Abstract
We have purified and characterized a 31-kDa protein named mapmodulin that binds to the microtubule-associated proteins (MAPs) MAP2, MAP4, and tau. Mapmodulin binds free MAPs in strong preference to microtubule-associated MAPs, and appears to do so via the MAP’s tubulin-binding domain. Mapmodulin inhibits the initial rate of MAP2 binding to microtubules, a property that may allow mapmodulin to displace MAPs from the path of organelles translocating along microtubules. In support of this possibility, mapmodulin stimulates the microtubule- and dynein-dependent localization of Golgi complexes in semi-intact CHO cells. To our knowledge, mapmodulin represents the first example of a protein that can bind and potentially regulate multiple MAP proteins.
Microtubules play an important role in cytoplasmic organization, determination of cell polarity, cellular locomotion, and chromosome segregation. The ability of microtubules to mediate these processes is aided by their dynamic instability (1). Microtubules undergo continual cycles of shrinkage and growth and may be stabilized at specific locations within the cell and/or at different points in the cell cycle. Once stabilized, they act as tracks along which the molecular motors dynein and kinesin move, carrying vesicular cargoes from one part of the cell to another.
Microtubule function is regulated by microtubule-associated proteins (MAPs). MAPs include the molecular motors, kinesin and dynein, as well as a group of proteins that can promote microtubule assembly and contribute to polymer stability in vitro, and in some cases in vivo (2–4). This latter group of assembly-promoting proteins includes MAP2 and tau that are both specific to neuronal cells. In contrast, MAP4 is the major MAP in nonneuronal mammalian cells (5). MAP2, MAP4, and tau share a homologous, C-terminal microtubule-binding domain that is comprised of three to five imperfect repeats of an 18-amino acid motif (6, 7). An adjacent, proline-rich region also contributes to microtubule binding.
Microtubule surfaces are essentially coated with MAPs in vivo, and the larger MAPs project a significant distance (≈90 nm) away from the microtubule surface (8). It has been estimated that MAP4 alone occurs once per ≈40–50 tubulin dimers, or ≈50 per μm of microtubule length (9, 10). Thus, motors moving organelles along microtubule tracks will encounter numerous bound MAPs as they move an organelle the equivalent of a single organelle diameter, because such organelles are usually larger than 0.1 μm.
Indications of MAP-mediated track blockage have come from in vitro motility assays. Paschal et al. (11) and Hagiwara et al. (12) have shown that MAP2 and tau inhibit kinesin and dynein-mediated microtubule gliding, possibly by competing with motors for the same or adjacent binding sites on the microtubules. López and Sheetz (13) found that MAP2 inhibited both microtubule gliding and in vitro membrane-bound organelle motility. Specifically, MAP2-decorated microtubules could not support the transport of vesicles in vitro in the presence of low concentrations of motor proteins, nor could they support formation of membranous networks (13). The large MAP mass was suggested to sterically inhibit motor binding and movement along microtubules. While the results have not always been identical, it does seem that microtubules bind coverslip-attached dynein to a greater extent in the absence of MAP2. Similar results have been seen for kinesin at low (12, 13) but not high (14) motor concentrations.
Factors are likely to exist that modulate MAP–microtubule interactions, thereby permitting motor-mediated transport. López and Sheetz (15) have suggested that MAP2 phosphorylation could be used to modulate microtubule association. We have taken the view that cells probably contain previously undiscovered accessory proteins that regulate MAP–microtubule association directly. Indeed, we report here the purification and characterization of a protein that interacts strongly with the microtubule-binding domains of MAP2, MAP4, and tau. We have named this protein mapmodulin because we show that it slows the rate of MAP binding to microtubules. Mapmodulin stimulates an in vitro assay (16) that reconstitutes the overall process of dynein-mediated, microtubule-based Golgi localization in broken cells. We propose a speculative model in which mapmodulin clears microtubule tracks like a train’s cowcatcher to permit microtubule-based motor-driven organelle motility.
MATERIALS AND METHODS
Mapmodulin Purification.
CHO cytosol was prepared as described by Balch et al. (17). Cytosol was desalted into cytosol buffer (25 mM Hepes, pH 7.5/50 mM KCl/2 mM MgCl2/0.5 mM EDTA/1 mM DTT) using Bio-Gel P6DG (Bio-Rad) and was immediately frozen in liquid nitrogen. Most preparations had protein concentrations of 4–6 mg/ml, as determined using Bio-Rad reagent and BSA as standard.
Desalted CHO cytosol was loaded onto a 20 ml Fast Flow Q (Pharmacia). The column was washed with 10 volumes, 0.35 M KCl (in 25 mM Tris, pH 7.6/1 mM DTT) and then eluted with a 200 ml gradient of 0.35 M to 0.7 M KCl. The active fractions (corresponding to 0.5 M KCl) were collected, concentrated on a 1 ml Fast Flow Q column, and loaded onto a 120 ml Superdex-200 fast protein liquid chromatography (FPLC) column in cytosol buffer. Gel filtration and chemical crosslinking revealed that mapmodulin is a dimer or trimer. Cytosol levels of the protein assumed a dimeric mass and cytosol concentration of 50 mg/ml in cells.
