Abstract
RNA interference (RNAi), a post-transcriptional gene silencing mechanism originally described in C. elegans, involves sequence-specific mRNA degradation mediated by double-stranded RNAs (dsRNAs). Passive dsRNA uptake has been uniquely observed in C. elegans due to the expression of systemic RNA interference defective-1 (SID-1). Here we investigated the ability of ectopic SID-1 expression to enable passive cellular uptake of short interfering RNA (siRNA) or double stranded RNA (dsRNA) in pluripotent mouse embryonic stem cells (mESCs). When SID-1-GFP and the Firefly luciferase reporter gene (luc Fir) were co-expressed in mESCs, lucFir activity could be suppressed by simple incubation with dsRNAs/siRNAs that were designed to specifically target lucFir. By contrast, suppression was not observed in mESCs expressing lucFir and GFP alone or when either GFP- or SID-1-GFP-expressing cells were incubated with control dsRNAs/siRNAs (non-silencing or Renilla luciferase-specific). These results may lead to high-throughput experimental strategies for studying ESC differentiation and novel approaches to genetically inhibit or eliminate the tumorigenicity of ESCs.
Keywords: SID-1, RNA interference, gene transfer, ectopic expression, embryonic stem cells
1. INTRODUCTION
RNA interference (RNAi), a post-transcriptional gene silencing mechanism originally described in C. elegans and plants, involves sequence-specific degradation of homologous messenger RNA (mRNA) mediated by double-stranded RNA (dsRNA) molecules [1]. Upon entering cells, dsRNAs are cleaved by the conserved Dicer family of RNase III enzymes to produce short interfering RNAs (siRNAs) that are typically 21–23 nucleotides in length. SiRNAs are then incorporated into the RNA-induced silencing complex (RISC) where the unzipped, antisense strand of the siRNA binds a complimentary sequence and subsequently cleaves the target mRNA resulting in gene-specific silencing [2; 3]. Although dsRNAs typically enter cells by natural viral infections (which in turn can lead to interferon responses in certain mammalian cell types) or other exogenous means, systemic dsRNA uptake has been uniquely observed in C. elegans [4] due to expression of systemic RNA interference defective-1 (SID-1), a 776-amino acid transmembrane channel protein [5; 6]. Interestingly, ectopic expression of the C. elegans protein SID-1 in Drosophila cells enables passive cellular uptake of soaking dsRNA or siRNA [5].
Although conventional methods such as injection, transient transfection (e.g. cationic lipid-based transfection reagent), electroporation and recombinant virus-mediated delivery of short hairpin RNA (shRNAs) are effective for inducing RNAi in mammalian cells, these approaches often require the generation and testing of multiple constructs, which can be labor-intensive. Here we investigated the ability of ectopic SID-1 expression to confer systemic dsRNA or siRNA uptake in pluripotent mouse embryonic stem cells (mESCs). Our experiments show that the simple incubation of SID-1-expressing mESCs in media containing dsRNAs or siRNAs is sufficient to cause gene-specific RNAi. These results may facilitate high-throughput silencing of multiple genes in future experiments as well as potential therapeutic applications.
2. MATERIALS AND METHODS
Cell culture
Human embryonic kidney 293T (HEK) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco BRL; Carlsbad, CA) supplemented with 10% fetal bovine serum (Gibco BRL), 2 mM L-glutamine, 0.1 mM nonessential amino acids and 500 μg/mL geneticin (Invitrogen; Carlsbad, CA).
For mESCs, the mouse ES-D3 line (ATCC; Manassas, VA) was used. Pluripotency was maintained by growing mESCs on an irradiated MEF feeder layer with 1000 U/ml leukemia inhibitory factor (LIF) (Chemicon; Temecula, CA) as we recently described [7].
