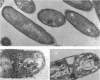
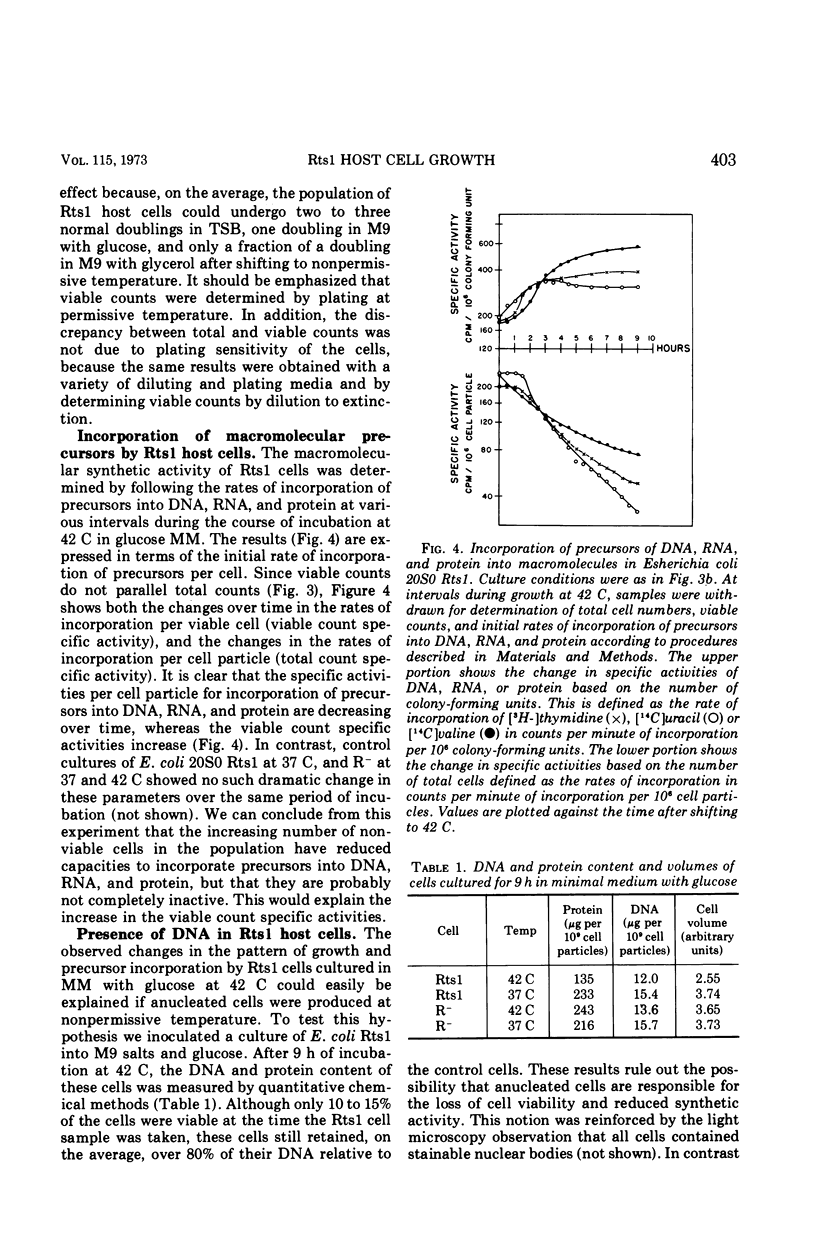
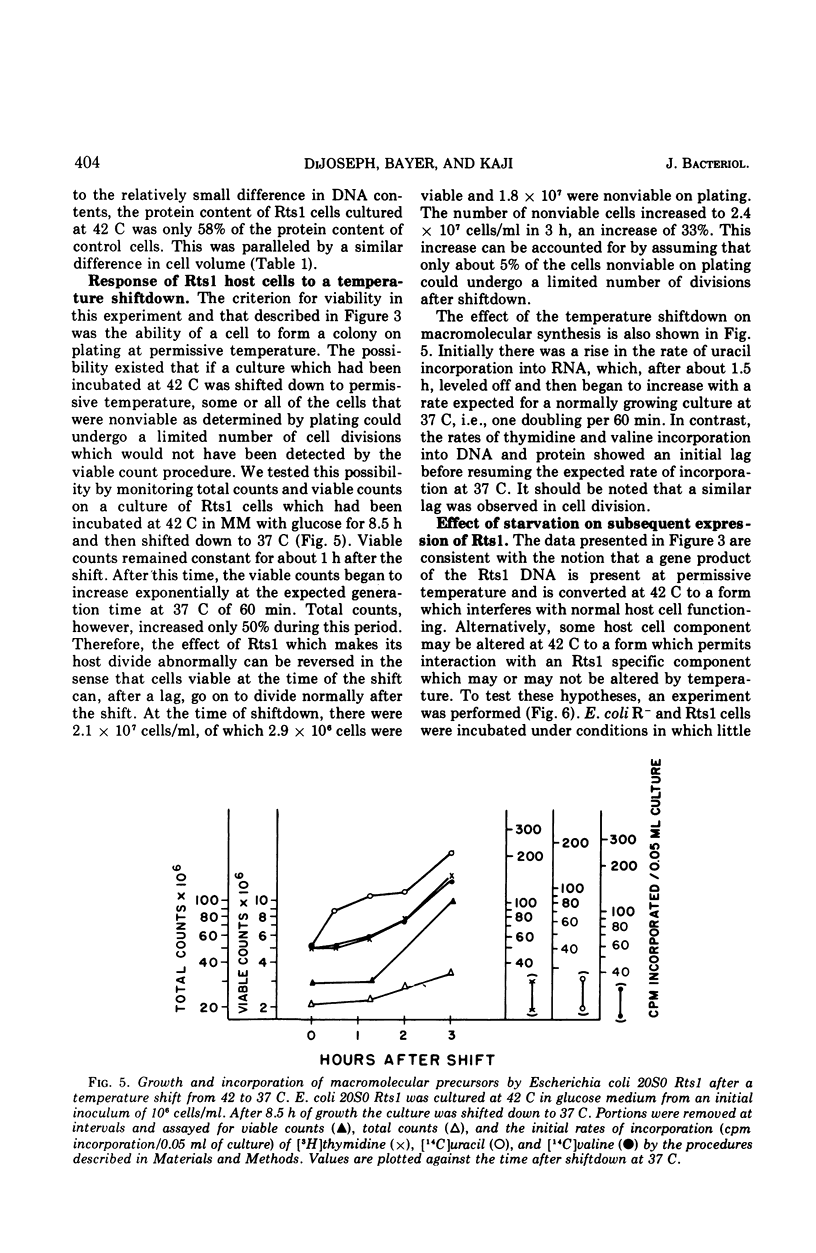
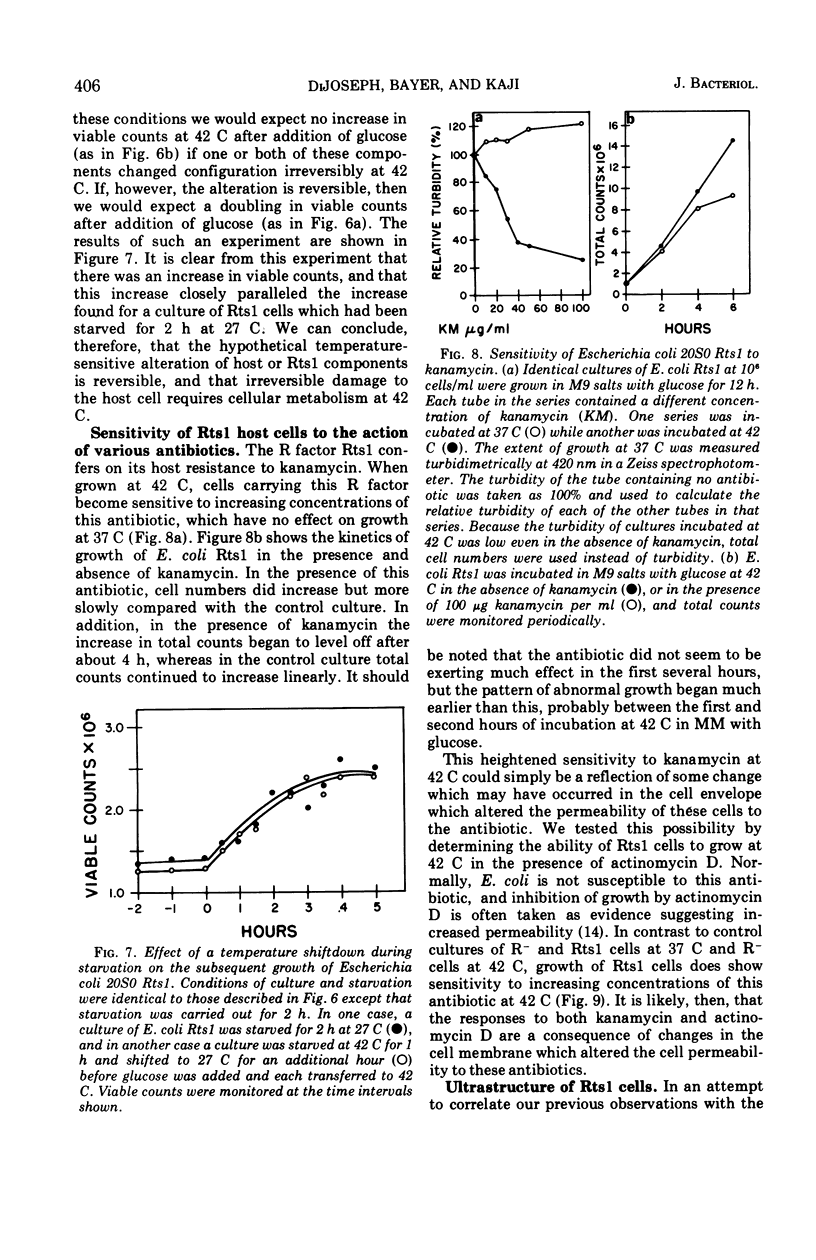
Abstract
We have confirmed and extended the observation of Terawaki et al. that the R factor, Rts1, alters the growth of its host at 42 C. In all media tested there was a period during which total cell numbers increased linearly, while viable counts remained constant. During this period the rate of precursor incorporation per cell particle into deoxyribonucleic acid, ribonucleic acid, and protein declined steadily. These patterns were a consequence of the accumulation of increasing numbers of cells which had lost colony-forming ability. A temperature shiftdown experiment showed that the colony formers could, after a lag, go on to divide normally, whereas most of the noncolony formers could not undergo even a limited number of divisions after shiftdown. The number of normal divisions which occurred after shiftup of Rts1 cells to 42 C was medium dependent. In rich medium there were, on the average, two or three doublings; in glucose medium, one; and in glycerol medium, only a fraction of a doubling. Even in glucose medium, however, no increase in viable counts was observed during growth at 42 C if the cells were first starved for glucose for 1 h at 42 C. A temperature shiftdown from 42 C to 27 C during