Bacterial Expression.
Mapmodulin is identical to a previously identified protein named PHAPI (putative histocompatibility antigen-associated protein I; see text and ref. 18). The full-length cDNA of PHAPI (18), cloned into pBlueScript (KpnI–SalI) and fused to the enterokinase recognition site at its N terminus, was subcloned into the pQE30 expression vector (Qiagen, Chatsworth, CA; KpnI–SalI). The His6-tagged protein was purified by standard methods. C-terminally truncated His-tagged mapmodulin/PHAPI was constructed by blunt-end ligation of the BsaAI–PstI fragment from pBlueScript-PHAPI into pQE30, deleting residues 164–249 which comprise the acidic tail.
Antibodies.
Polyclonal antibodies against CHO mapmodulin and recombinant mapmodulin/PHAPI were affinity purified using pure recombinant mapmodulin/PHAPI crosslinked to AffiGel-15 (Bio-Rad). Monoclonal anti-MAP2 was from Sigma; Texas red conjugated secondary antibodies were from The Jackson Laboratories.
Microtubule Protein Purification.
Tubulin was purified from bovine brain using two cycles of polymerization and depolymerization followed by a phosphocellulose column (19). MAP2 and tau were purified from bovine brain by stripping a twice-cycled microtubule pellet with 0.75 M NaCl and boiling for 5 min after adding 2% 2-mercaptoethanol. The samples were chilled on ice and then precipitates were sedimented at 150,000 × g for 20 min at 4°C. The heat-stable MAP supernatant was separated by FPLC on a Mono Q column (Pharmacia) using a 0–400 mM KCl gradient in 35 mM K-Pipes pH 7.2, 5 mM MgSO4, 1 mM EGTA, and 0.5 mM EDTA (13). The expression and purification of MAP4-C and MAP4-N were performed exactly according to Wang et al. (4).
Solid Phase Binding Assay.
Mapmodulin binding to microtubules and MAPs was carried out using a solid-phase immunoassay (20); all steps were at 22°C. Briefly, 50 μl of 0.1 mg/ml taxol stabilized, polymerized tubulin, or MAPs were added to microtiter wells (Falcon, Becton Dickenson) and incubated for 2 hr to allow binding to the plastic surface. After blocking residual binding sites with 200 μl of blocking buffer (5% skimmed milk/0.025% Tween 20/10 mM Tris, pH 7.4/150 mM NaCl) for 1 hr, wells were incubated with different concentrations of mapmodulin for 60 min. Wells were washed and incubated with anti-mapmodulin polyclonal antibody for 1 hr. After washing, horseradish peroxidase-labeled anti-rabbit IgG (1:2000) was added for 1 hr. Binding was detected using 128 μg/ml 3,3′,5,5′-tetramethylbenzidine and 0.05% H2O2 in 0.1 M NaOAc (pH 5.3); the reaction was terminated by adding 50 μl of 1 M H2SO4. Absorbance at 450 nm was quantified on a Molecular Devices microtiter plate reader.
Golgi Membranes.
Wild-type Golgi complexes were purified from rat liver according to Tabas and Kornfeld (21), yielding preparations with protein concentrations of 1.5–2.5 mg/ml. KCl-treated Golgi membranes were prepared as described (22).
Golgi Capture Assay.
The assay was carried out as described (16) with the following modification. N-acetylglucosamine (GlcNAc) transferase I activity was measured using the transfer of [3H]GlcNAc to the synthetic acceptor 3,6-di-O-(α-d-mannopyranosyl)-β-d-mannopyranoside (23). Assays were carried out for 40 min at 37°C with 1 μg of the synthetic acceptor in 50 μl. The labeled product was separated from the radioactive precursor by binding to Sep-Pak C-18-plus columns (Waters Chromatography), washing with 20 ml H2O, and eluted in 5 ml MeOH.
RESULTS
We previously described a system which reconstitutes the microtubule-dependent, cytoplasmic dynein-mediated localization of the Golgi complex in broken cells (16). Clone 15B CHO cells that lack the Golgi-specific enzyme, GlcNAc transferase I, are broken gently and incubated with isolated, wild-type Golgi complexes at 37°C. The “capture” of wild-type Golgi complexes by mutant CHO cells is then measured after differential centrifugation: semi-intact mutant cells sediment to the bottom of a sucrose cushion and carry with them bound, wild-type Golgi complexes. The presence of cell-associated Golgi complexes is detected by subjecting cell pellets to a GlcNAc transferase I enzyme assay. We have shown previously that this process leads to the correct localization of Golgi complexes to the microtubule organizing center (MTOC) of recipient cells, and requires ATP hydrolysis, cytosolic and peripheral membrane proteins, intact microtubules, and cytoplasmic dynein (16).
Treatment of Golgi complexes with 0.5–1 M KCl to remove peripherally associated proteins destroys their ability to be captured by semi-intact cells. However, if the released proteins are dialyzed into a physiological buffer and added back to the salt-washed Golgi complexes, Golgi capture activity is restored. CHO cytosol also restores activity to salt-treated Golgi complexes. These observations provided an assay for the purification of proteins needed for Golgi complex localization.
