Abstract
The thermal re-isomerization of retinal from the 13-cis to the all-trans state is a key step in the final stages of the photocycle of the light-driven proton pump, bacteriorhodopsin. This step is greatly slowed upon replacement of Leu-93, a residue in van der Waals contact with retinal. The most likely role of this key interaction is that it restricts the flexibility of retinal. To test this hypothesis, we have exchanged native retinal in Leu-93 mutants with bridged retinal analogs that render retinal less flexible by restricting free rotation around either the C10—C11 (9,11-bridged retinal) or C12—C13 (11,13-bridged retinal) single bonds. The effect of the analogs on the photocycle was then determined spectroscopically by taking advantage of the previous finding that the decay of the O intermediate in the Leu-93 mutants provides a convenient marker for retinal re-isomerization. Time-resolved spectroscopic studies showed that both retinal analogs resulted in a dramatic acceleration of the photocycling time by increasing the rate of decay of the O intermediate. In particular, exchange of native retinal in the Leu-93 → Ala mutant with the 9,11-bridged retinal resulted in an acceleration of the decay of the O intermediate to a rate similar to that seen in wild-type bacteriorhodopsin. We conclude that the protein-induced restriction of conformational flexibility in retinal is a key structural requirement for efficient protein–retinal coupling in the bacteriorhodopsin photocycle.
Bacteriorhodopsin is a retinal-based membrane protein which functions as a light-driven proton pump (1–3). Light absorption isomerizes retinal around the C13=C14 bond, resulting in the formation and decay of a series of optical intermediates. The best characterized of these intermediates (4, 5) are the K, L, M, N, and O intermediates, with the initial state being restored upon decay of the O intermediate. The changes in retinal configuration and protein conformation which occur during the photocycle can be grouped into three stages. In stage 1, retinal is isomerized by light from the all-trans to the 13-cis state, with no significant change in the protein conformation. In stage 2, protein conformational changes trigger the release of a proton on the extracellular side and the subsequent uptake of a proton on the cytoplasmic side. In stage 3, the photocycle is completed with the thermal re-isomerization of retinal to the starting all-trans configuration and the return of the protein to its initial state.
The protein-driven thermal re-isomerization of retinal during the photocycle is a unique feature of bacterial rhodopsins such as bacteriorhodopsin and halorhodopsin, and is not observed during steps in light transduction by either vertebrate or invertebrate rhodopsins. Determination of the structural basis of retinal re-isomerization is therefore at the heart of understanding energy transduction in this family of proteins. In previous spectroscopic studies (6–9), we identified Leu-93, a residue in close proximity (10) to the 13-methyl group of retinal, as a key residue which is required for the rapid completion of stage 3 of the photocycle. Mutants with smaller side chains at position 93 (Leu-93 → Ala, Leu-93 → Thr) displayed lower rates at which the photocycle was completed because of the formation of a long-lived intermediate having a red-shifted absorption maximum (λmax). Because this intermediate was kinetically formed from the M intermediate and had a red-shifted λmax relative to the initial state (6, 9), it was labeled the O intermediate in accordance with the nomenclature for the native photocycle (4). Detailed studies with the Leu-93 → Ala mutant (8) showed that the O intermediate was formed on the same time scale as proton uptake, and that conversion of retinal from the 13-cis to the all-trans state occurred with the decay of this O intermediate. These results led to the conclusion that retinal re-isomerization was the rate-limiting step in the completion of the photocycle of the Leu-93 → Ala mutant. The large kinetic defect observed in retinal re-isomerization in the Leu-93 mutants was not observed upon replacement of any other residue in van der Waals contact with retinal, or upon replacement of Leu-93 by Val.
