Abstract
During light-driven proton transport bacteriorhodopsin shuttles between two protein conformations. A large-scale structural change similar to that in the photochemical cycle is produced in the D85N mutant upon raising the pH, even without illumination. We report here that (i) the pKa values for the change in crystallographic parameters and for deprotonation of the retinal Schiff base are the same, (ii) the retinal isomeric configuration is nearly unaffected by the protein conformation, and (iii) preventing rotation of the C13—C14 double bond by replacing the retinal with an all-trans locked analogue makes little difference to the Schiff base pKa. We conclude that the direct cause of the conformational shift is destabilization of the structure upon loss of interaction of the positively charged Schiff base with anionic residues that form its counter-ion.
The functions of some transmembrane ion pumps, and receptors that transmit light-dependent or chemical signals, depend on global changes in protein structure upon a chemical reaction or ligand binding at the active site (1). In the light-driven proton pump, bacteriorhodopsin, such a conformational shift seems to be related to the change of the access of the buried proton binding group, the retinal Schiff base, from the extracellular to the cytoplasmic membrane surface (2–4). A mechanism based on such a two-state conformational change, and in particular on an alternating access of a single binding site, must account for transport in some of the more complex ion pumps also (5).
Bacteriorhodopsin is a small protein containing seven transmembrane helices A through G that enclose an all-trans retinal linked to Lys-216 via a protonated Schiff base (6). The reaction cycle that follows photoisomerization of the retinal to 13-cis,15-anti is described by the sequence of intermediate states J, K, L, M, N, and O, and their substates (for recent reviews, see refs. 7–13). The M state is formed when the Schiff base proton is transferred to Asp-85, near the extracellular surface. A proton from Glu-204 and probably bound water is then released to the extracellular surface. The N state is formed by reprotonation of the Schiff base from Asp-96, near the cytoplasmic surface. This is followed by proton uptake to Asp-96 from the cytoplasmic surface and reisomerization of the retinal to all-trans in the O state. The initial state recovers upon loss of proton from Asp-85. Maps of the changed structure of the M state obtained by diffraction have been interpreted as density increases at the cytoplasmic halves of helices B and G. When M is stabilized under various conditions this is seen to be followed by tilting of the cytoplasmic end of helix F away from the central cavity (14–16). The latter change persists in the N state stabilized in mutants (17, 18). A component of these large-scale structural changes, termed collectively the E to C shift (4), may be the basis of the extracellular-to-cytoplasmic change of the connectivity of the Schiff base, the postulated “reprotonation switch” (2–4). The outward tilt of helix F and its reversal, were shown to be linked to proton transfer from Asp-96 to the Schiff base (decay of M) and reprotonation of Asp-96 from the aqueous medium (decay of N), respectively (19). It has been argued that the appearance of a large negative amide I band in Fourier transform infared (FTIR) difference spectra, originating from changed hydrogen-bonding of the polypeptide chain, is another indication of the structural change (20–24).
Does photoisomerization of the retinal to 13-cis,15-anti trigger these protein conformational changes directly or indirectly? Analogously to the much discussed question of how energy is stored in this system after photoexcitation (25–27), there are three simple alternatives. First, the transition from a linear to a bent polyene chain could result in a poor fit of the retinal in its binding pocket, that forces the protein into a changed conformation that better accommodates it. Second, because the distal end of the retinal is fixed by three tryptophan rings (6), the acute bend in the polyene chain will displace its proximal end with the positively charged protonated Schiff base relative to the anionic Asp-85 and Asp-212, and nearby bound water (28, 29). This displacement may initiate long-range structural changes independent of the deprotonation of the Schiff base. Third, the Schiff base–counter-ion interaction becomes weaker once the proton is transferred to Asp-85 because the charges of both this residue and the Schiff base are eliminated. This may remove sufficient stabilization of the structure to allow its collapse into a more stable conformation. In the D85N mutant, where the counter-ion is weaker without a contribution from Asp-85, deprotonation of the Schiff base upon raising the pH from 7 to 11 without illumination resulted in a protein conformation similar to that which occurs normally only in the M state of photocycle (4). Diffraction measurements with the D85N/D96N double mutant revealed that the second mutation further destabilized the structure, so as to shift the conformation of the unphotolyzed protein substantially even without deprotonation of the Schiff base. These results tend to argue for the third alternative.
