Abstract
The E2F family of transcription factors plays a crucial role in cell cycle progression. E2F activity is tightly regulated by a number of mechanisms, which include the timely synthesis and degradation of E2F, interaction with retinoblastoma protein family members (“pocket proteins”), association with DP heterodimeric partner proteins, and phosphorylation of the E2F/DP complex. Here we report that another mechanism, subcellular localization, is important for the regulation of E2F activity. Unlike E2F-1, -2, or -3, which are constitutively nuclear, ectopic E2F-4 and -5 were predominantly cytoplasmic. Cotransfection of expression vectors encoding p107, p130, or DP-2, but not DP-1, resulted in the nuclear localization of E2F-4 and -5. Moreover, the transcriptional activity of E2F-4 was markedly enhanced when it was invariably nuclear. Conversely, it was reduced when the protein was excluded from the nucleus, implying that E2F-4 transcription function depends upon its cytological location. In keeping with this, the nuclear/cytoplasmic ratios of endogenous E2F-4 changed as cells exited G0, with high ratios in G0 and early G1 and a progressive increase in cytoplasmic E2F-4 as cells approached S phase. Thus, the subcellular location of E2F-4 is regulated in a cell cycle-dependent manner, providing another potential mechanism for its functional regulation.
The E2F family of transcription factors has been implicated in cell cycle control, and E2F DNA binding sites are present in the promoters of several growth regulating genes. These include enzymes required for nucleotide synthesis and DNA replication as well as known cell cycle regulatory proteins vital for the process of G1 exit and S phase entry and progression. Biochemical studies have shown that members of the E2F family function, at least in part, as heterodimers composed of an E2F and a DP subunit. Five members of the E2F family have been isolated and designated E2F-1 to E2F-5, and three heterodimeric partners, DP-1, -2, and -3 have been identified to date. The biological significance of the complexity of the E2F family is presently unclear (for reviews, see refs. 1–3).
Deregulated synthesis of E2F-1 to E2F-4 can lead to neoplastic transformation of certain immortalized cell lines (4–7), and E2F-1 induction in serum-deprived fibroblasts promoted S-phase entry followed by apoptosis (7–10). In contrast to overproduction studies, absence of E2F-1 synthesis in mice results in multiple somatic abnormalities including hyperplasia of certain lymphoid tissues within which a discrete defect in T cell apoptosis is demonstrated (11). Moreover, tumors of some epithelial organs occur in mature and late adulthood (12). These results imply that, in addition to its transforming activity when overproduced, E2F-1 also has a normal tumor-suppression function.
E2F activity is tightly regulated by a number of mechanisms during cell cycle progression. E2F/DP heterodimer formation facilitates binding to and negative regulation by pRb, the product of the retinoblastoma gene and the related pocket proteins, p107 and p130. Indeed, complexes of unphosphorylated pRb and E2F/DP act as transcriptional repressors which contribute to pRb-dependent G1 arrest. Once a pocket protein is phosphorylated by cyclin-dependent kinases (CDKs), E2F/DP/pocket protein complexes generally dissociate, resulting in free, transcriptionally active E2F/DP heterodimers. The combination of cessation of repression of some E2F-regulated genes and activation of others by the now activated transcription factor(s) constitutes a major step in promoting G1 exit (for reviews, see refs. 1 and 2).
The known E2F species can be grouped into two subfamilies based upon certain structural and functional characteristics. E2F-1, -2, and -3 are regulated exclusively by pRb, while E2F-4 and -5 are regulated primarily by p107 and p130 (4, 5, 13, 14). These differences in pocket protein/E2F binding specificity are not absolute, because E2F-4/pRb complexes have also been detected as relatively abundant late G1 and S phase elements (15). In addition, E2F-1, -2, and -3 each contains a dedicated cyclin binding sequence N terminal to its DNA binding domain, which mediates the stable binding of cyclin A/CDK2. This interaction results in late G1/S phase-mediated DP-1 phosphorylation which leads to loss of DNA binding (16–18). Failure of this reaction in cells overproducing E2F-1 translated into S phase delay and apoptosis (18, 19).
E2F-1 synthesis is cell cycle regulated in cells emerging from G0. In growth-arrested cells, E2F-1 synthesis is undetectable. Once they are induced to resume growth, E2F-1 levels rise during late G1 (20, 21). The E2F-1 promoter contains E2F DNA binding sites and is cell cycle regulated. Indeed, cell cycle-dependent modulation of its function depends upon the integrity of these sites (22–24). In marked contrast to E2F-1, E2F-4 and E2F-5 lack cyclin A binding sites. E2F-4 is present in growth-arrested cells, and its synthesis does not change significantly as cell progress through the cell cycle (4). Elucidating the significance of its continual synthesis is of special interest, since it accounts for the vast majority of ambient free E2F activity in asynchronous cultures (15).
It has recently been shown that overproduced E2F-4 is concentrated in the cytoplasm of transiently transfected cell lines and can be translocated to the nucleus in the presence of certain partner pocket and DP proteins (25). Here we report that the subcellular localization of endogenous E2F-4 changes in a cell cycle-dependent manner, as cultured cells emerge from G0. Our data further suggest that the regulated cytological location of this protein provides a measure of control of its biochemical function.