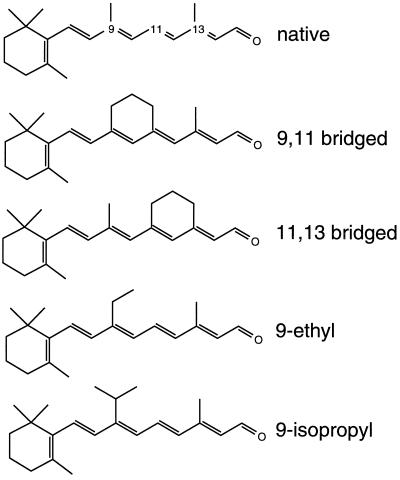
Why does the interaction of retinal with a nonpolar residue such as Leu affect the rate of re-isomerization, a process that is generally believed to involve charged residues in the binding pocket? Retinal is an inherently flexible molecule which can display rotation about several bonds. Upon binding to the protein, however, retinal is constrained to adopt specific conformations, restricting its flexibility. This loss of flexibility occurs because of van der Waals interactions between retinal and the amino acids which line the retinal binding pocket. One explanation for the remarkable effect of the Leu-93 mutations is that the presence of less bulky side chains at residue 93 increases retinal flexibility. The increased flexibility may prevent the population of a retinal–protein conformation that is necessary for rapid re-isomerization. This hypothesis is consistent with previous proposals that the rapid thermal isomerization of retinal requires specific placement of the polyene chain with respect to key charged residues (11–15). To test whether loss of the Leu-93/C13 methyl interaction leads to an increase in the flexibility of retinal, we have investigated the effects of bridged retinals on the photocycle in bacteriorhodopsin mutants (Leu-93 → Ala, Leu-93 → Thr, and the double mutant Leu-93 → Ala, Asp-96 → Asn) where stage 3 is slowed down. The 9,11- and 11,13-bridged retinals (Fig. 1) were selected because they restrict the flexibility of retinal, but have no significant effect on the photocycle of wild-type bacteriorhodopsin (16). Transient absorption spectroscopy was then used to follow the kinetics of the O intermediate, which is a marker for retinal re-isomerization in the photocycle in the Leu-93 mutants (8).
Figure 1.
All-trans retinal and the retinal analogs used in this study. The 9,11-bridged retinal restricts rotation around the C10—C11 single bond, and the 11,13-bridged retinal restricts rotation around the C12—C13 single bond. The 9-ethyl and 9-isopropyl analogs provide increasing amounts of bulk at the C9 position of retinal.
EXPERIMENTAL PROCEDURES
Generation of Reconstituted Pigments.
All-trans-E-11,19-ethanoretinal (9,11-bridged retinal) and all-trans-E-11,20-ethanoretinal (11,13-bridged retinal) were prepared as described elsewhere (16). The 9-ethyl and the 9-isopropyl retinal analogs were synthesized by condensation of β-ionone with either 0.5 (for 9-ethyl) or 2 (for 9-isopropyl) equivalents of methyl iodide in the presence of sodium amide (1 and 3 equivalents, correspondingly). Condensation of the resulting substituted β-ionone, to generate the retinal analog, was carried out by standard schemes. The purification of purple membranes containing the Leu-93 → Ala, Leu-93 → Val, Leu-93 → Thr, and Leu-93 → Ala, Asp-96 → Asn mutant proteins (8, 17) was carried out using methods previously described (18).
Replacement of native retinal in the wild-type and mutant bacteriorhodopsins was performed by first bleaching the bacteriorhodopsin membrane fragments with orange and red light in the presence of 1 M hydroxylamine at pH 8. The resulting retinal oximes were removed by washing the membranes four or five times with a 1% bovine serum albumin solution (19). Reconstitution studies of the retinal analogs with bacterio-opsin were carried out at 22°C in sodium phosphate buffer (10 mM) at pH 7 with 0.15 M KCl.
Spectroscopic Measurements.
Transient absorption spectroscopy was performed as described previously (8). In a typical experiment, 128 transient traces were averaged from a sample volume of 1 ml (absorbance ≈ 0.1–0.3 at the visible λmax). Spectroscopic studies of mutant opsins reconstituted with the analogs were carried out at 22°C in sodium phosphate buffer (10 mM) at pH 7 with 0.15 M KCl (unless otherwise stated).
RESULTS
Exchanging Native Retinal with the 9,11-Bridged Retinal Accelerates the Photocycle of the Leu-93 → Ala Mutant.