To succeed in the purification of a single protein that functions in a process requiring multiple proteins, reactions must contain limiting amounts of these other components. When cells are broken for the Golgi capture assay, endogenous cytosolic proteins (including cytoplasmic dynein) are diluted significantly, but are not removed. Thus, some Golgi capture is seen in the absence of additional cytosolic proteins; the process is stimulated at least 3-fold when reactions are supplemented with crude CHO cytosol added to a final concentration of 0.5–1 mg/ml. All of the results presented here reflect this stimulation; the level of Golgi capture detected in reactions lacking added cytosolic proteins has been subtracted throughout.
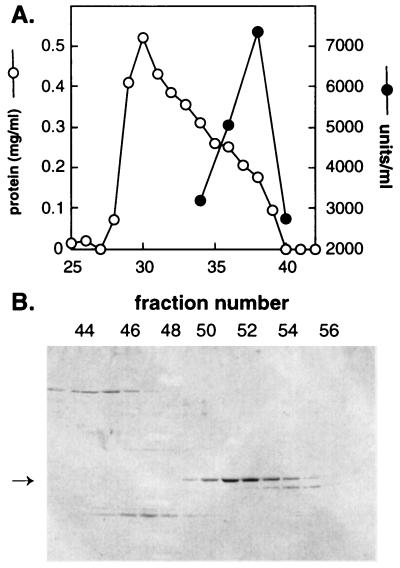
We first fractionated crude CHO cytosol by gel filtration to identify active components that were smaller than cytoplasmic dynein. As shown in Fig. 1A, a significant peak of activity chromatographed with an apparent mass of 68 kDa. This activity bound tightly to an anion-exchange resin, so we chose this as the first step for a subsequent large-scale purification scheme. The final procedure included differential centrifugation, followed by anion-exchange and Superdex S200 gel filtration chromatography, and resulted in an ≈800-fold purification of a protein that alone, added in nanogram quantities, had at least half the activity of total cytosol in terms of its ability to stimulate in vitro localization of Golgi complexes. Fig. 1B shows an SDS/PAGE analysis of the chromatographic profile of the final gel filtration step. Approximately 100 μg of this polypeptide was obtained from 100 mg of CHO cytosol.
Figure 1.
Mapmodulin purification. (A) Fractionation of total CHO cytosol (4 mg) by Sephacryl S100 (45 ml) gel filtration chromatography. Fractions (0.6 ml) were tested for Golgi capture activity. One unit is defined as the cpm obtained in a 60-min assay; 40 μl samples were tested. (B) Superdex S200 chromatography of Fast Flow Q fractions eluting at ≈0.5 M KCl. Fractions (1.5 ml) were collected; samples (20 μl) were analyzed by 12.5% SDS/PAGE and Coomassie blue staining. Golgi capture activity followed the polypeptide which peaked in fractions 51 and 52.
We named this protein mapmodulin because we show here that it has the capacity to bind MAPs and slow their rates of association with microtubules. Tryptic peptide sequences, 16 and 19 residues in length, were obtained from the purified protein by automated Edman degradation. A search of the database revealed identity of mapmodulin with a previously identified protein PHAPI. PHAPI had been purified from the cytosol of a human lymphoblastoid B cell line on the basis of its capacity to bind peptides representing the basic, cytosolic tail of HLA (18). A PHAPI cDNA clone (18) was used to generate a His-tagged version of PHAPI/mapmodulin. We also generated a His-tagged, C-terminally truncated version of PHAPI/mapmodulin (mapmodulinΔC).
PHAPI/Mapmodulin Facilitates Golgi Localization in Broken Cells.
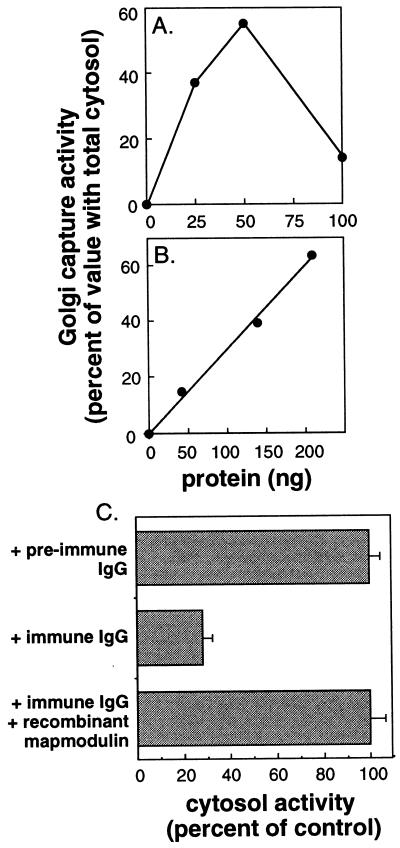
Addition of pure CHO mapmodulin to reactions containing KCl-treated Golgi complexes resulted in as much as 60% of the amount of Golgi localization obtained in reactions supplemented with total crude cytosol (Fig. 2A). Obviously, a number of other cytosol factors, including dynein, would be expected to be required for full cytosol activity. As shown in Fig. 2B, recombinant mapmodulin (PHAPI) was also active in the Golgi localization assay, and displayed activity similar to that observed for mapmodulin purified from CHO cells (Fig. 2B). The relative activities of recombinant and native mapmodulin varied between preparations, but were always roughly comparable; both showed inhibition when added at high concentrations. Mapmodulin was also active in reactions utilizing non-KCl-treated Golgi complexes (data not shown).