Ectopic SID-1 expression
For transgene delivery, we employed the same lentiviral vector [8] that we previously used for modifying human ESCs [7; 9; 10]. Briefly, the SID-1 transgene from pPACPl-SID-1::FLAG (a kind gift from Dr. Craig P. Hunter, Harvard University) was subcloned into pLV-CAG-GFP to generate pLV-CAG-SID-1-GFP, which directs the expression of the fusion protein SID-1-GFP under the control of CAG, an internal composite constitutive promoter containing the CMV enhancer and the β-actin promoter. Recombinant lentiviruses (LV) were generated using the 3-plasmid system [11] by co-transfecting HEK cells with pLV-CAG-GFP or pLV-CAG-SID-1-GFP, pMD.G and pCMV R8.91. For each 100-mm dish of HEK cells plated at 80–85% confluency, 5, 2.5, and 10 μg of pLV, pMD.G and pCMV R8.91 DNA were used for transfection, respectively. LVs were harvested by collecting the culture medium at 24 and 48 hours post-transfection. LVs generated using this protocol typically had titers in the range of 1×106 to 6×106 TU/mL, and were stored at -80°C before use. For transduction, purified LVs were added to cells at a final concentration of 10,000 TU ml−1 with 6 μg/mL polybrene. The multiplicity of infection (MOI) was ~5. Transduction was allowed to proceed for 12–16 hours.
dsRNA and siRNA
100bp dsRNAs that target against Firefly luciferase (lucFir) and Renilla luciferase (lucRen) were made by in vitro transcription using the Megascript RNAi kit (Ambion; Austin, TX). The templates for dsRNA synthesis were gel-purified PCR products from pGL3-lucFir or pRL-SV40-lucRen (Promega; Madison, WI), respectively. Primers for lucFir have been described [5], and those for lucRen are provided in Table 1. All primers were purchased from Invitrogen.
Table 1.
Renilla luciferase (lucRen) templates:
| 100mer | |
| Forward: | |
| lucRen 100 FORWARD T7: | TAATACGACTCACTATAGGGCGGCCTCT
TCTTATTTATGGC |
| lucRen 100 REVERSE: | GGGCTTGCCTGATTTGCCCATAC |
| Reverse: | |
| lucRen 100 REVERSE T7: | TAATACGACTCACTATAGGGCTTGCCTG
ATTTGCCCATAC |
| lucRen 100 FORWARD: | GGGCGGCCTCTTCTTATTTATGGC |
|
| |
| 500mer | |
| Forward: | |
| lucRen 500 FORWARD T7: | TAATACGACTCACTATAGGGCGGCC
TCTTCTTATTTATGGC |
| lucRen 500 REVERSE: | GGGCGCCATGATAATGTTGGAC |
| Reverse: | |
| lucRen 500 FORWARD: | TAATACGACTCACTATAGGGCGCCATGA
TAATGTTGGAC |
| lucRen 500 REVERSE T7: | GGGCGGCCTCTTCTTATTTATGGC |
For siRNA (Qiagen Inc.; Valencia, CA), the sense and antisense sequences were CUUACGCUGAGUACUUCGATT and UCGAAGUACUCAGCGUAAGTT for lucFir-siRNA, and UUCUCCGAACGUGUCACGUTT and ACGUGACACGUUCGGAGAATT for control (non-silencing) siRNA. All dsRNAs and siRNAs sequences were BLAST searched to ensure no sequence similarity to any known mammalian genes.
Transfection
Transfection was performed using Lipofectamine 2000 (Gibco BRL) according to the manufacturer’s protocol. Transfected cells were incubated at 37°C for 24 hours for protein expression before lucFir activity was assessed.
Flow Cytometry
Flow cytometry analysis was performed on a FACSCalibur instrument (BD Pharmingen; San Diego, CA) and analyzed using FlowJo software (Tree Star, San Carlos, CA). For assessing apoptosis, Annexin V-Phycoerythrin (Annexin V-PE) and 7-Amino-actinomycin (7-AAD) were employed (BD Pharmingen). Briefly, cells were washed with cold PBS, resuspended in Annexin V-binding buffer (BD Pharmingen) at a concentration of 1 × 106 cells/mL, followed by incubating with 5μL of Anexin V-PE and 7-AAD for 15 minutes at room temperature in the dark prior to flow cytometry analysis. For fluorescence-activated cell sorting (FACS), FACS Vantage instrument (BD Pharmingen) was used.