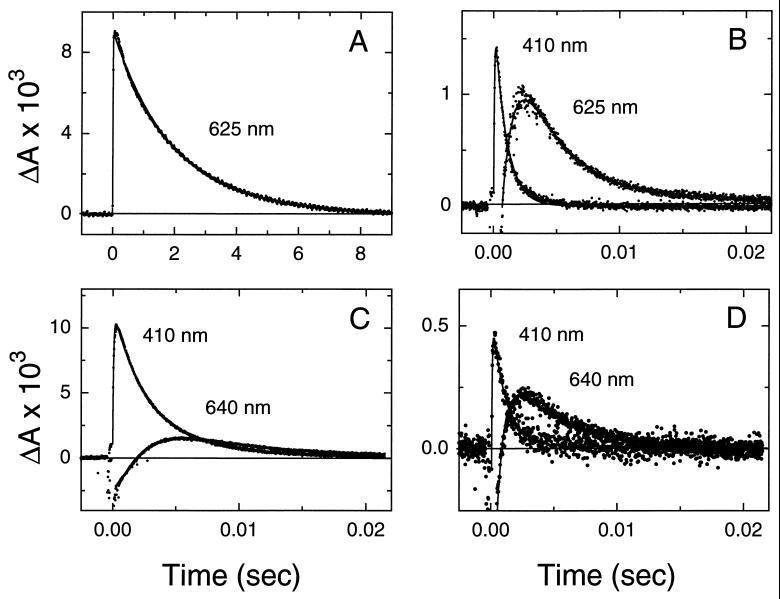
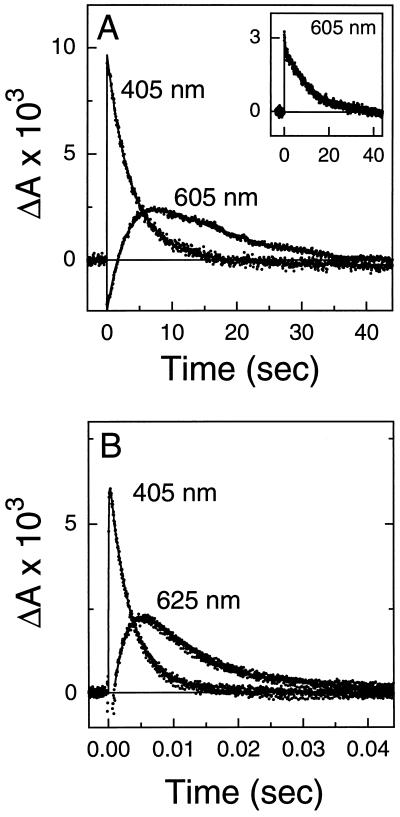
Exchange of retinal in the Leu-93 → Ala mutant with the 9,11-bridged retinal did not alter the visible λmax (Table 1). Retinal extraction experiments indicated that the 9,11-bridged retinal was predominantly in the all-trans form in the reconstituted pigment (J.K.D. and S.S, unpublished results). The analog slightly accelerated (≈3-fold) the decay of the M intermediate in the Leu-93 → Ala mutant (Table 1). However, a dramatic acceleration (≈550-fold) in the decay of the O intermediate was observed (Fig. 2 A and B; Table 1). These results demonstrate that bridging the polyene chain between the C9 to C11 regions with a six-membered ring is sufficient to restore the rapid completion of the photocycle, and therefore rapid retinal re-isomerization in the presence of the Leu-93 → Ala mutation. Control experiments with wild-type bacteriorhodopsin showed that exchanging native retinal with the 9,11-bridged retinal did not significantly alter the visible λmax or steps in the photocycle (ref. 16, Fig. 2 C and D, and Table 1). The small (≈2-fold) increase in the time constant of decay of the M intermediate is similar to that observed with the Leu-93 → Ala mutant (Table 1).
Table 1.
Effects of the bridged retinals on the λmax and photocycle kinetics
| Protein | Retinal | Visible λmax, nm | M decay, ms | O decay, ms |
|---|---|---|---|---|
| Wild type | Native | 568 | 2.5* | 7 |
| Wild type | 9,11-Bridged | 567 | 1.1 | 5 |
| Wild type | 11,13-Bridged | 556 | 0.6 | ≈4 |
| Leu-93 → Ala | Native | 539 | 3.0 | 1900 |
| Leu-93 → Ala | 9,11-Bridged | 538 | 0.9 | 3.4 |
| Leu-93 → Ala | 11,13-Bridged | 530 | 0.7 | ≈15* |
| Leu-93 → Thr | Native | 541 | 17 | 1100 |
| Leu-93 → Thr | 9,11-Bridged | 540 | 90 | Not obs. |
| Leu-93 → Thr | 11,13-Bridged | 532 | 60 | Not obs. |
| Leu-93 → Val | Native | 564 | 2.4* | 13 |
| Leu-93 → Val | 9,11-Bridged | 564 | 2.2* | 6.2 |
| Leu-93 → Val | 11,13-Bridged | 552 | 1.0 | 5.3 |
Represents decay of dominant component.