MATERIALS AND METHODS
Cell Culture and Synchronization.
Cells were grown at 37°C in a 10% CO2-containing atmosphere. U20S cells were maintained in DMEM supplemented with 10% fetal calf serum (HyClone) and NIH 3T3 cells in DMEM supplemented with 5% bovine calf serum (BCS; HyClone). NIH 3T3 cells were growth arrested by incubation for 72 h in DMEM containing 0.5% BCS. Cells were stimulated by addition of BCS to a final concentration of 10% and harvested at appropriate times. Fluorescence-activated cell sorter analysis was carried out as described (4).
Construction of Plasmids.
To generate pCMV-HA-E2F-4Δ18, a PCR product was first generated from cDNA and the oligonucleotides 5′-GGTGGATC-CGCGATGGCGGAGGCCGGG-3′ and 5′-GGGGAATTCTCAGAGGTTGAGAACAGGCACGTCTCCCGGGGGTGGAGA-3′. It was then subcloned into the BamHI–EcoRI sites of pcDNA1-HA-E2F-1 to replace E2F-1. Similarly, for pCMV-HA-E2F-4Δ1–81, a PCR product was first generated using a pCMV-E2F-4 template and the following oligonucleotides: 5′-GGTGGATCCACCATGAAGGGTGTGGGGCCTGGCTGC-3′ and 5′-GGGGAATTCTCAGAGGTTGAGAACAGG-3′ and then subcloned into the BamHI–EcoRI sites of pCMV-HA-E2F-4 to replace the wild-type (wt) E2F-4 sequences.
pcDNA3-HA-E2F-4 was constructed by subcloning the HindIII–EcoRI insert from pCMV-HA-E2F-4 into pcDNA3. To construct pcDNA3-HA-E2F-5, mouse E2F-5 cDNA was generated by PCR from pPC67 (a kind gift from R. Bernards, Netherlands Cancer Institute, Amsterdam) using the oligonucleotides: 5′-CGGGATCCATGGCGGCGGCGGAGCCCACG-3′ and 5′-CCCGAATTCTAATAATTTAGTATCTGAACATC-3′. The E2F-5 BamHI–EcoRI product was then inserted in pcDNA3-HA-E2F-4 to replace E2F-4.
pcDNA3-HA-E2F-4-NLS and pcDNA3-HA-E2F-4-NES contain sequences from the nuclear localization sequence (NLS) of simian virus 40 (SV40) large T antigen and the nuclear export sequence (NES) of HIV-1 Rev, respectively (26). These localization sequences were inserted between an N-terminal HA-tag and the E2F-4 open reading frame. They were constructed by generating PCR products using the NLS and NES oligonucleotides, 5′-CGCGGATCCGCGATGCCAAAAAAGAAGAGAAAGGTAATGGCGGAGGCCGGGCCAC-3′ and 5′-CGCGGATCCGCGATGCTGCCCCCCCTGGAAAGACTGACCCTGATGGCGGAGGCCGGGCCAC-3′, respectively, with 5′-GCCTGATGGGGCCCGGATGGCC-3′ and the pcDNA3-HA-E2F-4 template. The BamHI–SfiI PCR products were then substituted for that fragment in pcDNA3-HA-E2F-4. Verification of PCR product sequences was performed using an Applied Biosystems automated sequencer.
Other expression plasmids have been described: pcDNA1-HAE2F-1, pcDNA1-HAE2F-2, pcDNA1-HAE2F-3 (19), pCMV-HA-E2F-4 (4), pCMV-Rb (27), pCMV-p107 (28), pcDNA-HAp130 (29), pcDNAI-DP-1 and pcDNAI-HA-DP-1 (30), pCMV-HA-DP-2 (31), E2F-luciferase reporter plasmid, and 3×WT-E2F-Luc and pCMV-β-Gal (30).
Transient Transfections and Reporter Assays.
Transfections were performed using a modified calcium phosphate method. Transfections for immunofluorescence studies included E2F, pocket protein, or DP-containing expression plasmid (2 μg each). For luciferase reporter assays, transfections included the amounts of expression vector indicated together with 2 μg of reporter plasmid, 3×WT-E2F-Luc, and 2 μg pCMV-β-Gal. Cells were harvested 36–48 h following transfection. Cell lysis, β-galactosidase, and luciferase assays were carried out as described (32).
Cell Extracts and Immunoblotting.
Cells were briefly trypsinized and washed in PBS. Samples of whole cell extracts were lysed in RIPA buffer (50 mM Tris, pH 8.1/150 mM NaCl/0.2% SDS/1% sodium deoxycholate/1% Nonidet P-40/5 mM EDTA) containing protease inhibitors (9 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride). For nuclear and cytoplasmic extracts, cells were suspended in hypotonic RSB buffer (10 mM Hepes, pH 6.2/10 mM NaCl/1.5 mM MgCl2) with protease inhibitors for 30 min, and homogenized using a Dounce homogenizer until disruption of the plasma membrane was confirmed by staining in 0.25% trypan blue dye in PBS. Following centrifugation, the supernatant, which contained the cytoplasmic fraction, was concentrated using a Millipore low binding regenerated cellulose filter unit (10,000 NMWL; Amersham) and adjusted to 150 mM NaCl. The nuclear pellet was washed once in hypotonic buffer and then lysed with RIPA buffer. Extracts were quantitated with the Bio-Rad protein assay kit.