Western blot
Cells were harvested in a lysis buffer containing 20 mM HEPES (pH 7.4), 2 mM EGTA, 50 mM β-glycerophosphate, 1 mM Na3VO4, 5 mM NaF, 1% Triton X-100, 10% glycerol, 2 mM dithiothreitol, 2 mM phenylmethylsulfonyl fluoride, 2 μg of leupeptin/ml, and 2 μg of aprotinin/ml, vortexed, and centrifuged at 25,000 × g for 15 min. Whole-cell protein aliquots (30 μg) were size-fractionated by SDS-PAGE and transferred to nitrocellulose membranes (Schleicher & Schuell; Keene, NH). Protein signals were detected using Western Lightning Chemiluminescence Reagent Plus (Perkin Elmer Life Sciences; Boston, MA).
Luciferase assay
4 × 104 cells per well were plated on a solid black 96-well plate (Corning). Luciferase activity was measured using the Steady-Glo luciferase assay system (Promega; Madison, WI) on a microplate reader.
Statistical analysis
All data reported are means ± S.E.M. Statistical analysis was performed using one-way ANOVA and Tukey’s HSD post hoc test with p<0.05 being considered as statistically significant.
3. RESULTS
Generation of a human cell line that stably expresses SID-1-GFP
HEK 293T cells were transduced with LV-CAG-SID-1-GFP (Figure 1A) and upon initial transduction, yielded ~ 30% GFP-positive cells (vs. 0% for non-transduced control cells; Figure 1B). Cells expressing the highest GFP signal (8.23% of the original population) were FACS-sorted, followed by culturing for recovery. Recovered, stably LV-CAG-SID-1-GFP-transduced HEK 293T cells displayed normal morphology (Figure 1C), remained green for >4 months (longer periods were not monitored) while proliferating indistinguishably from non-transduced control cells (data not shown). Furthermore, GFP epifluorescence was limited to the membrane surface, consistent with the notion that SID-1 is a transmembrane protein [5]. Western blot analysis of stably LV-CAG-SID-1-GFP-transduced HEK 293T cells with a monoclonal antibody against GFP generated a detectable band at ~116 kDa consistent with the expected size of SID-1-GFP (Figure 1D). Transient pLV-CAG-SID-1-GFP transfection of HEK 293T cells yielded a similar result.
Figure 1.
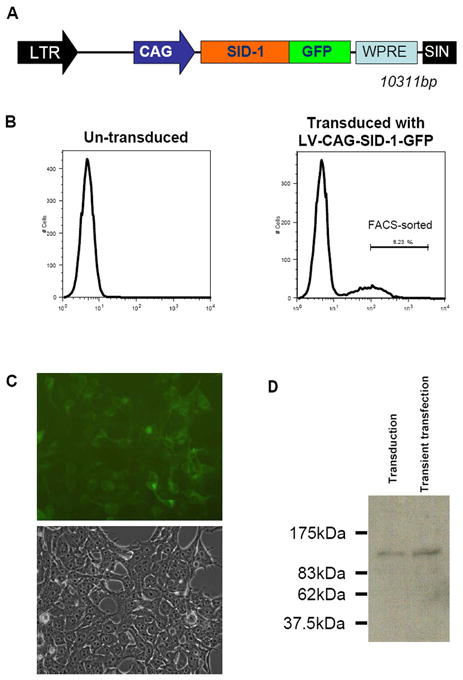
A) Schematic representation of LV-CAG-SID-1-GFP
B) FACS analysis of control HEK cells (left) or LV-CAG-SID-1-GFP transduced (right).