Figure 2.
The 9,11-bridged retinal acts to rescue the long-lived O intermediate in the Leu-93 → Ala mutant. (A) Transient changes in absorbance of the Leu-93 → Ala mutant (with native retinal) to follow decay of the O intermediate (measured at 625 nm). Note the long time scale for the decay. (B) Same as in A except native retinal was replaced by the 9,11-bridged retinal. Changes in absorbance at 410 nm (M intermediate) and at 625 nm (O intermediate) are shown. (C) Transient changes in absorbance of wild-type bacteriorhodopsin (with native retinal) showing the kinetics of the M (measured at 410 nm) and O (measured at 640 nm) intermediates. (D) Same as in C except native retinal was replaced by the 9,11-bridged retinal. Comparison of the decay of the O intermediate in the different panels shows that the 9,11-bridged retinal acts to accelerate the decay of the O intermediate by ≈550-fold in the Leu-93 → Ala mutant while leaving it nearly unchanged in wild-type bacteriorhodopsin.
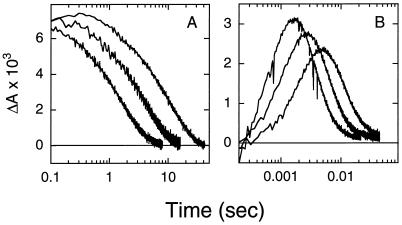
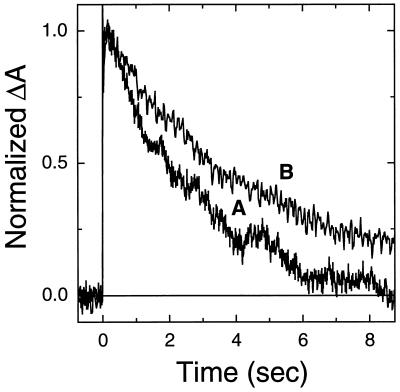
The acceleration of the photocycle of the Leu-93 → Ala mutant by the 9,11-bridged retinal is also associated with a change in the temperature dependence of the peak amplitude of the O intermediate (Fig. 3). The maximal amount of the O intermediate decreased with decreasing temperature in the pigment reconstituted with the 9,11-bridged retinal, in contrast to the near temperature-independent behavior observed with native retinal. Further, replacement of native retinal in the Leu-93 → Ala mutant by the 9,11-bridged retinal led to a decrease in the activation energy for the decay of the O intermediate from ≈77 kJ/mol to ≈54 kJ/mol. Interestingly, both features (i.e., the temperature dependence of the amplitude of the O intermediate, and the activation energy) observed with the analog-reconstituted mutant are closer to that of wild-type bacteriorhodopsin than to the Leu-93 → Ala mutant (8, 20).
Figure 3.
The 9,11-bridged retinal acts to reverse the effects of the Leu-93 → Ala mutant on the temperature dependence of the O intermediate. (A) Transient changes in absorbance of the Leu-93 → Ala mutant (with native retinal) measured at 600 nm to follow decay of the O intermediate. Measurements were made at 25.5, 17, and 9 ± 1°C. (B) Same as in A except native retinal was replaced by the 9,11-bridged retinal. Measurements were made at 25.5, 17, and 9 ± 1°C.
Reconstitution of Other Leu-93 Mutants with the 9,11-Bridged Retinal also Accelerates the Photocycle.