Descriptions of the complex and interdependent chromophore and protein changes during the photochemical cycle do not clearly distinguish between what is cause and what is effect, and thus do not provide an unambiguous answer. The D85N mutant, in which the conformation changes occur independent of photoexcitation of the retinal, is a simpler system. Experiments directed to three questions in this recombinant protein will decide whether deprotonation of the Schiff base during the photocycle is indeed the cause of the E to C conformational shift: (i) Does the structural change in D85N strictly correlate with deprotonation of the Schiff base? The appearance of the FTIR amide band after photoexcitation of the D85N mutant with unprotonated Schiff base at pH 9.4 suggested that it might not (30). If the amide band originates from the structural shift, it cannot have occurred at this pH before photoexcitation even though the Schiff base is deprotonated. It was therefore suggested that the C structure correlates instead with another FTIR change of the polypeptide backbone, observed (31) at much higher pH. (ii) Does the retinal configuration change along with the protein conformation? The D85N and D85N/D96N mutants provide the means to test this without complication from other events in the photocycle. The all-trans and 13-cis,15-syn retinal configurations, that equilibrate thermally, have roughly the same linear shape. However, it is possible that the retinal is forced by the changed protein conformation into the strongly bent 13-cis,15-anti configuration that is produced normally only upon photoisomerization. The possibility that such a configuration can be assumed in the absence of illumination was raised from solid-state 13C NMR measurements of wild-type bacteriorhodopsin at a pH low enough to protonate Asp-85 (32). (iii) Finally, is retinal isomerization necessarily coupled to deprotonation of the Schiff base? As judged from the effect of deuterium substitution at C15 on the C—C vibrational bands of the retinal, once the Schiff base of D85N deprotonated at elevated pH the C13—C14 bond configuration is mostly trans (30), but according to analysis of extracted retinal the 13-cis to all-trans ratio increases rather than decreases with pH (33).
METHODS
Structure Determination.
X-ray diffraction measurements were carried out with the MUSCLE Diffractometer at BL-15A in the Photon Factory at Tsukuba, Japan, as described (15). The purple membranes were suspended in 100 mM KCl buffered at the pH indicated, up to A570 or A600 = 20. Of this suspension, 100 μl was dropped on a piece of aluminum foil, incubated at a relative humidity of 95% for 3 days, and another drop was layered on to the resulting wet film. The layering procedure was repeated four to five times to obtain a thick wet, well-hydrated film. The analysis of the intensities of the reflections was as before (4).
Fourier Transform Raman (FT-Raman) Spectroscopy.
Instrument used was a Bruker IFS66/S-FRA106/S, with 180° backscatter, at 2 cm−1 resolution. The excitation energy of the Nd-yttrium/aluminum garnet (Yag) laser was 300 mW. The samples were membrane suspensions in a 4 mm path length quartz cuvette.
Retinal Reconstitution.
The samples were bleached by illuminating with hydroxylamine (34). After removal of the retinal oxime by washing three times with 1% bovine serum albumin, retinal or its analogue was added. The chromophore regenerated in a few hours with authentic retinal, and in about 2 weeks with the analogue.
RESULTS
pH Dependence of the Crystallographic Structure of D85N Bacteriorhodopsin.