C) Representative images (top, green fluorescence; bottom, phase contrast) of LV-CAG-SID-1-GFP-transduced HEK cells after recovery from sorting (from B). Note that GFP epifluorescence was limited to the membrane surface.
D) Western blot analysis of CAG-SID-1-GFP transduced (lane 1) and transient transfected (lane 2) HEK 293 cells. Specific bands at ~116 kDa consistent with the expected size of SID-1-GFP were observed when probed with an anti-GFP antibody.
dsRNA- and siRNA-induced luciferase suppression was sequence-specific
We next verified the efficacy of our siRNA and dsRNA constructs to induce gene-specific RNAi by assessing their ability to suppress the transiently expressed reporter lucFir activity. Figure 2A shows that co-transfection of control HEK 293T cells with pGL3-lucFir (which encodes for the Firefly luciferase (lucFir)) and lucFir-siRNA, or pGL3-lucFir and 100bp lucFir-dsRNA, significantly suppressed lucFir activity 24 hours after transfection (relative to control experiments performed without lucFir-siRNA or lucFir-dsRNA; p< 0.05). By contrast, control non-silencing siRNA, and 100bp lucRen-dsRNA (targets Renilla luciferase (lucRen)) had no effect on lucFir activity (p>0.05), suggesting that the suppressive effects observed with lucFir-siRNA and 100bp lucFir-dsRNA were sequence- and gene-specific.
Figure 2.
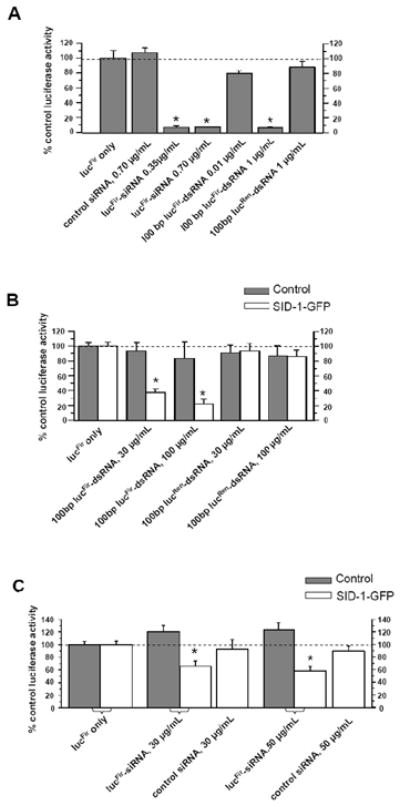
A) Bar graph summarizing the effects of transient transfection of various siRNA or dsRNA constructs in HEK cells on lucFir activity. lucFir expression levels were normalized to that of control cells (lucFir only). Transfection with lucFir-siRNA (0.35 and 0.7μg/ml) and 100bp lucFir-dsRNA significantly suppressed lucFir activity. By contrast, control non-silencing siRNA, and 100bp lucRen-dsRNA had no effect
B) Soaking of SID-1-GFP-expressing HEK cells in 100bp (30 and 100μg/ml) lucFir-dsRNA significantly suppressed lucFir activity. By contrast, control lucRen-dsRNA at the same concentrations did not exert any lucFir suppressive effect.
C) Soaking of SID-1-GFP-expressing HEK cells in lucFir-siRNA (30 and 50μg/ml) significantly suppressed lucFir activity. By contrast, control non-silencing siRNA did not exert any lucFir suppressive effect.
Data presented were averages from 3–5 experiments. *, p<0.05.