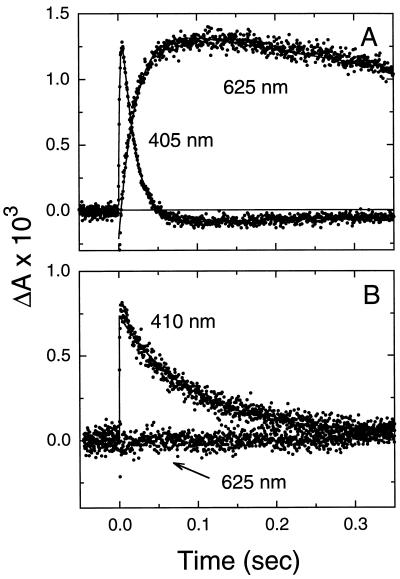
Previous studies have shown that other replacements at Leu-93 also slow the completion of the photocycle, but to differing extents (6). For example, the loss of one CH2 group in the Leu-93 → Val mutant has little effect on the lifetime of the O intermediate (ref. 6 and Table 1), whereas the replacement of Leu-93 by Thr results in an ≈150-fold increase in the lifetime of the O intermediate (refs. 6 and 17 and Fig. 4A). We therefore investigated the effects of exchanging native retinal in these mutants with the 9,11-bridged retinal.
Figure 4.
The 9,11-bridged retinal acts to rescue the long-lived O intermediate in the Leu-93 → Thr mutant. (A) Transient changes in absorbance of the Leu-93 → Thr mutant (with native retinal) showing the kinetics of the M (measured at 410 nm) and O (measured at 625 nm) intermediates. (B) Same as in A except native retinal was replaced by the 9,11-bridged retinal. Transient absorbance changes at 410 nm are due to the formation and decay of the M intermediate, but unlike with native retinal (A) no absorbance changes that can be assigned to the O intermediate are observed at 625 nm.
Replacement of native retinal with the 9,11-bridged retinal in the Leu-93 → Val and the Leu-93 → Thr mutants did not shift the visible λmax. As expected, the effect of the retinal analog on the Leu-93 → Val mutant was similar to that seen for wild-type bacteriorhodopsin, and only an ≈2-fold acceleration was observed in the decay of the O intermediate (Table 1). However, in the photocycle of the Leu-93 → Thr mutant, introduction of the 9,11-bridged retinal resulted in no net accumulation of the O intermediate (Fig. 4B). Completion of the photocycle occurred with the decay of the M intermediate, indicating that decay of the O intermediate was accelerated by at least 12-fold. Together, these findings strongly suggest that the acceleration of decay of the O intermediate by the 9,11-bridged retinal is independent of the nature of the amino acid replacement at residue 93.
To further investigate the ability of the 9,11-bridged retinal to accelerate decay of the O intermediate, we studied the effect of this retinal on the photocycle of the Leu-93 → Ala, Asp-96 → Asn double mutant. Previous studies have shown that the decays of the M and O intermediates are considerably slower in this double mutant than in wild-type bacteriorhodopsin (Fig. 5A and ref. 9). Moreover, these mutations act separately in the photocycle, with the long-lived M intermediate arising from the Asp-96 → Asn mutation and the long-lived O intermediate originating from the Leu-93 → Ala mutation.
Figure 5.
The 9,11-bridged retinal acts to rescue the long-lived O intermediate in the double mutant Leu-93 → Ala, Asp-96 → Asn independent of the lifetime of the M intermediate. (A) Transient changes in absorbance with native retinal showing the presence of long-lived M (measured at 405 nm) and O (measured at 605 nm) intermediates. The increased lifetime of the M intermediate is due to the Asp-96 → Asn mutation. (Inset) The addition of ≈4 mM sodium azide accelerates the decay of the M intermediate and the rise of the O intermediate, but not the decay of the O intermediate. (B) Same as in A except native retinal was replaced by the 9,11-bridged retinal and sodium azide (≈4 mM) was added.
Replacement of native retinal in the double mutant with the 9,11-bridged retinal resulted in a photocycle where no accumulation of the O intermediate was observed, and decay of the M intermediate was the rate-limiting step in the completion of the photocycle (data not shown). To determine the extent of acceleration of the decay of the O intermediate by the bridged retinal, photocycle kinetics were measured in the presence of azide. Azide selectively accelerates decay of the M intermediate without affecting decay of the O intermediate in the double mutant (refs. 9 and 21; Fig. 5A Inset). In the presence of azide, the decay of the O intermediate in the analog-reconstituted Leu-93 → Ala, Asp-96 → Asn mutant was accelerated by ≈1000-fold (τ ≈ 9 ms; Fig. 5B). These results demonstrate that the effect of the 9,11-bridged retinal on the acceleration of decay of the O intermediate occurs under conditions where decay of the M intermediate is variable by up to three orders of magnitude.