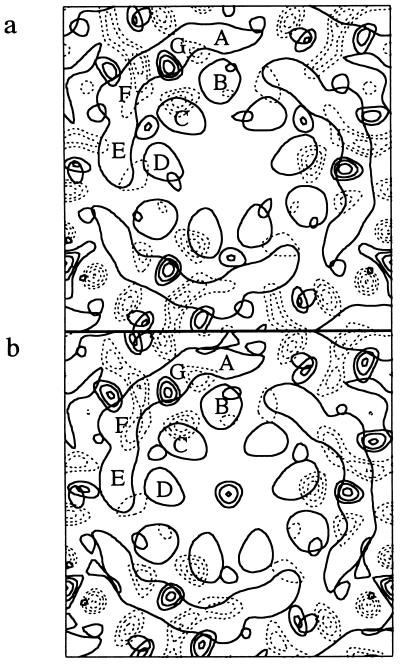
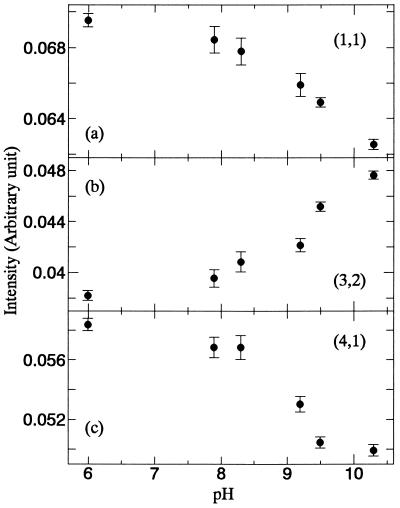
The pKa of the protonated Schiff base in D85N is lowered from >13 in the wild type (35) to about 9 (4, 33, 36, 37). We measured x-ray diffraction at numerous pH values between pH 6 and 11. Fig. 1 shows difference Fourier transform projection maps between the structures upon raising the pH from 6.0 to 9.2 and 6.0 to 10.4. The density increase at helices G and a pair of negative and positive density differences at helix F resemble the changes in the M state. The near identity of the difference maps below and above the pKa indicates that essentially only one kind of structural change occurs. Fig. 2 shows the integrated intensities of reflections (1,1), (3,2), and (4,1) as functions of pH. The changes in reflections (3,2) and (4,1), are the most characteristic of the E to C conformational transition (4, 38). The same dependence of the crystallographic parameters on pH as the deprotonation of the Schiff base strongly implicates the protonation state of the Schiff base in the structural change. The other reflections were nearly unaffected by pH (data not shown).
Figure 1.
Difference projection maps of structural changes in D85N bacteriorhodopsin upon raising the pH from 6.0 to 9.2 (a) or 10.4 (b). Bold contour lines indicate density increase, dashed lines density decrease. Although the changes in b are larger than in a, they are represented with equal amplitude. End-on views of transmembrane helices B, C, and D, and the smeared outline of the more tilted helices A, G, F, and E of one bacteriorhodopsin in the trimer are superimposed.
Figure 2.
Integrated intensities of reflections (1,1), (3,2), and (4,1) in a, b, and c, respectively vs. pH. Conditions are the same as in Fig. 1.
The results raise a question about the origin of protein conformation change detected by FTIR. Are the structural changes of the polypeptide backbone that give rise to the amide I band in the MN (24) and N (21–23) photointermediates the same as those described by diffraction in M and N? Since it is observed after photoexcitation of D85N with unprotonated chromophore (30), the FTIR band seems not to be linked to the structural change that we detect upon deprotonation of the Schiff base. We must conclude that what is measured by diffraction methods and FTIR are distinct from one another.
Relationship of Retinal Isomeric State and Global Protein Conformation.