LV-CAG-SID-1-GFP-transduced cells uniquely enabled passive entry of soaking dsRNAs/siRNAs and subsequent gene-specific RNAi
To investigate whether SID-1 overexpression enables passive dsRNA/siRNA entry into mammalian cells, control (non-transduced) and LV-SID-1-GFP-transduced HEK 293T cells were transfected with pGL3-lucFir, followed by simple soaking with dsRNAs (Figure 2B) or siRNAs (Figure 2C) for 48 hrs before measuring lucFir activity. While control cells were insensitive to either lucFir-dsRNA or lucFir-siRNA soaking (solid bars) at all the concentrations tested, LV-SID-1-GFP-transduced HEK 293T cells exhibited significantly suppressed lucFir activity (open bars) under the same conditions: 30 and 100μg/mL 100bp lucFir-dsRNA dose-dependently decreased lucFir activities to 37.4 ± 5.2% and 22.2 ± 5.9%, respectively (p<0.05; Figure 2B).
Similarly, soaking of SID-1-GFP-expressing cells in lucFir-siRNAs also led to reduced lucFir activity (66.3 ± 8.0% and 58.4 ± 7.3% for 30μg/mL and 50μg/mL, respectively; p<0.05; Figure 2C) although the extents of suppression were smaller than those induced by longer lucFir-dsRNAs at comparable concentrations. Of note, lucFir activity suppression was not observed in the absence of lucFir-dsRNAs or –siRNAs, or when either control or LV-SID-1-GFP-transduced cells were soaked in control, lucRen-dsRNA or non-silencing siRNA, suggesting that the SID-1-mediated RNAi effects observed were sequence and gene specific.
Functional consequences of SID-1 overexpression in mouse embryonic stem cells
To explore the use of SID-1 to confer on pluripotent mESCs the ability to passively uptake dsRNA/siRNA so that gene-specific RNAi can be induced by simple soaking, we co-expressed lucFir and either GFP or SID-1-GFP in mESCs (Figures 3A & B). Similar to our HEK 293T cell experiments, only GFP-positive mESCs were FACS-sorted for experiments (Figures 3C and D). As anticipated, GFP signals were found throughout the cytoplasm of GFP-expressing mESCs; by contrast, epifluorescence was limited to the membrane surface for mESCs expressing the fusion protein SID-1-GFP.
Figure 3.
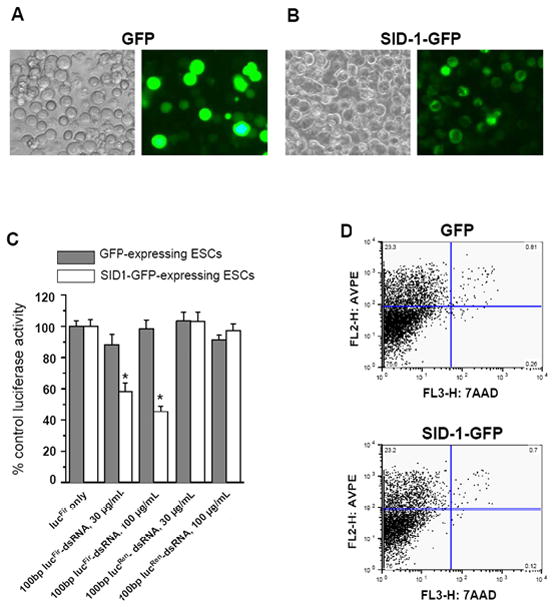
Representative images of A) GFP- and B) SID-1-GFP-expressing mESCs (left, phase-contrast; right, fluorescence). C) Soaking of mESCs co-expressing SID-1-GFP+ lucFir (but not GFP+ lucFir) in 100 bp (30 and 100μg/ml) lucFir-specific dsRNA led to suppression of lucFir activity. No suppression was observed for SID-1-GFP+ lucFir and GFP+ lucFir cells soaked with control lucRen-dsRNAs, indicating that RNAi was sequence-specific
Data presented were averages from 7–14 experiments. *, p<0.05.
D) FACS analysis of GFP- or SID-1-GFP- expressing mESCs indicates that ectopic expression of SID-1 did not alter the percentage of early apoptotic (Annexin V-PE positive, 7-AAD negative) and dead cells (Annexin V-PE positive, 7-AAD positive).