glucose starvation reversed the effect of starvation at 42 C alone. These results are consistent with the hypothesis that the thermosensitive Rts1 component(s) responsible for the host effects is present at permissive temperature, but can undergo a reversible temperature-induced alteration which then interferes with some essential host function. The detrimental effects of this R factor on its host were also reflected in a heightened sensitivity to kanamycin and actinomycin D at 42 C. Electron microscope observations revealed changes in the appearance of the cell membrane. Membranous invaginations were noted at discrete sites in the cell.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayer M. E. Areas of adhesion between wall and membrane of Escherichia coli. J Gen Microbiol. 1968 Oct;53(3):395–404. doi: 10.1099/00221287-53-3-395. [DOI] [PubMed] [Google Scholar]
- Capaldo-Kimball F., Barbour S. D. Involvement of recombination genes in growth and viability of Escherichia coli K-12. J Bacteriol. 1971 Apr;106(1):204–212. doi: 10.1128/jb.106.1.204-212.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee J. N., Datta N., Hedges R. W. R factors from Proteus rettgeri. J Gen Microbiol. 1972 Oct;72(3):543–552. doi: 10.1099/00221287-72-3-543. [DOI] [PubMed] [Google Scholar]
- Goto N., Yoshida Y., Terawaki Y., Nakaya R., Suzuki K. Base composition of deoxyribonucleic acid of the temperature-sensitive kanamycin-resistant R factor, Rts1. J Bacteriol. 1970 Mar;101(3):856–859. doi: 10.1128/jb.101.3.856-859.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota Y., Ryter A., Jacob F. Thermosensitive mutants of E. coli affected in the processes of DNA synthesis and cellular division. Cold Spring Harb Symp Quant Biol. 1968;33:677–693. doi: 10.1101/sqb.1968.033.01.077. [DOI] [PubMed] [Google Scholar]
- Koch A. L. The adaptive responses of Escherichia coli to a feast and famine existence. Adv Microb Physiol. 1971;6:147–217. doi: 10.1016/s0065-2911(08)60069-7. [DOI] [PubMed] [Google Scholar]
- Kogut M., Prizant E. The accumulation of "run-off" ribosomes during treatment of Escherichia coli with dihydrostreptomycin in vivo. J Gen Microbiol. 1972 May;70(3):567–570. doi: 10.1099/00221287-70-3-567. [DOI] [PubMed] [Google Scholar]
- Kontomichalou P., Mitani M., Clowes R. C. Circular R-factor molecules controlling penicillinase synthesis, replicating in Escherichia coli under either relaxed or stringent control. J Bacteriol. 1970 Oct;104(1):34–44. doi: 10.1128/jb.104.1.34-44.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- MASTER R. W. POSSIBLE SYNTHESIS OF POLYRIBONUCLEOTIDES OF KNOWN BASE-TRIPLET SEQUENCES. Nature. 1965 Apr 3;206:93–93. doi: 10.1038/206093b0. [DOI] [PubMed] [Google Scholar]
