oxidation of methanol, formaldehyde and formate by methanol-grown Pseudomonas AM-1. Biochem J. 1964 Nov;93(2):281–290. doi: 10.1042/bj0930281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp M. B., Quayle J. R. Incorporation of C1 units into allulose phosphate by methane-grown Pseudomonas methanica. Biochim Biophys Acta. 1965 Aug 24;107(1):174–176. doi: 10.1016/0304-4165(65)90415-0. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Patel R. N., Bose H. R., Mandy W. J., Hoare D. S. Physiological studies of methane- and methanol-oxidizing bacteria: comparison of a primary alcohol dehydrogenase from Methylococcus capsulatus (Texas strain) and Pseudomonas species M27. J Bacteriol. 1972 May;110(2):570–577. doi: 10.1128/jb.110.2.570-577.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R. N., Hoare D. S. Physiological studies of methane and methanol-oxidizing bacteria: oxidation of C-1 compounds by Methylococcus capsulatus. J Bacteriol. 1971 Jul;107(1):187–192. doi: 10.1128/jb.107.1.187-192.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbons D. W., Harrison J. E., Wadzinski A. M. Metabolism of single carbon compounds. Annu Rev Microbiol. 1970;24:135–158. doi: 10.1146/annurev.mi.24.100170.001031. [DOI] [PubMed] [Google Scholar]
- Ribbons D. W. Oxidation of C1 Compounds by Particulate fractions from Methylococcus capsulatus: distribution and properties of methane-dependent reduced nicotinamide adenine dinucleotide oxidase (methane hydroxylase). J Bacteriol. 1975 Jun;122(3):1351–1363. doi: 10.1128/jb.122.3.1351-1363.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahm H., Wagner F. Microbial assimilation of methanol. The ethanol- and methanol-oxidizing enzymes of the yeast Candida boidinii. Eur J Biochem. 1973 Jul 2;36(1):250–256. doi: 10.1111/j.1432-1033.1973.tb02907.x. [DOI] [PubMed] [Google Scholar]
- Shapiro A. L., Viñuela E., Maizel J. V., Jr Molecular weight estimation of polypeptide chains by electrophoresis in SDS-polyacrylamide gels. Biochem Biophys Res Commun. 1967 Sep 7;28(5):815–820. doi: 10.1016/0006-291x(67)90391-9. [DOI] [PubMed] [Google Scholar]
- Whittenbury R., Phillips K. C., Wilkinson J. F. Enrichment, isolation and some properties of methane-utilizing bacteria. J Gen Microbiol. 1970 May;61(2):205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]