Figure 3C shows that soaking SID-1-GFP- but not GFP-expressing mESCs in lucFir-dsRNA significantly suppressed lucFir activity: 30μg/mL and 100μg/mL 100bp lucFir-dsRNAs suppressed the lucFir activity of SID-1-GFP-expressing mESCs to 58.3 ± 5.4%, 45.4 ± 3.5%, respectively (p < 0.05). By contrast, no suppression was observed for soaking SID-1-GFP-expressing mESCs 1) without any lucFir-dsRNA or 2) with 100bp lucRen-dsRNA. In addition, control GFP-expressing cells soaked in lucFir-dsRNA also showed no change in lucFir reporter activity, suggesting that the RNAi-suppressed lucFir activity observed was SID-1-mediated and sequence-specific. Examination for apoptosis and cell viability by Annexin V-PE and 7-AAD staining revealed no significant differences between GFP- and SID-1-GFP-expressing mESCs (Figure 3D).
4. DISCUSSION
SID-1, originally identified in C. elegans, belongs to a novel, uncharacterized gene family that is also present in mammals [5; 6]. Although the structure-function properties are as yet unknown, a recent report demonstrates that overexpression of a human SID-1 homolog in pancreatic ductal adenocarcinoma cells enhances the passive uptake of siRNAs [12]. The expression levels of the mammalian homologues have not been determined, but our present results indicate that the endogenous activity is insignificant in HEK 293T and mESCs because their incubation with dsRNAs or siRNAs alone did not suppress the targeted gene unless SID-1 is also expressed. The lack of detectable endogenous activity might be due to the presence of non-functional SID-1 homologues. Alternatively, it is possible that functional SID-1 homologues are simply not expressed in the cell types investigated. Our present experiments do not allow us to distinguish between these possibilities. Nonetheless, we have demonstrated that ectopic SID-1 expression in HEK and mES cells enables passive uptake of dsRNAs and siRNAs, which in turn can suppress sequence-specific targets without detrimental effects, at least over the time course of our experiments where parameters such as proliferation rate, morphology and apoptosis, were assessed.
The accumulation of long dsRNAs (>30bp), commonly present in numerous viral life cycles, in certain mammalian cells can lead to an immune response and the induction interferon. This subsequently blocks protein translation in a non-specific manner via protein kinase-dependent inactivation of elongation factors, and ultimately leads to apoptosis [4; 13–15]. Indeed, the non-specificity of such interferon-meditated effects, which can mask the specific RNAi of interest, have been a major limitation for many RNAi experiments in mammalian cells [16–20]. In contrast, long dsRNA duplexes can efficiently induce specific RNAi in organisms or systems that lack interferon responses [1; 4; 21–33]. For instance, long dsRNA (typically ~ 500–1500 bp) can readily induce gene-specific silencing in oocytes [28], preimplantation embryos [27; 32], embryonal teratocarcinoma cells [25], and ESCs [26; 33]. Taken collectively, our observations of gene-specific RNAi with SID-1-expressing mESCs are consistent with the refractoriness of interferons in undifferentiated mESCs to induction [26; 33].
In addition to their promising therapeutic potential, ESCs are also useful models for studying pluripotency, self-renewal and differentiation. Indeed, a better understanding of these important properties and processes of ESCs is critical for the development of ESC-based therapies. Loss-of-function studies have been proven to shed functional and mechanistic insights into various important developmental and differentiation processes. However, high-throughput genomic screening is largely limited by tedious experimental procedures (e.g. homologous recombination to knock-out a specific gene) and gene-specific RNAi may be an alternative for applicable cell types. Transfection or transduction (for transfection-resistant cells such as human ESCs [9; 10] and other terminally-differentiated cells such as neurons and cardiomyocytes) is commonly exploited to introduce dsRNAs/siRNAs/shRNAs into cells. However, unless delivery vehicles such as LVs and adeno-associated viruses are employed, the resultant RNAi activity is often transient and short-lived. Our present results suggest that SID-1-expressing ESCs may conveniently enable high-throughput silencing of multiple genes for studying ESC differentiation. The ability to suppress genes specifically may also lead to the development of novel strategies to inhibit or eliminate the oncogenic potential of ESC for therapeutic applications (e.g. suppression of such gene products as ion channels that modulate cell proliferation [7; 9; 10]).