Boiled extracts in sample buffer were subjected to electrophoresis (30 μg of cellular protein per lane unless otherwise stated) in a 10% SDS/polyacrylamide gel and transferred to nitrocellulose by standard procedures. Membranes were blocked with 5% nonfat milk in Tris buffered saline (TBS; pH 8.0) with 0.05% Tween 20 and incubated with primary antibody. Endogenous E2F-4 was detected using the anti-E2F-4 mAb, GG22, hybridoma tissue culture supernatant (4) at 1:10 dilution in blocking buffer. E2F-1 was detected using mAb KH20 (a kind gift from E. Harlow, Massachusetts General Hospital Cancer Center, Charlestown, MA) at 1:20 dilution, and β-tubulin using mAb KMX-1 at 0.5 μg/ml (Boehringer Mannheim). After washing with 0.05% Tween 20 in TBS, immunoblots were incubated with horseradish-peroxidase-conjugated rabbit-anti-mouse-IgG (Amersham), and proteins were visualized using enhanced chemiluminescence according to the manufacturer’s instructions (Amersham). Relative intensities of bands were scored by digital scanning and image software for the Macintosh (National Institutes of Health).
Immunofluorescence Staining.
Cells were plated onto coverslips in 6 cm Petri dishes prior to transient transfection and were washed 12–16 h following transfection. For immunostaining, cells were fixed 24–36 h later with 3% paraformaldehyde and 2% sucrose in PBS, permeablized by a short treatment with 0.2% Triton X-100 in PBS, and then incubated with primary antibody at room temperature for 1–2 h. Primary mAbs included those which recognized E2F-1 (SQ41) (20), the HA-epitope tagged to E2F-2 or E2F-3 (12CA5), E2F-4 (GG22), E2F-5 (MH-5; Santa Cruz Biotechnology), pRb (PMG3–245; PharMingen), or p107 (SD6, generously provided by N. Dyson, Massachusetts General Hospital Cancer Center). Primary affinity-purified polyclonal antibodies included p130 (C-20; Santa Cruz Biotechnology), E2F-4 (C-108; Santa Cruz Biotechnology), DP-1 (K-20; Santa Cruz Biotechnology), and DP-2 (C-20; Santa Cruz Biotechnology). Secondary antibody conjugated to either fluorescein isothiocyanate (FITC) or rhodamine (Sigma or Boehringer Mannheim) was then applied at a dilution of 1:200. Nuclei were counterstained with 1 μg/ml Hoechst 33258 (Sigma).
RESULTS
Subcellular Localization of Endogenous E2F-4 Changes During the Cell Cycle.
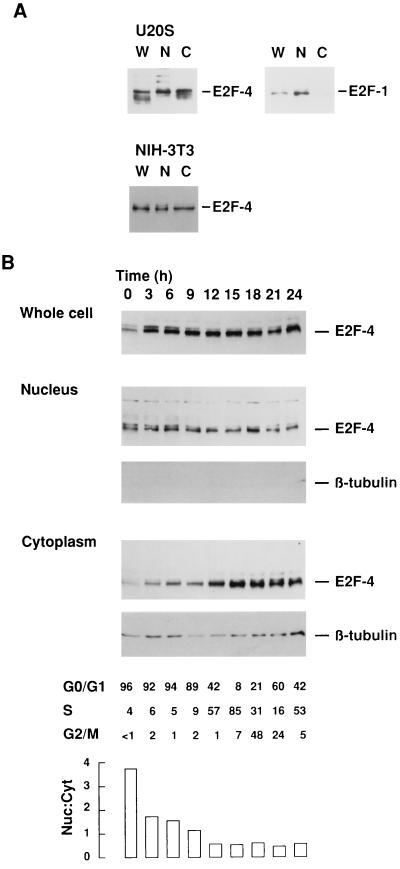
We first analyzed the subcellular compartmentalization of endogenous E2F-4 in different cell lines. Nuclear and cytoplasmic extracts prepared from asynchronously growing U20S cells were assessed by Western blot analysis using mouse mAbs, GG22 and KH20, which are specific for E2F-4 and E2F-1, respectively. Endogenous E2F-4 was detected in similar levels in both the nuclear and cytoplasmic fractions (Fig. 1A Upper Left). In marked contrast, endogenous E2F-1 was primarily detected in the nuclear fraction of asynchronously growing U2OS cells (Fig. 1A Upper Right). Thus, endogenous E2F-1 and E2F-4 show distinctly different patterns of subcellular localization. Endogenous E2F-4 was also seen in cytoplasmic extracts of immortal, but otherwise untransformed NIH 3T3 cells (Fig. 1A Lower), indicating that cytoplasmic concentration of E2F-4 is not confined to transformed cell lines.
Figure 1.