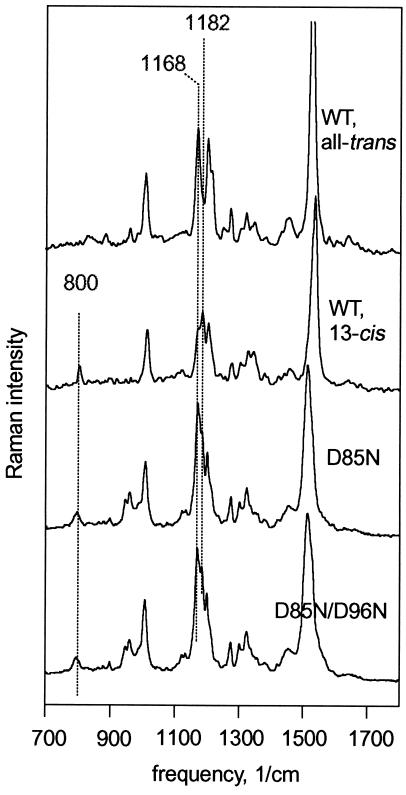
Extraction of retinal and analysis by chromatography is often used to determine isomeric composition. HPLC of the extracted retinal gave 40 and 55% 13-cis content for dark-adapted D85N and D85N/D96N, respectively, but could give no information on the configuration of the C15=N bond (37). Recently FT-Raman (39, 40) became the method of choice for determining the configuration of retinal because (i) it does not perturb the protein, (ii) unlike resonance Raman it utilizes 1.064 μm excitation that has virtually no effect on the isomeric state, and (iii) it can distinguish between 13-cis,15-anti and 13-cis,15-syn. When the protonation state of the Schiff base is unchanged, the amplitudes of several vibrational modes determine the configuration of the bonds between C13 and C15 unambiguously. Fig. 3 contains FT-Raman spectra for the wild-type all-trans and 13-cis,15-syn chromophores. The C14—C15 stretch band at 1168 cm−1 is prominent in all-trans but weak in 13-cis, while the opposite is true for the C10—C11 band at 1182 cm−1. For reasons of steric constraint in the binding pocket, the C14 hydrogen-out-of-plane band at 800 cm−1 is strong in 13-cis,15-syn retinal but has negligible amplitude in all-trans or 13-cis,15-anti (41–43). Fig. 3 shows that D85N near neutral pH, with the E protein conformation, contains a mixture of all-trans and 13-cis retinals, consistent with chemical analysis (37) and resonance Raman (44). In D85N/D96N at the same pH, but with a mostly C protein conformation (4), the ratio of the 1182 to the 1168 cm−1 band is slightly higher, but the 800 cm−1 band is hardly changed. Thus, the isomeric state of the retinal is nearly unaffected by the global protein conformation, and the conformational shift produces no detectable isomerization to 13-cis,15-anti.
Figure 3.
FT-Raman spectra of wild-type, D85N, and D85N/D96N bacteriorhodopsins. Conditions are 100 mM NaCl, pH 6.6, at 22°C. Wild-type spectra are for all-trans (light-adapted) and 13-cis (from a dark-adapted sample that contains 33% all-trans) chromophores. These are essentially the same as reported before (39) and shown as controls only. For the mutants spectra of dark-adapted samples are shown. Amplitudes of the two wild-type spectra adjusted (together) for best comparison with the mutants.
Relationship of Retinal Isomeric State and Deprotonation of the Schiff Base.
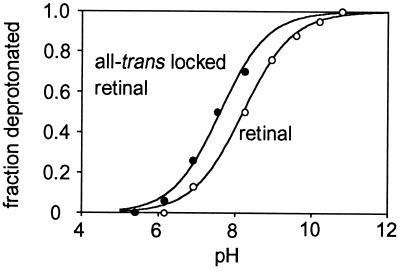
According to these results, in these mutants the isomeric state of the retinal should not be linked to the deprotonation of the Schiff base since the latter is associated with the protein conformation (Figs. 1 and 2). If it were, making isomerization impossible would substantially raise the observed pKa. In 13-demethyl 12–14 ethano retinal (45, 46), the C13—C14 double bond is locked in the all-trans configuration. Fig. 4 shows titration of the Schiff base of D85N containing authentic retinal and the locked analogue. Eliminating the possibility of isomerization has little effect on the apparent pKa in D85N. Thus, when residue 85 is uncharged, deprotonation of the Schiff base (and therefore the global conformation change it triggers) is at most weakly coupled to the isomeric state of the retinal.
Figure 4.
Titration of the retinal Schiff base in D85N bacteriorhodopsin. Deprotonation was determined from the amplitudes of the shift of the maximum from near 600 nm to 410 nm. Reconstitution of the hydroxylamine-bleached chromophores was with either authentic (○) or 13-demethyl 12–14 ethano (•) retinal. Titration was in 2 M NaCl, where the apparent pKa of the Schiff base is somewhat lower than at lower NaCl concentrations (4, 33, 37).