Although conceptually attractive, a number of hurdles need to be overcome before SID-1 or its homologues can be broadly applied for high-throughput silencing in ESCs. For instance, we attempted but were unable to establish mouse and human ESC lines that stably expressed SID-1-GFP. Although mESCs could be readily transiently transfected with SID-1-GFP without apparent ill effects (when apoptosis and cell viability were assessed) during the time course of our experiments (i.e. 48–72 hrs), stable transduction of either human or mouse ESCs with LV-SID-1-GFP led to cell death typically 4–5 days after the GFP signals initially appeared. Given such short time windows, we were unable to reproducibly and statistically suppress endogenous gene expression such as that of β–integrin (as assayed by quantitative RT-PCR) via SID-1-mediated dsRNA uptake. The basis of this time-dependent cytotoxic effect of SID-1 on ESCs is not known. Perhaps, human and mouse ESCs would be more tolerant to ectopic expression of mammalian homologues of SID-1 (rather than from c. elegans). Alternatively, it is possible that constitutive expression of SID-1 per se is detrimental. If this is the case, inducible expression of SID-1 for a brief period for incubation with dsRNA or siRNAs can be a potential solution but such conditional gene expression systems for human ESCs have not yet been described.
Hunter and colleagues (2003) reported that longer the dsRNAs, the better their SID-1-mediated uptake [5]. Thus, we have also investigated the effect of 500bp lucFir-dsRNAs (Primer sequences for lucFir from [5]and lucRen shown in Table 1) in addition to the 100bp constructs reported here. Although some suppressive effect was also observed when SID-1-GFP expressing HEK 293T and mESC were incubated in media containing 30 μg/ml 500bp dsRNA targeting LucFir, a dose-dependent response similar to that of our 100bp lucFir-dsRNAs was not observed. Furthermore, higher concentrations led to non-specific suppression. The reason for this non-specific effect at higher concentrations is not known, but was observed when using dsRNAs targeting either lucFir or lucRen. A better understanding of the basic biology (e.g. structure-function properties) of SID-1 will be crucial for overcoming the above mentioned limitations. Nevertheless, our present results shed insights into the development of high-throughput experimental strategies for studying ESC differentiation.
Acknowledgments
We thank James Flook for expert technical assistance with FACS analysis.
This work was supported by grants from the NIH (R01 HL-52768 and R01 HL72857 to R.A.L. and F32 HL078330 to J.C.M), the Stem Cell Program of the University of California School of Medicine at Davis (to R.A.L.), and the Shriners Hospital for Children of North America (to C.W.C). S.Y.T. was supported by a post-doctoral fellowship award from the Croucher Foundation.