Location of endogenous E2F-4 is cell cycle-dependent. (A) Western blot analysis of whole cell (W), nuclear (N) and cytoplasmic (C) protein extracts from asynchronously growing U2OS (Upper) and NIH 3T3 cells (Lower) using mAbs to E2F-4 (GG22) and E2F-1 (KH20) to detect endogenous protein. (B) NIH 3T3 cells were serum-starved for 72 h in 0.5% serum and synchronously released using 10% serum-containing medium. Whole cell, nuclear, and cytoplasmic protein extracts were collected at the times shown above and subject to Western blot analyses. Blots were probed with mAb to E2F-4 or the cytoplasmic protein, β-tubulin, respectively. The cell cycle status of each sample is shown below, together with the relative nuclear:cytoplasmic levels of E2F-4 protein. A total of 100 μg of cellular protein was loaded per lane for U2OS cells, and 30 μg for NIH 3T3 cells.
To determine whether E2F-4 nuclear/cytoplasmic ratios change during the cell cycle, NIH 3T3 cells were arrested in G0 by serum starvation for 72 h and then released into the cycle by refeeding with 10% serum for 24 h. Synchronous release and cell-cycle progression was confirmed by flow cytometric analysis at three hourly intervals. Whole cell, nuclear, and cytoplasmic preparations were analyzed by Western blotting using mAbs to E2F-4 and cytoplasmic β-tubulin, the latter serving as a control for the effectiveness of fractionation (Fig. 1B). The E2F-4 levels in whole cell extracts did not change significantly following release from G0, although levels were demonstrably lower in serum-starved cells, as reported previously (4). However, when subcellular fractions were analyzed, it appeared that E2F-4 was largely confined to the nuclear fraction during G0, and levels progressively accumulated in the cytoplasm thereafter. Interestingly, the nuclear levels of E2F-4 did not change appreciably. The relative levels of nuclear E2F-4 exceeded cytoplasmic levels in the G0/G1 population, whereas cytoplasmic levels exceeded nuclear levels in S phase and G2/M. Thus, endogenous E2F-4 is detectable in both the nucleus and cytoplasm and the relative levels in these fractions change significantly as cells exit G0 and proceed into and through the cycle.
Subcellular Localization of Ectopic E2F-4 and -5 Is Altered by the Cotransfection of p107, p130, or DP-2.
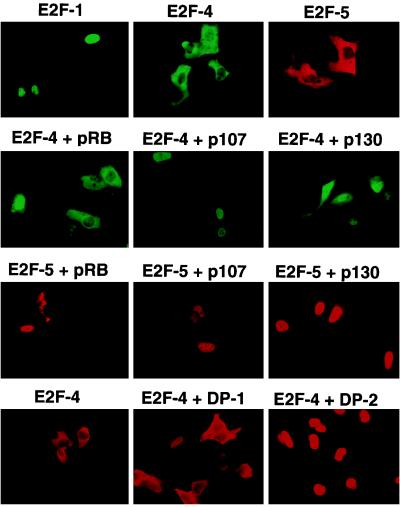
To analyze whether various ectopically expressed E2Fs exhibited differences in their subcellular localization, U2OS osteogenic sarcoma cells were examined by immunostaining 40 h following transient transfection with expression vectors encoding certain E2F family members (Fig. 2, top row). Ectopic E2F-1 localized to the nucleus of transfected cells. E2F-2 and E2F-3, which share greatest homology with E2F-1, were also detected in the nuclei of transfected cells (data not shown). In marked contrast to E2F-1, E2F-4 and E2F-5 (which shows the greatest homology with E2F-4 among the known E2F family members) were predominantly cytoplasmic. Thus, the differential localization of ectopic E2F-1 and E2F-4, determined by immunofluorescence staining, correlated with the subcellular localization of endogenous E2F described above.
Figure 2.
Subcellular localization of E2F-4 and -5 is affected by coexpression of either the p107 or p130 pocket proteins or DP-2. Asynchronously growing U20S cells were transiently transfected with expression vectors encoding the proteins shown above each panel. After 40 h, overproduced E2F protein was detected in fixed cells by indirect immunofluorescence using the corresponding primary antibody and a FITC- or rhodamine-conjugated secondary antibody. In each case, location of the cotransfected protein was also determined (data not shown). Nuclei were revealed by Hoechst staining (data not shown).
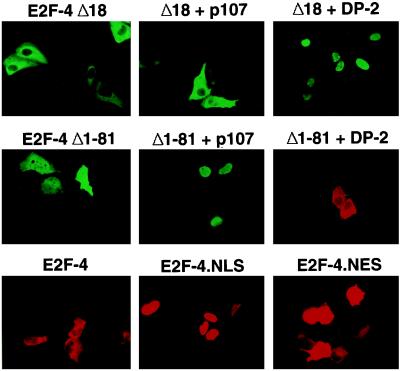
E2F species are known to complex with pocket proteins, which are by themselves nuclear (28, 33, 34). To study the effect of this protein–protein interaction on the location of ectopic E2F-4 and E2F-5, U2OS cells were cotransfected with either E2F-4 or -5 and pRb, p107, or p130 expression vectors, and the subcellular localization of these E2F proteins was again determined by immunostaining. In keeping with the prior results of Magae et al. (25), synthesis of both p107 and p130, and to a much lesser extent pRb, led to an apparent translocation of both E2F-4 and E2F-5 to the nucleus (Fig. 2, second and third rows). To determine whether nuclear localization of E2F-4 and -5 was the product of an indirect effect of p107, we examined the effect of p107 cotransfection on the localization of E2F-4Δ18, an E2F-4 mutant that cannot directly bind to p107. When transfected alone into cells, E2F-4Δ18, like wt-E2F-4, localized to the cytoplasm (Figs. 3 and 4 Top). However, unlike wt-E2F-4, E2F-4Δ18 remained in the cytoplasm when cotransfected with p107. As expected, p107 itself had a nuclear subcellular location (data not shown). Thus, the p107-binding region of E2F-4 was necessary for the p107-promoted nuclear localization of E2F-4. This suggests that p107/E2F-4 complex formation is essential for E2F-4 nuclear translocation under the conditions which were utilized.