DISCUSSION
We report here on conditions where a conformational shift similar to the large-scale structural change of bacteriorhodopsin in the photocycle occurs, but in the absence of photoexcitation. The structural change detected is nearly or completely indifferent to the isomeric state of the retinal and depends instead on deprotonation of the retinal Schiff base. If this applies to the photocycle as well, we would argue that the main significance of the photoisomerization is that it allows proton transfer from the Schiff base to Asp-85. It may be then the resulting abolition of the charge-pair at the active site, rather than the isomerization directly, that allows the protein to assume its alternative conformation and to facilitate the reprotonation of the Schiff base by Asp-96. Inasmuch as the pKa of Asp-96, and therefore the role of this residue as proton donor, depends on the conformation changes at the cytoplasmic surface (19, 47), this seems reasonable. From different evidence, it has been argued, however, that deprotonation of the Schiff base and the reprotonation switch are both direct consequences of the isomerization (48). Deciding between these alternatives will require a better understanding of the relationship between protein conformation changes, both local and global, and the accessibility of protons to the Schiff base.
In similar experiments with unphotolyzed visual rhodopsin, Schiff base–counter-ion interaction and retinal isomeric configuration both affected protein conformation. From a survey of the signaling state in a large number of mutated rhodopsins (49) it appears that there is an electrostatic as well as a steric component to what determines the active receptor conformation. Thus, for example, removal of the ion pair at the active site of vertebrate rhodopsin, by replacing Glu-113, caused shift to the postulated transducin-binding conformation only in the retinal-free opsin (50), and in a mutant with weakened counter-ion interaction photoisomerization was alone sufficient for transducin binding (51).
The results reported here emphasize the role of the Schiff base–Asp-85 proton donor-acceptor pair as the active site in bacteriorhodopsin. This is in keeping with the findings that the D85T bacteriorhodopsin mutant transports not protons but chloride like halorhodopsin (52), and halorhodopsin, which lacks the equivalent of Asp-85, transports not chloride but protons like bacteriorhodopsin when an artificial proton acceptor is bound near the Schiff base (53).
Acknowledgments
This work was supported partly by grants from the Department of Energy (DEFG03-86ER13525) and the National Institutes of Health (GM29498).
ABBREVIATIONS
- FTIR
Fourier transform infared
- FT-Raman
Fourier transform Raman
- HPLC
high pressure liquid chromatography. Bacteriorhodopsin mutants are designated with the single-letter code, the wild-type residue code followed by the residue number, and the mutant residue code
References
- 1.Spudich J L, Lanyi J K. Curr Opin Cell Biol. 1996;8:452–457. doi: 10.1016/s0955-0674(96)80020-2. [DOI] [PubMed] [Google Scholar]
- 2.Fodor S P, Ames J B, Gebhard R, van der Berg E M, Stoeckenius W, Lugtenburg J, Mathies R A. Biochemistry. 1988;27:7097–7101. doi: 10.1021/bi00418a064. [DOI] [PubMed] [Google Scholar]