Figure 2.
Mapmodulin purified from either CHO cells (A) or bacteria (B) drives the dynein-mediated, microtubule-based localization of Golgi complexes. Reactions contained 5 μg of KCl-stripped Golgi membranes and also contained 0.2 mg/ml CHO cytosol (B). Golgi capture activity is presented as the percentage of maximum cytosolic activity (245 cpm). Background due to the added, limiting cytosol (39 cpm) was subtracted. The average of duplicates is shown; standard errors were 10% and 6% for A and B, respectively. (C) Anti-mapmodulin antibodies inhibit Golgi capture. CHO cytosol (200 μg) was pre-incubated with anti-mapmodulin IgG or preimmune IgG (9 μg) for 30 min at 37°C in the presence or absence of 280 ng purified recombinant mapmodulin. Golgi capture activity is presented relative to the preimmune-treated cytosol. Bars represent the standard error of the mean for duplicate samples.
To further establish the role of mapmodulin in Golgi localization, we tested if anti-mapmodulin antibodies would block this process. As shown in Fig. 2C, incubation of CHO cytosol with anti-mapmodulin antibodies significantly inhibited Golgi capture (60–80% reduction in activity); activity could be restored in full by adding pure, recombinant mapmodulin to the assay. These experiments demonstrate that mapmodulin activity is due to the protein encoded by the PHAPI gene, and that mapmodulin facilitates microtubule-based localization of Golgi complexes. Since mapmodulin is also localized to the endoplasmic reticulum (see below), it may very well modulate endoplasmic reticulum localization, and need not be restricted to dynein-dependent processes.
Mapmodulin Binds MAPs.
Because mapmodulin was discovered using an assay that depends (at least in part) upon microtubule-based organelle motility, we explored its possible interaction with microtubules or MAPs. For this purpose, we utilized a solid phase immunoassay (20). Wells were coated with either taxol-stabilized bovine microtubules bearing native MAPs, or with MAPs released from microtubules by 0.5 M KCl treatment. Unoccupied sites were blocked, and then increasing concentrations of mapmodulin were added. Bound mapmodulin was detected using anti-mapmodulin polyclonal antibodies and a colorimetric, antibody sandwich detection system. Control experiments using anti-MAP2 antibodies showed that the wells containing either microtubules bearing MAPs or crude, non-microtubule-associated MAPs contained essentially the same amounts of MAP2 protein (OD = 0.34 vs. 0.37, respectively).
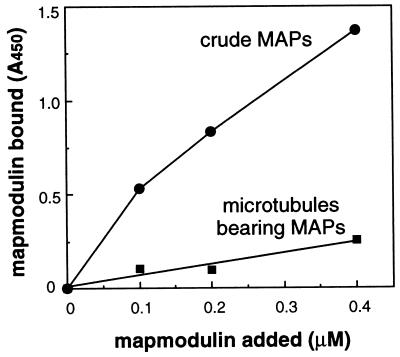
As shown in Fig. 3, a low level of mapmodulin bound to bovine brain microtubules bearing native MAPs. In contrast, mapmodulin showed substantially more binding to non-microtubule-associated brain MAPs. No binding was seen with pure, taxol stabilized, polymerized tubulin. Because mapmodulin showed a roughly 5-fold preference for MAPs not associated with microtubules, these data show that mapmodulin binds released MAPs in preference to microtubule-bound MAPs.
Figure 3.
Mapmodulin binds to MAPs. Microtiter wells were coated with either taxol-stabilized microtubules bearing native MAPs (0.1 mg/ml) or 0.5 M KCl-released, crude MAPs (0.1 mg/ml). After blocking wells, mapmodulin was added (50 μl). Bound mapmodulin was detected using anti-mapmodulin antibodies. Results represent the mean of duplicate wells.
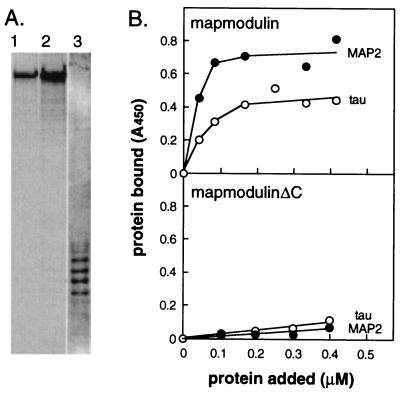
MAP2 and tau proteins are heat stable proteins, and mapmodulin bound to total heat stable MAP preparations (data not shown). Thus, we purified MAP2 and tau proteins to homogeneity from total heat stable MAP preparations to identify mapmodulin’s binding partner(s) (Fig. 4A). As shown in Fig. 4B Upper, mapmodulin bound to both highly purified MAP2 and tau. Binding was saturable, and 50% binding was observed when reactions contained ≈0.1 μM mapmodulin protein. Since mapmodulin is present in CHO cytosol at ≈0.8 μM, this binding is likely to be of physiological significance. In addition, MAP binding required the presence of mapmodulin’s C terminus (Fig. 4B Lower).