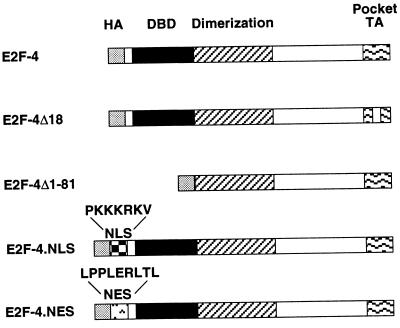
Figure 3.
Wild-type and mutant E2F-4 species. E2F-4Δ18 lacks pocket protein binding sequences, E2F-4Δ1–81 lacks the DNA-binding domain. The NLS from SV40 T antigen NLS and the NES from the HIV-1 Rev protein were inserted at the N terminus in E2F-4.NLS and E2F-4.NES, respectively. HA, hemagglutinin epitope; DBD, DNA-binding domain; dimerization domain; TA, pocket protein-binding and transactivation domain.
Figure 4.
Effects of p107 and DP-2 on the nuclear localization of E2F-4 mutants. U2OS cells were transiently cotransfected with plasmids encoding either a mutant deficient in the ability to bind pocket proteins (E2F-4Δ18) or to bind DNA (E2F-4Δ1–81) and p107 or DP-2, as indicated above each panel (Top and Middle). E2F-4 was detected by immunofluorescence using mAb GG22 and a FITC- or rhodamine-conjugated secondary antibody. Nuclear location of coexpressed p107 or DP-2 was confirmed in each case (data not shown). (Bottom) The location of E2F-4 in U2OS cells transiently transfected with wt-E2F-4, E2F-4.NLS, or E2F-4.NES is shown.
Because E2F species function as heterodimers, each composed of an E2F and a DP subunit, it was plausible that their interaction might affect E2F subcellular localization. When cotransfected with DP-1, which by itself localizes to the cytoplasm (data not shown), both E2F-4 and DP-1 were detected in the cytoplasm (Fig. 2 Bottom). However, cotransfection of DP-2, which by itself localized to the nucleus (data not shown), and E2F-4 resulted in efficient E2F-4 nuclear staining. In a parallel cotransfection experiment, DP-2 induced nuclear translocation of the E2F-4Δ18 mutant that was incapable of binding to p107 (Figs. 3 and 4 Top). This suggests that DP-2 and p107 can independently promote the nuclear location of E2F-4.
To search for any relevance between E2F-4 DNA binding activity and its ability to concentrate in the nucleus, an E2F-4 mutant lacking its N-terminal 81 residues, E2F-4Δ1–81, was generated (Fig. 3). This mutant lacks both the DNA-binding domain and sequences that were previously shown to be required for interaction of E2F-1 with its DP partner (30, 35). When synthesized alone, E2F-4Δ1–81 was present in both the cytoplasm and the nucleus (Fig. 4 Middle). Cotransfection of p107 resulted in its apparent nuclear concentration, whereas cotransfection of DP-2 had no effect. These data suggest that DNA binding is not essential for E2F-4 nuclear localization and further support the notion that, at least in part, p107 and DP-2 can independently induce E2F-4 nuclear translocation.
Nuclear Localization of E2F-4 Correlates with its Transcriptional Activity.
E2F-4 lacks a consensus NLS. To establish that exclusion of E2F-4 from the nucleus is due to lack of this structural element, a heterologous sequence encoding the canonical NLS sequence from SV40 T antigen was fused to E2F-4 to generate E2F-4.NLS (Fig. 3). When transfected into U2OS cells, E2F-4.NLS protein did, indeed, localize to the nucleus, suggesting that E2F-4 is inherently excluded from the nucleus due to the absence of a native NLS (Fig. 4 Bottom). As expected, E2F-4 remained cytoplasmic following the incorporation of a NES derived from the HIV Rev protein (26) (Figs. 3 and 4 Bottom).