Addition of Bulk at the C9 Position of Retinal Does Not Accelerate the Photocycle of the Leu-93 → Ala Mutant.
The introduction of the 9,11-bridging group may introduce more bulk in the region of the C9 position of retinal than is present in native retinal (Fig. 1). To determine if the acceleration of the photocycle is due to the added bulk at the C9 position, native retinal was replaced by an analog containing either an ethyl or an isopropyl group at the C9 position. The bulk provided by the ethyl group at the C9 position is similar to that in the 9,11-bridged retinal, whereas the bulk provided by the isopropyl group exceeds that of the 9,11-bridged retinal. As shown in Fig. 6, no acceleration of the photocycle was observed with either the 9-ethyl or the 9-isopropyl analog, suggesting that the mere presence of bulk at the C9 position is not sufficient to accelerate the Leu-93 → Ala mutant photocycle.
Figure 6.
Increasing the bulk of the substituent at the C9 position of retinal does not accelerate the decay of the O intermediate in the photocycle of the Leu-93 → Ala mutant. Transient absorbance traces were measured at 625 nm in the Leu-93 → Ala mutant with 9-ethylretinal (trace A) or 9-isopropylretinal (trace B). The kinetics were measured at 22°C in sodium phosphate buffer (10 mM) at pH 7 with 0.15 M KCl and 3 M NaCl, because formation of a chromophore with a single visible absorbance peak was observed only at high ionic strengths.
The 11,13-Bridged Retinal Analog also Accelerates Decay of the O Intermediate in the Leu-93 Mutants.
To test whether retinal analogs containing other bridging groups would also accelerate completion of the photocycle of the Leu-93 mutants, we studied the effects of exchanging native retinal with 11,13-bridged retinal, in which the bridging group spans the C11 to C13 portion of the polyene chain. Exchange of native retinal in wild-type bacteriorhodopsin with the 11,13-bridged retinal does not alter the photocycle, but it does result in an ≈12-nm blue shift of the visible absorbance peak (ref. 16; Table 1). However, when incorporated into the Leu-93 → Val and Leu-93 → Ala mutants, the 11,13-bridged retinal accelerates the decay of the O intermediate by ≈2-fold and ≈120-fold, respectively. Similarly, the 11,13-bridged retinal accelerates decay of the O intermediate in the Leu-93 → Thr mutant sufficiently to make the photocycle complete with the decay of the M intermediate. Since retinal flexibility is restricted by introduction of the bridging groups, the results with the 9,11- and 11,13-bridged retinals point to an important role for the Leu-93/C13 retinal methyl interaction in constraining motion within the polyene chain to aid in its rapid re-isomerization.
DISCUSSION
The rate of thermal retinal re-isomerization in the bacteriorhodopsin photocycle is expected to depend on both electrostatic and steric factors (12). Varying electrostatic interactions between the protonated Schiff base and neighboring negative charges (e.g., Asp-85) can directly lower the barrier for thermal isomerization by increasing the π-electron delocalization along the polyene chain (11–15). Independent of these electrostatic effects, steric interactions within the binding pocket may also regulate the barriers to isomerization, for example, by placing retinal in a conformation that favors efficient re-isomerization. Thus, the protein can “drive” retinal re-isomerization by ensuring that a retinal–protein conformation is adopted which ultimately results in the lowering of the barrier to isomerization. Since retinal is a flexible molecule, the lowering of these barriers most likely originates from specific van der Waals interactions between retinal and amino acids in the binding pocket.
Systematic mutagenesis studies of each of the residues in the binding pocket have identified a critical role for Leu-93 in the thermal re-isomerization of retinal during the photocycle. Leu-93 is in contact with the C13 methyl group and is in close proximity to the C13=C14 bond, which is the site of isomerization (10). The replacement of Leu-93 by less bulky amino acids should effectively create a cavity within the protein. Even though some re-organization of the binding pocket may occur in such mutants, the replacement of Leu-93 results in the loss of a key retinal–protein interaction, which is essential for rapid re-isomerization. If the loss of this interaction specifically increases the flexibility of the bound retinal, it should be possible, in principle, to compensate for the loss of the interaction by independently restricting the flexibility of retinal.