Figure 4.
Mapmodulin’s C terminus is required for binding to purified MAP2 and tau. (A) SDS/PAGE (8%) and Coomassie blue staining of purified bovine brain MAPs. MAP2 (lanes 1 and 2, 2 and 4 μg) and tau (lane 3, ≈2 μg) were purified by salt-stripping a twice-cycled bovine microtubule pellet, followed by heat treatment and fractionation on Mono Q. (B) Mapmodulin binding to purified MAP2 and tau. The procedure was the same as in Fig. 3 except wells were coated with 10 μg/ml of either purified tau or MAP2. (Upper) Intact mapmodulin. (Lower) MapmodulinΔC.
While MAP2 and tau are believed to be neuron-specific, MAP4 is abundant in a wide variety of cell and tissue types (5). MAP2, MAP4, and tau interact with microtubules by virtue of a homologous, repeated basic sequence near their C termini (reviewed in ref. 7). These basic residues interact with the acidic domain of tubulin. Because mapmodulin showed affinity for both MAP2 and tau, it was of interest to determine whether it could also bind MAP4. Indeed, MAP4 purified from HeLa cells was found to bind mapmodulin at a level comparable to MAP2 (data not shown; see below).
Mapmodulin Binds the Tubulin-Binding Domain of MAP4.
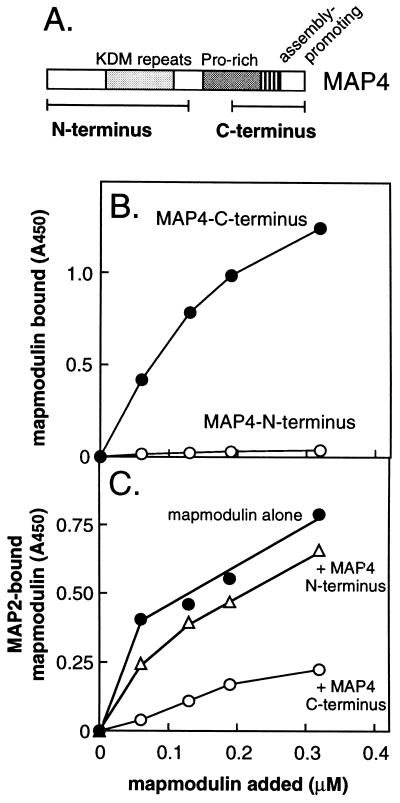
We reasoned that if mapmodulin bound preferentially to MAPs when they were not bound to microtubules, mapmodulin might bind to MAPs via their homologous tubulin-binding domains. To test this directly, we expressed fragments representing the N- and C-terminal domains of MAP4, respectively, fused to glutathione S-transferase (GST) using the constructs described by Wang et al. (4; see Fig. 5A). The C-terminal portion contains the microtubule binding repeat region and also the proline rich-region, both of which have been shown to contribute significantly to MAP4’s microtubule binding capacity (24). The N-terminal fragment comprises the projection domain and represents a large proportion of the mass of MAP4.
Figure 5.
(A) Schematic diagram of MAP4 constructs, redrawn from ref. 4. The KDM repeats are repetitive motifs of 14 residues; the C-terminal half of the proline-rich region and assembly-promoting region together comprise the microtubule binding domain (24). (B) Mapmodulin binds to MAP4’s microtubule binding, C-terminal domain but not to its N-terminal projection domain. Wells were coated with saturating concentrations of the MAP4 C-terminal domain GST fusion protein or GST–MAP4 N-terminal domain fusion protein. After blocking, mapmodulin was added (50 μl). Bound mapmodulin was detected using anti-mapmodulin antibodies. (C) The MAP4 C-terminal domain interferes with mapmodulin binding to MAP2. Wells were coated with 0.15 mg/ml MAP2, excess binding sites were blocked, and different concentrations of mapmodulin were added together with 0.06 μM MAP4 N-terminal domain or 0.05 μM MAP4 C-terminal domain.
Consistent with our prediction, mapmodulin failed to bind the N-terminal portion of MAP4, but showed a high level of binding to the tubulin-binding, C-terminal portion (Fig. 5B). Moreover, the C-terminally truncated form of mapmodulin failed to bind the C-terminal portion of MAP4 (data not shown), consistent with the inability of the C-terminally truncated protein to mediate MAP2 and tau binding. Finally, the ability of mapmodulin to bind MAP2 was inhibited significantly by the presence of the MAP4 C-terminal fragment but not the MAP4 N-terminal fragment (Fig. 5C). These experiments demonstrate directly that mapmodulin binds the tubulin-binding domain of MAP4 and likely also, MAP2. Further experiments will be needed to confirm the precise binding sites on MAP2 and tau.
Mapmodulin Modulates MAP Binding to Microtubules.