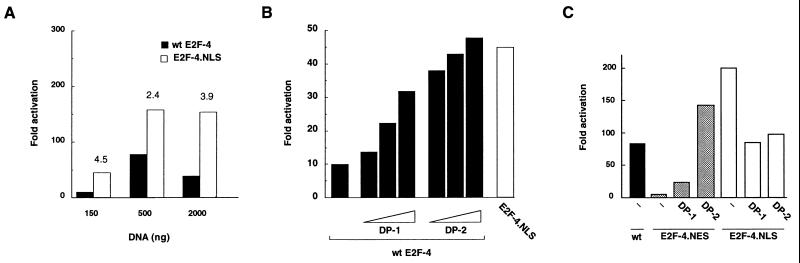
The ability of wt-E2F-4, E2F-4 linked to nuclear localization sequence (E2F-4.NLS) and E2F-4 fused to the HIV Rev nuclear export sequence (E2F-4.NES) to transactivate E2F promoter sequences was assessed. This was carried out by cotransfection of U2OS cells with expression vectors for the above proteins and a reporter plasmid containing three E2F DNA-binding sites linked to a luciferase cDNA sequence. E2F-4.NLS consistently exhibited 2- to 5-fold higher luciferase activity than wt-E2F-4 at all concentrations tested (Fig. 5). Conversely, E2F-4.NES proved a much weaker trans-activator than wt-E2F-4 (Fig. 5C). Western blot analysis confirmed that the amount of ectopically expressed wt-E2F-4 was equivalent to E2F-4.NES protein and even exceeded E2F-4.NLS levels (data not shown). Because differences in the synthesis levels of these various ectopic species cannot account for the observed differences in transcription function and because E2F-4 functions as a heterodimer, the levels of E2F activity in the various E2F-4-transfected cells likely reflect the formation of functional complexes between overproduced E2F-4 protein and endogenous DP protein(s). Thus, the greater transcription activity achieved by E2F-4.NLS over wt-E2F-4, and by wt-E2F-4 over E2F-4.NES, probably reflects the size of the pool of nuclear E2F that is available for dimerization to endogenous DP protein.
Figure 5.
Nuclear import enhances E2F-4 transcriptional activity. (A) U20S cells were transfected with 2 μg of E2F-luciferase, 2 μg of pCMV-β-Gal, and 150, 500, or 2,000 ng of either wt-E2F-4 or E2F-4.NLS. The resulting cell extracts were analyzed by luciferase reporter assay and values normalized to β-galactosidase units. Fold-activation over wt-E2F-4 is indicated above the E2F-4.NLS bars. (B) Transcriptional activity 150 ng of wt-E2F-4 alone versus wt-E2F-4 cotransfected with 100, 500, or 1,000 ng of either DP-1 or DP-2. A total of 150 ng of the E2F-4.NLS expression vector induced the transactivation noted in the far right bar of this panel. (C) Luciferase reporter assay using 2 μg of either wt-E2F-4, E2F-4.NES or E2F-4.NLS expression vector, alone, or after cotransfection with 1 μg of DP-1 or DP-2.
DP-1 has been shown previously to enhance the trans-activation potential of E2F-4. However, it is not clear whether DP-1 or DP-2 (or both) represent the physiologic partners of E2F-4 in the nucleus. We therefore compared the transactivation potential of HA-tagged DP-1 and DP-2 on E2F-4 (Fig. 5B). DP-2 proved to be more effective than DP-1 at all concentrations tested. Again, Western blot analysis using the 12CA5 HA-antibody confirmed that the levels of ectopically expressed DP-1 and DP-2 were equivalent when similar concentrations of plasmid were used in the transfection (data not shown). A role for the DP-2 protein in the nuclear import of E2F-4 was further supported by cotransfection studies using E2F-4.NES and E2F-4.NLS (Fig. 5C). Specifically, we asked whether DP-1 or DP-2 would enhance E2F-4.NES and E2F-4.NLS transcriptional activity. Whereas DP-1 led to a minimal effect on E2F-4.NES transactivation function, cotransfection of DP-2 restored its trans-activation potential to wild-type levels. In contrast, neither DP-1 nor DP-2 augmented the transcriptional activity of E2F-4.NLS. In fact, in some experiments transcriptional repression was seen. The reason for this is not clear. Thus, nuclear import of E2F-4 resulting from DP-2 binding was dominant over any nuclear export function imparted by the NES motif. Immunofluorescence staining of transfected cells supported this conclusion, since ectopic E2F-4.NES was cytoplasmic before and nuclear after U2OS cells were cotransfected with DP-2 (data not shown). These findings suggest that the transcriptional activity of E2F-4 may be restricted, in part, by its cytoplasmic location and that DP-2 (but not DP-1) normally present in limiting quantities, plays a role in the nuclear localization and functional activation of E2F-4.
DISCUSSION
There is by now considerable evidence to suggest that E2F species play a pivotal role in cell cycle control. Deregulated synthesis of E2F can result, in some cases, in S-phase entry followed by apoptosis, and, in others, neoplastic transformation. Because overproduction of E2F can have a deleterious effect on cell growth and survival, it is not surprising that E2F activity is tightly regulated. Indeed, multiple regulatory mechanisms affect E2F behavior. E2F species interact with and are negatively regulated by pocket proteins. At a later time in the cell cycle, certain E2F/DP heterodimers interact with and are negatively regulated by cyclin A/CDK2 complexes. Furthermore, certain E2F species are actively degraded by the ubiquitin-proteasome pathway, and their synthesis is cell cycle regulated (36, 37).