The introduction of a bridging group along the polyene chain is expected to restrict retinal flexibility whether or not it is bound to the protein. This intrinsic reduction in flexibility alone may be sufficient to account for the observed acceleration of decay of the O intermediate. In addition, it is possible that new steric interactions which are introduced between retinal and the amino acids in the binding pocket may also restrict its flexibility and contribute to accelerating the decay of the O intermediate. However, the acceleration cannot be attributed to the re-establishment of the C13/Leu-93 interaction, since neither of the bridged retinals used here provides additional bulk at the C13 position to re-establish the van der Waals interaction with Leu-93. Similarly, the results with the C9-substituted analogs (Fig. 6) show that acceleration of the decay of the O intermediate by the bridged retinals cannot simply be a consequence of additional bulk at the C9 position. Thus, while we cannot exclude the possibility that the introduction of a new steric interaction at a position other than C9 or C13 may be important for the observed effects of the bridged retinals, the results presented here strongly argue that the restriction of retinal flexibility by its van der Waals interaction with Leu-93 is necessary for the its rapid thermal re-isomerization during the photocycle.
The conclusion that the bridged analogs specifically rescue the loss of the Leu-93–retinal van der Waals interaction is further underscored by recent studies with the Glu-204 → Gln mutant. As in the case of the Leu-93 → Ala mutant, a long-lived O intermediate is observed in the photocycle of this mutant (22). However, unlike Leu-93, Glu-204 is not in van der Waals contact with retinal, and the presence of the long-lived O intermediate has been attributed to a delay in the deprotonation of Asp-85. Time-resolved spectroscopic measurements show that the decay of the O intermediate in the Glu-204 → Gln mutant is not accelerated when it is reconstituted with the 9,11-bridged retinal (J.K.D. and S.S., unpublished data). These experiments show that the bridged retinals do not nonspecifically accelerate the late stages of the photocycle in all cases where a long-lived O intermediate is observed, but that they specifically compensate for the absence of Leu-93.
The O intermediate populated in the Leu-93 → Ala mutant appears to be significantly different from the O intermediate populated in the photocycle of wild-type bacteriorhodopsin (8, 9). Several lines of evidence demonstrate that the O intermediate in the Leu-93 → Ala mutant contains retinal in the 13-cis configuration, in contrast to the all-trans configuration (23) observed in wild-type bacteriorhodopsin. As discussed previously (8, 9) the 13-cis O intermediate could be a precursor for a transient all-trans O intermediate which does not accumulate during the photocycle of the Leu-93 → Ala mutant. Interestingly, incorporation of the 9,11-bridged analog not only accelerates completion of the photocycle but also appears to restore wild-type-like properties to the O intermediate that accumulates during the photocycle (Figs. 2 and 3). Thus, it is conceivable that an all-trans O intermediate is accumulated in the photocycle of the Leu-93 → Ala mutant reconstituted with the 9,11-bridged retinal.
Rapid thermal re-isomerization of retinal is a common feature of the photocycles of the archaebacterial rhodopsins bacteriorhodopsin and halorhodopsin and of sensory rhodopsins I and II. Sequence alignments indicate that in almost all members of this family sequenced so far, a Leu or Ile side chain is present at residue 93 (24, 25), suggesting that similar van der Waals interactions may be critical for efficiently coupling protein conformational changes during the photocycle to the rapid thermal re-isomerization of retinal. More generally, our results suggest that regulation of ligand conformation by van der Waals interactions with buried nonpolar residues may also be a critical element in signal transduction by seven-helix proteins that function as G protein-coupled receptors.
Acknowledgments
We thank Ms. K. Turbedsky for assistance with preparing some of the samples used for spectroscopic experiments and Drs. J. Lanyi and R. Needleman for gift of the Glu-204 → Gln mutant. This work was supported by grants from the National Eye Institute and the Searle Scholars Program/Chicago Community Trust (to S.S.), and by a National Eye Institute National Research Service Award Fellowship (to J.K.D.).