MAP proteins are found predominantly in association with microtubules at steady state, but they are known to be in equilibrium with a soluble pool, and their rate of exchange on microtubules is altered in a cell cycle-dependent manner (25). Having shown that mapmodulin binds MAPs at their microtubule-binding sites, we then asked whether such interaction might modulate MAP association with microtubule polymers.
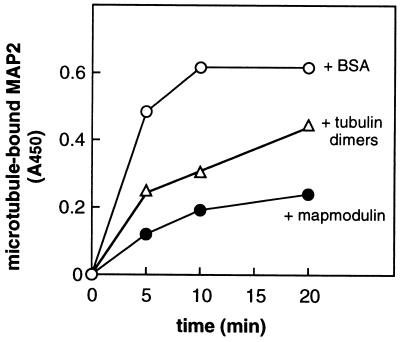
Purified MAP2 was preincubated with a 20-fold molar excess of either BSA, phosphocellulose-purified tubulin α/β heterodimers, or mapmodulin. We then examined the ability of the preincubated MAP2 to bind taxol-stabilized microtubules (Fig. 6). Tubulin dimers slowed the rate of MAP2 association with microtubules. However, mapmodulin led to an almost 4-fold decrease in the initial rate of MAP2 binding to microtubules. This indicates that mapmodulin’s interaction with MAP2 is at least as strong as that of tubulin dimers, which enables it to compete with microtubule polymers for the binding of MAP2. However, MAP2 bound polymerized microtubules with higher affinity than to mapmodulin, consistent with the steady-state distributions of these molecules in living cells. In summary, these data show that mapmodulin can influence the rate of association of a soluble MAP with polymerized microtubules.
Figure 6.
Mapmodulin competes with microtubules for binding to MAP2. MAP2 (0.015 μM) was preincubated with either BSA (0.3 μM), tubulin dimers (0.27 μM), or mapmodulin (0.3 μM) in 80 mM K-Pipes (pH 6.9), 1 mM EGTA, and 1 mM MgSO4 for 1 hr at 22°C. The different complexes were added to microtiter wells precoated with 0.1 mg/ml taxol-stabilized microtubules. The reaction was stopped at the indicated times by washing with blocking buffer. Bound MAP2 was detected using monoclonal anti-MAP2 antibody (diluted 1:600).
Mapmodulin in Cultured Cells.
Indirect immunofluorescence of CHO cells and immunoblot analyses of fractionated CHO cells revealed that mapmodulin is highly abundant in cytosol and is also present on endoplasmic reticulum and Golgi membranes as well as endosomes (N.U., unpublished data). The ability of mapmodulin to interact with MAPs when they are not microtubule-bound was consistent with the subcellular localization observed; microtubule association would not have been expected.
DISCUSSION
We have reported here, to our knowledge, the first example of a protein (other than tubulin) that can bind to the microtubule-binding domains of the MAPs, MAP2, MAP4, and tau. This protein, named mapmodulin, is relatively abundant; it is present in the cytosol and to a variable extent, broadly distributed on the membranes of a wide variety of cell and tissue types. We have shown that mapmodulin binds free MAPs in strong preference to microtubule-bound MAPs, and does so via the MAP’s microtubule-binding domain. Binding to each of these MAPs requires the C-terminal portion of mapmodulin, and decreases the rate with which MAPs can subsequently re-bind to microtubules.
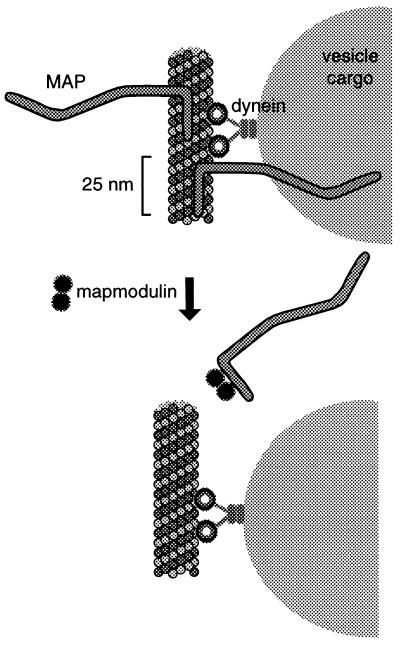
Fig. 7 summarizes our speculative working model for how mapmodulin may function. Membrane-bound organelles are translocated along microtubule tracks by the motors dynein and kinesin (data not shown). Membrane-bound vesicles have a minimum average diameter of ≈100 nm and will include much larger tubules of endoplasmic reticulum and Golgi cisternae. In nonneuronal cells, membrane cargo will encounter at least five MAP4 molecules per vesicle diameter, and are likely to encounter other MAPs as well. In other words, without even moving, the vesicles will be surrounded by MAPs. Even if the motor has the capacity to move around MAPs along the microtubule surface, the large MAP-projection domains are likely to extend as much as 90 nm away from the microtubule surface (8, 26) and will surely contribute drag in terms of vesicle motility. As discussed earlier, direct evidence for actual MAP-mediated interference has been obtained by several groups (11–13).
Figure 7.