Here we describe a mechanism that controls the abundance of transcriptionally active E2F-4 (and presumably, E2F-5). This new mechanism regulates the subcellular localization of E2F-4 and E2F-5. When overproduced, these two transcription factor were located mainly in the cytoplasm in marked contrast to E2F-1, -2, and -3, which are nuclear. It is likely that E2F-4 (and perhaps E2F-5) are inherently cytoplasmic and lack functional NLSs, since addition to E2F-4 of such a motif, derived from the SV40 T antigen, resulted in efficient nuclear import. Moreover, wt-E2F-4 and -5 were translocated to the nucleus upon interaction with p107 or p130 pocket proteins or the DP-2 heterodimeric partner. These overexpression studies provide evidence for a model in which p107/p130 or DP-2 play an important role in nuclear localization of E2F-4 and/or -5, presumably by contributing NLSs in trans. Our data demonstrate that induced E2F-4 nuclear translocation requires interaction(s) with proteins such as p107/p130 or DP-2. The relative influence of DP-2, under physiologic conditions, upon E2F4/5 nuclear translocation is currently unclear, since it has not been possible for us to detect endogenous DP-2 using the antibodies currently available. Nevertheless, nuclear concentration of E2F-4 did correlate with its transcriptional activity while nuclear exclusion had a negative effect, as if cytological trafficking of this protein could be an important step in the mechanism which allows cells to experience the full transcription regulation effect(s) of this protein. In keeping with this notion, E2F-4.NLS appeared to be a more potent transactivator than wt-E2F-4, and its activity was not increased by DP-2.
Moreover, the fact that E2F-4.NLS was transcriptionally active and not further activated by ectopic DP protein means that the cells under study here were not likely defective in DP protein for functional heterodimerization. Hence, the activating effect of DP-2 on E2F-4 transactivating function depended upon its specific E2F-4/5 nuclear import function.
Because levels of E2F-4 present after transfection were significantly higher than endogenous levels, localization of ectopically expressed protein might not accurately reflect that of the endogenous protein. To address this question, we studied the location of endogenous E2F-4 in U2OS and NIH 3T3 cells. Endogenous E2F-4 could be detected in both the nucleus and cytoplasm and relative levels in these two compartments changed significantly as cells progressed through the cell cycle. When cells were arrested in G0 or early G1 the vast majority of E2F-4 was nuclear. As cells progressed toward S phase, more E2F-4 was detected in the cytoplasm. One might speculate, then, that maintenance of G0 depends on concentration of E2F-4 in the nucleus, while progression through the cell cycle may require its exclusion. Mechanisms that might explain the presence of E2F-4 in the cytoplasm include active nuclear export or even cytoplasmic retention by association with an anchoring protein. Whether nuclear E2F-4 acts as a transcriptional activator or as part of a repressor complex in different stages of the cell cycle remains to be determined. In this context, our observations that cotransfected p107 or p130 can contribute to E2F-4 and -5 nuclear localization supports the notion that they play an important role in E2F-4 or -5 mediated transcriptional repression.
The E2F family can be subgrouped into two classes (E2F-1, -2, and -3; and E2F-4 and -5), based on structural homology and on their interaction with pocket proteins. Results presented in this paper, together with previously published data (4, 5, 14, 31), support this stratification and indicate that the cell cycle activity of these transcription factors may be achieved by different mechanisms. For example, E2F-1 levels rise and fall during the cell cycle although, once produced, this protein is efficiently concentrated in the nucleus. Conversely E2F-4 is continuously present, is inherently cytoplasmic, and its concentration in the nucleus is cell cycle-dependent. Unlike E2F-1 nuclear entry, E2F-4 nuclear concentration appears to be regulated, in part, by association with certain partner proteins. Moreover, since E2F-4 is constitutively present during the cycle, one wonders whether it could modulate downstream target gene expression more readily than E2F-1 in response to relevant exogenous stimuli delivered at times when E2F-1 levels are relatively low.
Acknowledgments
We are grateful to all our divisional colleagues for numerous helpful discussions. This work was supported by funds provided to D.M.L. by the National Cancer Institute and the Dana–Farber/Sandoz Drug Discovery Program. G.J.L. is a recipient of a Neil–Hamilton Fairley Postdoctoral Fellowship from the National Health and Medical Research Council of Australia. S.G. is supported by a European Molecular Biology Organization Fellowship and D.G. was a recipient of a postdoctoral fellowship the the Damon Runyon–Walter Winchell Cancer Research Fund (DRG-1202) and currently is a recipient of a Senior Research Fellowship from the Leukemia Society of America.