References
- 1.Stoeckenius W, Lozier R H, Bogomolni R A. Biochim Biophys Acta. 1979;505:215–278. doi: 10.1016/0304-4173(79)90006-5. [DOI] [PubMed] [Google Scholar]
- 2.Khorana H G. J Biol Chem. 1988;263:7439–7442. [PubMed] [Google Scholar]
- 3.Lanyi J K. Biochim Biophys Acta. 1993;1183:241–261. doi: 10.1016/0005-2728(93)90226-6. [DOI] [PubMed] [Google Scholar]
- 4.Lozier R H, Bogomolni R A, Stoeckenius W. Biophys J. 1975;15:955–962. doi: 10.1016/S0006-3495(75)85875-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebrey T G. In: Thermodynamics of Membrane Receptors and Channels. Jackson M, editor. Boca Raton, FL: CRC; 1993. pp. 353–387. [Google Scholar]
- 6.Subramaniam S, Greenhalgh D A, Rath P, Rothschild K J, Khorana H G. Proc Natl Acad Sci USA. 1991;88:6873–6877. doi: 10.1073/pnas.88.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenhalgh D A, Farrens D L, Subramaniam S, Khorana H G. J Biol Chem. 1993;268:20305–20311. [PubMed] [Google Scholar]
- 8.Delaney J K, Schweiger U, Subramaniam S. Proc Natl Acad Sci USA. 1995;92:11120–11124. doi: 10.1073/pnas.92.24.11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delaney J K, Subramaniam S. Biophys J. 1996;70:2366–2372. doi: 10.1016/S0006-3495(96)79803-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson R, Baldwin J M, Ceska T A, Zemlin F, Beckmann E, Downing K H. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 11.Orlandi G, Schulten K. Chem Phys Lett. 1979;64:370–374. [Google Scholar]
- 12.Sheves M, Baasov T. J Am Chem Soc. 1984;106:6840–6841. [Google Scholar]
- 13.Tavan P, Schulten K, Oesterhelt D. Biophys J. 1985;47:415–430. doi: 10.1016/S0006-3495(85)83933-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milder S J. Biophys J. 1991;60:440–446. doi: 10.1016/S0006-3495(91)82070-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balashov S P, Govindjee R, Kono M, Imasheva E S, Lukashev E, Ebrey T G, Crouch R K, Menick D R, Feng Y. Biochemistry. 1993;32:10331–10343. doi: 10.1021/bi00090a008. [DOI] [PubMed] [Google Scholar]
- 16.Albeck A, Friedman N, Sheves M, Ottolenghi M. J Am Chem Soc. 1986;108:4614–4618. [Google Scholar]
- 17.Cao Y, Brown L S, Needleman R, Lanyi J K. Biochemistry. 1993;32:10239–10248. doi: 10.1021/bi00089a046. [DOI] [PubMed] [Google Scholar]
- 18.Oesterhelt D, Stoeckenius W. Methods Enzymol. 1974;31:667–678. doi: 10.1016/0076-6879(74)31072-5. [DOI] [PubMed] [Google Scholar]
- 19.Katre N V, Wolber P K, Stoeckenius W, Stroud R M. Proc Natl Acad Sci USA. 1981;78:4068–4072. doi: 10.1073/pnas.78.7.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vàrò G, Lanyi J K. Biochemistry. 1991;30:5016–5022. doi: 10.1021/bi00234a025. [DOI] [PubMed] [Google Scholar]
- 21.Tittor J, Wahl M, Schweiger U, Oesterhelt D. Biochim Biophys Acta. 1994;1187:191–197. [Google Scholar]
- 22.Brown L S, Sasaki J, Kandori H, Maeda A, Needleman R, Lanyi J K. J Biol Chem. 1995;270:27122–27126. doi: 10.1074/jbc.270.45.27122. [DOI] [PubMed] [Google Scholar]
- 23.Smith S O, Pardoen J A, Mulder P P J, Curry B, Lugtenburg J, Mathies R. Biochemistry. 1983;22:6141–6148. [Google Scholar]
- 24.Mukohata Y. Biophys Chem. 1994;50:191–201. doi: 10.1016/0301-4622(94)85031-3. [DOI] [PubMed] [Google Scholar]
- 25.Seidel R, Scharf B, Gautel M, Kleine K, Oesterhelt D, Engelhard M. Proc Natl Acad Sci USA. 1995;92:3036–3040. doi: 10.1073/pnas.92.7.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]