Speculative model for the role of mapmodulin in modulating MAP-microtubule interaction. Dynein-based vesicle translocation is inhibited by the presence of MAPs. By binding to the microtubule-binding domain of a MAP, mapmodulin may influence the association rate of MAPs onto microtubules, and thereby facilitate the translocation of motors and cargo. Such activity might also facilitate kinesin-driven motility. Mapmodulin is found both in soluble and membrane-bound forms. An attractive scenario would be that it is the membrane bound form that provides local MAP release in conjunction with organelle translocation.
Membrane-associated mapmodulin could lead to the local release of MAPs, thereby enhancing organelle motility. Alternatively, cytosolic mapmodulin may be recruited transiently by molecular motors (or their associated proteins) to permit MAP circumnavigation. Full release might not be required; mapmodulin could help to slide a MAP to the opposite face of a microtubule to permit organelle translocation.
We first discovered and purified mapmodulin based on its ability to stimulate dynein-mediated, microtubule-dependent Golgi localization in broken cells. This provides functional evidence of mapmodulin’s ability to facilitate vesicle movements within a physiological context. Our assay reconstitutes the centrosomal localization of Golgi complexes, and we have shown that this process is absolutely dependent on the presence of cytoplasmic dynein and functional microtubules (16). Both native mapmodulin purified from CHO cells and recombinant mapmodulin obtained from Escherichia coli showed significant activity compared with total cytosol in terms of their ability to stimulate Golgi localization in semi-intact cells.
The sequence of mapmodulin revealed that it was identical to a previously identified protein termed PHAPI (18, 27, 28). The activity of recombinant mapmodulin (PHAPI) in Golgi capture reactions, and its ability to relieve the inhibitory effect of anti-mapmodulin antibodies, confirmed that mapmodulin activity is due to the protein encoded by the gene encoding PHAPI.
After completion of this study, it was reported that PHAPI is a potent and specific inhibitor of protein phosphatase 2A (29). These workers utilized boiling and trichloroacetic acid precipitation steps during their purification and obtained a protein that chromatographed as a large molecular aggregate. We have shown that mapmodulin occurs as a dimer or trimer of 31-kDa subunits in CHO cytosol, and the protein we have purified also chromatographs as a dimer or trimer after purification. This gives us confidence that we are studying the native form of this protein. Of course, it is entirely possible that mapmodulin has dual functions. While proof of mapmodulin’s role(s) in living cells will require further study, in our hands, native mapmodulin was unable to inhibit protein phosphatase 2A (N.U., unpublished data).
As described earlier, MAP2, tau and MAP4 have homologous tubulin-binding domains (7). These basic regions interact with the highly acidic C terminus of tubulin. Several studies indicate that the MAP binding site on tubulin is comprised of the sequence GEFEEEEGE(ED) (30) or EEGEE (11), although additional upstream sequences may also be important (7). It is noteworthy that mapmodulin contains repeats of similar sequences DEDEEEEGEEED, VSGEEEEDEEGY, and DEEDEEELGEEE near its C terminus, and we have shown here that this domain is essential for MAP binding.
There is growing evidence that accessory factors enhance the function of microtubule-based molecular motors. Dynein-based motility surely involves a variety of intracellular components that include membrane-associated receptors and cytosolic regulatory factors. In vitro studies of cytoplasmic dynein-mediated vesicle transport have revealed the importance of the dynactin complex, a 20S multiprotein assembly composed of eight different proteins (31). Another, yet uncharacterized 9S activating factor was found to increase both the frequency and velocity of vesicle transport and also supported plus end-directed motor movements (32). Factors may also exist to modulate MAP–microtubule interactions, thereby permitting motor-mediated transport. We propose that mapmodulin represents a novel class of proteins that functions to displace MAPs transiently from microtubules, thereby permitting organelle translocation. A future goal will be to test this hypothesis directly.
Acknowledgments
We are grateful to Drs. Hartmut Kratzin and Norbert Hilschmann for providing us with the PHAPI cDNA clone, Dr. Gary Borisy for providing the MAP4 GST-constructs, and Dr. Ole Hindsgaul for supplying us with oligosaccharide substrate for GlcNAc transferase assays. This research was funded by National Institutes of Health Grant GM49843. N.U. and M.H. were recipients of postdoctoral fellowships from the American Heart Association and the Leukemia Society, respectively.