- 3.Henderson R, Baldwin J M, Ceska T A, Zemlin F, Beckmann E, Downing K H. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 4.Kataoka M, Kamikubo H, Tokunaga F, Brown L S, Yamazaki Y, Maeda A, Sheves M, Needleman R, Lanyi J K. J Mol Biol. 1994;243:621–638. doi: 10.1016/0022-2836(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 5.Lanyi J K. Nature (London) 1995;375:461–463. doi: 10.1038/375461a0. [DOI] [PubMed] [Google Scholar]
- 6.Grigorieff N, Ceska T A, Downing K H, Baldwin J M, Henderson R. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 7.Mathies R A, Lin S W, Ames J B, Pollard W T. Annu Rev Biophys Biophys Chem. 1991;20:491–518. doi: 10.1146/annurev.bb.20.060191.002423. [DOI] [PubMed] [Google Scholar]
- 8.Rothschild K J. J Bioenerg Biomembr. 1992;24:147–167. doi: 10.1007/BF00762674. [DOI] [PubMed] [Google Scholar]
- 9.Oesterhelt D, Tittor J, Bamberg E. J Bioenerg Biomembr. 1992;24:181–191. doi: 10.1007/BF00762676. [DOI] [PubMed] [Google Scholar]
- 10.Ebrey T G. In: Thermodynamics of Membranes, Receptors and Channels. Jackson M, editor. New York: CRC; 1993. pp. 353–387. [Google Scholar]
- 11.Khorana H G. Proc Natl Acad Sci USA. 1993;90:1166–1171. doi: 10.1073/pnas.90.4.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanyi J K. Biochim Biophys Acta. 1993;1183:241–261. doi: 10.1016/0005-2728(93)90226-6. [DOI] [PubMed] [Google Scholar]
- 13.Lanyi J K, Váró G. Isr J Chem. 1995;35:365–386. [Google Scholar]
- 14.Nakasako M, Kataoka M, Amemiya Y, Tokunaga F. FEBS Lett. 1991;292:73–75. doi: 10.1016/0014-5793(91)80837-s. [DOI] [PubMed] [Google Scholar]
- 15.Subramaniam S, Gerstein M, Oesterhelt D, Henderson R. EMBO J. 1993;12:1–8. doi: 10.1002/j.1460-2075.1993.tb05625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han B-G, Vonck J, Glaeser R M. Biophys J. 1994;67:1179–1186. doi: 10.1016/S0006-3495(94)80586-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vonck J. Biochemistry. 1996;35:5870–5878. doi: 10.1021/bi952663c. [DOI] [PubMed] [Google Scholar]
- 18.Kamikubo H, Kataoka M, Váró G, Oka T, Tokunaga F, Needleman R, Lanyi J K. Proc Natl Acad Sci USA. 1996;93:1386–1390. doi: 10.1073/pnas.93.4.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown L S, Váró G, Needleman R, Lanyi J K. Biophys J. 1995;69:2103–2111. doi: 10.1016/S0006-3495(95)80081-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothschild K J, Zagaeski M, Cantore W A. Biochem Biophys Res Commun. 1981;103:483–489. doi: 10.1016/0006-291x(81)90478-2. [DOI] [PubMed] [Google Scholar]
- 21.Braiman M S, Bousché O, Rothschild K J. Proc Natl Acad Sci USA. 1991;88:2388–2392. doi: 10.1073/pnas.88.6.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerwert K, Souvignier G, Hess B. Proc Natl Acad Sci USA. 1990;87:9774–9778. doi: 10.1073/pnas.87.24.9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfefferlé J-M, Maeda A, Sasaki J, Yoshizawa T. Biochemistry. 1991;30:6548–6556. doi: 10.1021/bi00240a027. [DOI] [PubMed] [Google Scholar]
- 24.Sasaki J, Shichida Y, Lanyi J K, Maeda A. J Biol Chem. 1992;267:20782–20786. [PubMed] [Google Scholar]
- 25.Honig B, Ebrey T, Callender R H, Dinur U, Ottolenghi M. Proc Natl Acad Sci USA. 1979;76:2503–2507. doi: 10.1073/pnas.76.6.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalisky O, Ottolenghi M, Honig B, Korenstein R. Biochemistry. 1981;20:649–655. doi: 10.1021/bi00506a031. [DOI] [PubMed] [Google Scholar]
- 27.Honig B, Ebrey T G. Methods Enzymol. 1982;88:462–470. [Google Scholar]
- 28.Brown L S, Gat Y, Sheves M, Yamazaki Y, Maeda A, Needleman R, Lanyi J K. Biochemistry. 1994;33:12001–12011. doi: 10.1021/bi00206a001. [DOI] [PubMed] [Google Scholar]
- 29.Gat Y, Sheves M. J Am Chem Soc. 1993;115:3772–3773. [Google Scholar]