- Rownd R., Nakaya R., Nakamura A. Molecular nature of the drug-resistance factors of the Enterobacteriaceae. J Mol Biol. 1966 Jun;17(2):376–393. doi: 10.1016/s0022-2836(66)80149-3. [DOI] [PubMed] [Google Scholar]
- Singh A. P., Cheng K. J., Costerton J. W., Idziak E. S., Ingram J. M. Sensitivity of normal and mutant strains of Escherichia coli to actinomycin-D. Can J Microbiol. 1972 Jun;18(6):909–915. doi: 10.1139/m72-139. [DOI] [PubMed] [Google Scholar]
- Terawaki Y., Kakizawa Y., Takayasu H., Yoshikawa M. Temperature sensitivity of cell growth in Escherichia coli associated with the temperature sensitive R(KM) factor. Nature. 1968 Jul 20;219(5151):284–285. doi: 10.1038/219284a0. [DOI] [PubMed] [Google Scholar]
- Terawaki Y., Rownd R. Replication of the R factor Rts1 in Proteus mirabilis. J Bacteriol. 1972 Feb;109(2):492–498. doi: 10.1128/jb.109.2.492-498.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terawaki Y., Takayasu H., Akiba T. Thermosensitive replication of a kanamycin resistance factor. J Bacteriol. 1967 Sep;94(3):687–690. doi: 10.1128/jb.94.3.687-690.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T., Kanamaru Y., Mori R., Akiba T. Recombination between a thermosensitive kanamycin resistance factor and a nonthermosensitive multiple-drug resistant factor. J Bacteriol. 1969 Jun;98(3):863–873. doi: 10.1128/jb.98.3.863-873.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]