ABBREVIATIONS
- CDK
cyclin-dependent kinase
- NLS
nuclear localization sequence
- SV40
simian virus 40
- NES
nuclear export sequence
- FITC
fluorescein isothiocyanate
- wt
wild type
References
- 1.Adams P D, Kaelin W G. Curr Top Microbiol Immunol. 1996;208:79–93. doi: 10.1007/978-3-642-79910-5_4. [DOI] [PubMed] [Google Scholar]
- 2.Cobrinik D. Curr Top Microbiol Immunol. 1996;208:31–61. doi: 10.1007/978-3-642-79910-5_2. [DOI] [PubMed] [Google Scholar]
- 3.Slansky J E, Farnham P J. Curr Top Microbiol Immunol. 1996;208:1–30. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- 4.Ginsberg D, Vairo G, Chittenden T, Xiao Z-X, Xu G, Wydner K L, DeCaprio J A, Lawrence J B, Livingston D M. Genes Dev. 1994;8:2665–2679. doi: 10.1101/gad.8.22.2665. [DOI] [PubMed] [Google Scholar]
- 5.Beijersbergen R L, Kerkhoven R M, Zhu L, Carlee L, Voorhoeve P M, Bernards R. Genes Dev. 1994;8:2680–2690. doi: 10.1101/gad.8.22.2680. [DOI] [PubMed] [Google Scholar]
- 6.Singh P, Wong S H, Hong W. EMBO J. 1994;13:3329–3338. doi: 10.1002/j.1460-2075.1994.tb06635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu G, Livingston D M, Krek W. Proc Natl Acad Sci USA. 1995;92:1357–1361. doi: 10.1073/pnas.92.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson D G, Schwarz J K, Cress W D, Nevins J R. Nature (London) 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 9.Qin X Q, Livingston D M, Kaelin W G, Adams P D. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shan B, Lee W-H. Mol Cell Biol. 1994;14:8166–8173. doi: 10.1128/mcb.14.12.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Field S J, Tsai F-Y, Kuo F, Zubiaga A M, Kaelin W G, Livingston D M, Orkin S H, Greenberg M E. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 12.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson N J. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 13.Hijmans E M, Voorhoeve P M, Beijersbergen R L, van’t Veer L J, Bernards R. Mol Cell Biol. 1995;15:3082–3089. doi: 10.1128/mcb.15.6.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sardet C, Vidal M, Cobrinik D, Geng Y, Onufryk C, Chen A, Weinberg R A. Proc Natl Acad Sci USA. 1995;92:2403–2407. doi: 10.1073/pnas.92.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moberg K, Starz M A, Lees J A. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dynlacht B D, Flores O, Lees J A, Harlow E. Genes Dev. 1994;8:1772–1786. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- 17.Xu M, Sheppard K-A, Peng C-Y, Yee A S, Piwnica-Worms H. Mol Cell Biol. 1994;14:8420–8431. doi: 10.1128/mcb.14.12.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krek W, Xu G, Livingston D M. Cell. 1995;83:1149–1158. doi: 10.1016/0092-8674(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 19.Krek W, Ewen M E, Shirodkar S, Arany Z, Kaelin W G, Livingston D M. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 20.Kaelin W G, Krek W, Sellers W R, DeCaprio J A, Ajchenbaum F, Fuchs C S, Chittenden T, Li Y, Farnham P J, Blanar M A, Livingston D M, Flemington E K. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 21.Slansky J E, Li Y, Kaelin W G, Farnham P J. Mol Cell Biol. 1993;13:1610–1618. doi: 10.1128/mcb.13.3.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsiao K-M, McMahon S L, Farnham P J. Genes Dev. 1994;8:1526–1537. doi: 10.1101/gad.8.13.1526. [DOI] [PubMed] [Google Scholar]
- 23.Johnson D G, Ohtani K, Nevins J R. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 24.Neuman E, Flemington E K, Sellers W R, Kaelin W G. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magae J, Wu C-L, Illenye S, Harlow E, Heintz N H. J Cell Sci. 1996;109:1717–1726. doi: 10.1242/jcs.109.7.1717. [DOI] [PubMed] [Google Scholar]
- 26.Gerace L. Cell. 1995;82:341–344. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- 27.Qin X-Q, Chittenden T, Livingston D M, Kaelin W G. Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 28.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D M, Dyson N, Harlow E. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]
- 29.Vairo G, Livingston D M, Ginsberg D. Genes Dev. 1995;9:869–881. doi: 10.1101/gad.9.7.869. [DOI] [PubMed] [Google Scholar]
- 30.Krek W, Livingston D M, Shirodkar S. Science. 1993;262:1557–1560. doi: 10.1126/science.8248803. [DOI] [PubMed] [Google Scholar]
- 31.Wu C-L, Zukerberg L R, Ngwu C, Harlow E, Lees J A. Mol Cell Biol. 1995;15:2536–2546. doi: 10.1128/mcb.15.5.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current Protocols in Molecular Biology. New York: Greene/Wiley-Interscience; 1990. , Vols. 1 and 2. [Google Scholar]
- 33.Lee W-H, Shew J-Y, Hong F D, Sery T W, Donoso L A, Young L J, Bookstein R, Lee E Y. Nature (London) 1987;329:642–645. doi: 10.1038/329642a0. [DOI] [PubMed] [Google Scholar]
- 34.Baldi A, De Luca A, Claudio P P, Baldi F, Giordano G G, Tommasino M, Paggi M G, Giordano A. J Cell Biochem. 1995;59:402–408. doi: 10.1002/jcb.240590311. [DOI] [PubMed] [Google Scholar]
- 35.Helin K, Wu C L, Fattaey A R, Lees J A, Dynlacht B D, Ngwu C, Harlow E. Genes Dev. 1993;7:1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- 36.Hateboer G, Kerkhoven R M, Shvarts A, Bernards R, Beijersbergen R L. Genes Dev. 1996;10:2960–2970. doi: 10.1101/gad.10.23.2960. [DOI] [PubMed] [Google Scholar]
- 37.Hofmann F, Martelli F, Livingston D M, Wang Z. Genes Dev. 1996;10:2949–2959. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]