- 30.Nilsson A, Rath P, Olejnik J, Coleman M, Rothschild K J. J Biol Chem. 1995;270:29746–29751. doi: 10.1074/jbc.270.50.29746. [DOI] [PubMed] [Google Scholar]
- 31.Száraz S, Oesterhelt D, Ormos P. Biophys J. 1994;67:1706–1712. doi: 10.1016/S0006-3495(94)80644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Groot H J M, Smith S O, Courtin J, Van den Berg E, Winkel C, Lugtenburg J, Griffin R G, Herzfeld J. Biochemistry. 1990;29:6873–6883. doi: 10.1021/bi00481a017. [DOI] [PubMed] [Google Scholar]
- 33.Turner G J, Miercke L J W, Thorgeirsson T E, Kliger D S, Betlach M C, Stroud R M. Biochemistry. 1993;32:1332–1337. doi: 10.1021/bi00056a019. [DOI] [PubMed] [Google Scholar]
- 34.Oesterhelt D, Schuhmann L. FEBS Lett. 1974;44:262–265. doi: 10.1016/0014-5793(74)81153-1. [DOI] [PubMed] [Google Scholar]
- 35.Druckmann S, Ottolenghi M, Pande A, Pande J, Callender R H. Biochemistry. 1982;21:4953–4959. doi: 10.1021/bi00263a019. [DOI] [PubMed] [Google Scholar]
- 36.Brown L S, Bonet L, Needleman R, Lanyi J K. Biophys J. 1993;65:124–130. doi: 10.1016/S0006-3495(93)81064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tittor J, Schweiger U, Oesterhelt D, Bamberg E. Biophys J. 1994;67:1682–1690. doi: 10.1016/S0006-3495(94)80642-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dencher N A, Dresselhaus D, Zaccai G, Büldt G. Proc Natl Acad Sci USA. 1989;86:7876–7879. doi: 10.1073/pnas.86.20.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawatzki J, Fischer R, Scheer H, Siebert F. Proc Natl Acad Sci USA. 1990;87:5903–5906. doi: 10.1073/pnas.87.15.5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rath P, Krebs M P, He Y, Khorana H G, Rothschild K J. Biochemistry. 1993;32:2272–2281. doi: 10.1021/bi00060a020. [DOI] [PubMed] [Google Scholar]
- 41.Braiman M S, Mathies R A. Biochemistry. 1980;19:5421–5428. doi: 10.1021/bi00564a042. [DOI] [PubMed] [Google Scholar]
- 42.Smith S O, Pardoen J A, Lugtenburg J, Mathies R A. J Phys Chem. 1987;91:804–819. [Google Scholar]
- 43.Althaus T, Eisfeld W, Lohrmann R, Stockburger M. Isr J Chem. 1995;35:227–251. [Google Scholar]
- 44.Rath P, Marti T, Sonar S, Khorana H G, Rothschild K J. J Biol Chem. 1993;268:17742–17749. [PubMed] [Google Scholar]
- 45.Bhattacharya S, Marti T, Otto H, Heyn M P, Khorana H G. J Biol Chem. 1992;267:6757–6762. [PubMed] [Google Scholar]
- 46.Rousso I, Friedman N, Sheves M, Ottolenghi M. Biochemistry. 1995;34:12059–12065. doi: 10.1021/bi00037a049. [DOI] [PubMed] [Google Scholar]
- 47.Váró G, Needleman R, Lanyi J K. Biophys J. 1996;70:461–467. doi: 10.1016/S0006-3495(96)79589-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haupts U, Tittor J, Bamberg E, Oesterhelt D. Biochemistry. 1997;36:2–7. doi: 10.1021/bi962014g. [DOI] [PubMed] [Google Scholar]
- 49.Fahmy K, Siebert F, Sakmar T P. Biophys Chem. 1995;56:171–181. doi: 10.1016/0301-4622(95)00030-2. [DOI] [PubMed] [Google Scholar]
- 50.Cohen G B, Oprian D D, Robinson P R. Biochemistry. 1992;31:12592–12601. doi: 10.1021/bi00165a008. [DOI] [PubMed] [Google Scholar]
- 51.Zvyaga T A, Fahmy K, Sakmar T P. Biochemistry. 1994;33:9753–9761. doi: 10.1021/bi00198a046. [DOI] [PubMed] [Google Scholar]
- 52.Sasaki J, Brown L S, Chon Y-S, Kandori H, Maeda A, Needleman R, Lanyi J K. Science. 1995;269:73–75. doi: 10.1126/science.7604281. [DOI] [PubMed] [Google Scholar]
- 53.Váró G, Brown L S, Needleman R, Lanyi J K. Biochemistry. 1996;35:6604–6611. doi: 10.1021/bi9601159. [DOI] [PubMed] [Google Scholar]