ABBREVIATIONS
- MAP
microtubule-associated protein
- PHAPI
putative histocompatibility antigen-associated protein I
- GlcNAc
N-acetylglucosamine
- GST
glutathione S-transferase
References
- 1.Kirschner M, Mitchison T. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- 2.Chapin S J, Bulinski J C. Cell Motil Cytoskeleton. 1992;23:236–243. doi: 10.1002/cm.970230403. [DOI] [PubMed] [Google Scholar]
- 3.Thaler C D, Haimo L T. Int Rev Cytol. 1996;164:269–327. doi: 10.1016/s0074-7696(08)62388-8. [DOI] [PubMed] [Google Scholar]
- 4.Wang X M, Peloquin J G, Zhai Y, Bulinski J C, Borisy G G. J Cell Biol. 1996;132:345–357. doi: 10.1083/jcb.132.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulinski J C, Borisy G G. J Biol Chem. 1980;255:11570–11576. [PubMed] [Google Scholar]
- 6.Hirokawa N. Curr Opin Cell Biol. 1994;6:74–81. doi: 10.1016/0955-0674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 7.Maccioni R B, Cambiazo V. Physiol Rev. 1995;75:835–864. doi: 10.1152/physrev.1995.75.4.835. [DOI] [PubMed] [Google Scholar]
- 8.Voter W A, Erickson H P. J Ultrastruct Res. 1982;80:374–382. doi: 10.1016/s0022-5320(82)80051-8. [DOI] [PubMed] [Google Scholar]
- 9.Barlow S, Gonzalez-Garay L, West R R, Olmsted J B, Cabral F. J Cell Biol. 1984;126:1017–1029. doi: 10.1083/jcb.126.4.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapin S J, Bulinski J C. Cell Motil Cytoskeleton. 1994;27:133–149. doi: 10.1002/cm.970270205. [DOI] [PubMed] [Google Scholar]
- 11.Paschal B M, Obar R A, Vallee R B. Nature (London) 1989;342:569–572. doi: 10.1038/342569a0. [DOI] [PubMed] [Google Scholar]
- 12.Hagiwara H, Yorifuji H, Sato-Yoshitake R, Hirokawa N. J Biol Chem. 1994;269:3581–3589. [PubMed] [Google Scholar]
- 13.López L A, Sheetz M P. Cell Motil Cytoskeleton. 1993;24:1–16. doi: 10.1002/cm.970240102. [DOI] [PubMed] [Google Scholar]
- 14.von Massow A, Mandelkow E-M, Mandelkow E. Cell Motil Cytoskeleton. 1989;14:562–571. doi: 10.1002/cm.970140413. [DOI] [PubMed] [Google Scholar]
- 15.López L A, Sheetz M P. J Biol Chem. 1995;270:12511–12517. doi: 10.1074/jbc.270.21.12511. [DOI] [PubMed] [Google Scholar]
- 16.Corthésy-Theulaz I, Pauloin A, Pfeffer S R. J Cell Biol. 1992;118:1333–1345. doi: 10.1083/jcb.118.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balch W E, Dunphy W G, Braell W A, Rothman J E. Cell. 1984;39:405–416. doi: 10.1016/0092-8674(84)90019-9. [DOI] [PubMed] [Google Scholar]
- 18.Vaesen M, Barnikol-Watanabe S, Götz H, Awni L A, Cole T, Zimmermann B, Kratzin H D, Hilschmann N. Biol Chem Hoppe-Seyler. 1994;375:113–126. doi: 10.1515/bchm3.1994.375.2.113. [DOI] [PubMed] [Google Scholar]
- 19.Vallee R B. Methods Enzymol. 1986;134:89–115. doi: 10.1016/0076-6879(86)34078-3. [DOI] [PubMed] [Google Scholar]
- 20.Pedrotti B, Colombo R, Islam K. Biochemistry. 1994;33:8798–8806. doi: 10.1021/bi00195a023. [DOI] [PubMed] [Google Scholar]
- 21.Tabas I, Kornfeld S. J Biol Chem. 1979;254:11655–11663. [PubMed] [Google Scholar]
- 22.Clary D O, Rothman J E. J Biol Chem. 1990;265:10109–10117. [PubMed] [Google Scholar]
- 23.Kaur K J, Hindsgaul O. Glycoconjugate J. 1991;8:90–94. doi: 10.1007/BF00731017. [DOI] [PubMed] [Google Scholar]
- 24.Aizawa H Y, Emori A, Mori H, Murofushi H, Sakai H, Suzuki K. J Biol Chem. 1991;266:9841–9846. [PubMed] [Google Scholar]
- 25.Olmsted J B, Stemple D L, Saxton W M, Neighbors B W, McIntosh J R. J Cell Biol. 1989;109:211–223. doi: 10.1083/jcb.109.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottlieb R A, Murphy D B. J Cell Biol. 1985;101:1782–1789. doi: 10.1083/jcb.101.5.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuoka K, Taoka M, Satozawa N, Nakayama H, Ichimura T, Takahashi N, Yamakuni T, Song S-Y, Isobe T. Proc Natl Acad Sci USA. 1994;91:9670–9674. doi: 10.1073/pnas.91.21.9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fink T M, Vaesen M, Kratzin H D, Lichter P, Zimmer M. Genomics. 1995;29:309–310. doi: 10.1006/geno.1995.1257. [DOI] [PubMed] [Google Scholar]
- 29.Li M, Makkinje A, Damuni Z. Biochemistry. 1996;35:6998–7002. doi: 10.1021/bi960581y. [DOI] [PubMed] [Google Scholar]
- 30.Littauer U Z, Giveon D, Thierauf M, Ginzburg I, Ponstingl H. Proc Natl Acad Sci USA. 1986;83:7162–7166. doi: 10.1073/pnas.83.19.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gill S R, Schroer T A, Szilak I, Steuer E R, Sheetz M P, Cleveland D W. J Cell Biol. 1991;155:1639–1650. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroer T A, Sheetz M P. J Cell Biol. 1991;115:1309–1318. doi: 10.1083/jcb.115.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]