Abbreviations
- SID-1
systemic RNA interference defective-1
- RNAi
RNA interference
- dsRNA
double-stranded RNA
- siRNA
short interfering RNA
- HEK
human embryonic kidney
- ESCs
embryonic stem cells
- mESCs
mouse embryonic stem cells
- LV
lentiviral vector
- RISC
RNA-induced silencing complex
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Hannon GJ. RNA interference. Nature. 2002;418:244–51. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 3.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–9. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 4.Svoboda P. Long dsRNA and silent genes strike back:RNAi in mouse oocytes and early embryos. Cytogenet Genome Res. 2004;105:422–34. doi: 10.1159/000078215. [DOI] [PubMed] [Google Scholar]
- 5.Feinberg EH, Hunter CP. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301:1545–7. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- 6.Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–9. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- 7.Wang C, Xue T, Tsang SY, Wong J, Cheng L, Zhang J, Li G, Lau CP, Tse HF, Li RA. Electrophysiological properties of pluripotent human and mouse embryonic stem cells. Stem Cells. 2005 doi: 10.1634/stemcells.2004-0299. In Press. [DOI] [PubMed] [Google Scholar]
- 8.Trono D. Lentiviral vectors. Spring-Verlag Berlin Heidelberg; New York: 2002. [Google Scholar]
- 9.Xue T, Cho HC, Akar FG, Tsang SY, Jones SP, Marban E, Tomaselli GF, Li RA. Functional integration of electrically active cardiac derivatives from genetically engineered human embryonic stem cells with quiescent recipient ventricular cardiomyocytes: insights into the development of cell-based pacemakers. Circulation. 2005;111:11–20. doi: 10.1161/01.CIR.0000151313.18547.A2. [DOI] [PubMed] [Google Scholar]
- 10.Moore JC, van Laake LW, Braam SR, Xue T, Tsang SY, Ward D, Passier R, Tertoolen LL, Li RA, Mummery CL. Human embryonic stem cells: Genetic manipulation on the way to cardiac cell therapies. Reprod Toxicol. 2005 doi: 10.1016/j.reprotox.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 11.Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–80. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duxbury MS, Ashley SW, Whang EE. RNA interference: a mammalian SID-1 homologue enhances siRNA uptake and gene silencing efficacy in human cells. Biochem Biophys Res Commun. 2005;331:459–63. doi: 10.1016/j.bbrc.2005.03.199. [DOI] [PubMed] [Google Scholar]
- 13.Kaufman RJ. Double-stranded RNA-activated protein kinase mediates virus-induced apoptosis: a new role for an old actor. Proc Natl Acad Sci U S A. 1999;96:11693–5. doi: 10.1073/pnas.96.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Der SD, Yang YL, Weissmann C, Williams BR. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc Natl Acad Sci U S A. 1997;94:3279–83. doi: 10.1073/pnas.94.7.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sledz CA, Williams BR. RNA interference and double-stranded-RNA-activated pathways. Biochem Soc Trans. 2004;32:952–6. doi: 10.1042/BST0320952. [DOI] [PubMed] [Google Scholar]
- 16.Persengiev SP, Zhu X, Green MR. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) Rna. 2004;10:12–8. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834–9. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 18.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34:263–4. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 19.Moss EG, Taylor JM. Small-interfering RNAs in the radar of the interferon system. Nat Cell Biol. 2003;5:771–2. doi: 10.1038/ncb0903-771. [DOI] [PubMed] [Google Scholar]
- 20.Kim DH, Longo M, Han Y, Lundberg P, Cantin E, Rossi JJ. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat Biotechnol. 2004;22:321–5. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 21.Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–26. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 22.Misquitta L, Paterson BM. Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): a role for nautilus in embryonic somatic muscle formation. Proc Natl Acad Sci U S A. 1999;96:1451–6. doi: 10.1073/pnas.96.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caplen NJ, Fleenor J, Fire A, Morgan RA. dsRNA-mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene. 2000;252:95–105. doi: 10.1016/s0378-1119(00)00224-9. [DOI] [PubMed] [Google Scholar]
- 24.Ngo H, Tschudi C, Gull K, Ullu E. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci U S A. 1998;95:14687–92. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Billy E, Brondani V, Zhang H, Muller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci U S A. 2001;98:14428–33. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang S, Tutton S, Pierce E, Yoon K. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol Cell Biol. 2001;21:7807–16. doi: 10.1128/MCB.21.22.7807-7816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wianny F, Zernicka-Goetz M. Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol. 2000;2:70–5. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- 28.Svoboda P, Stein P, Hayashi H, Schultz RM. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development. 2000;127:4147–56. doi: 10.1242/dev.127.19.4147. [DOI] [PubMed] [Google Scholar]
- 29.Gan L, Anton KE, Masterson BA, Vincent VA, Ye S, Gonzalez-Zulueta M. Specific interference with gene expression and gene function mediated by long dsRNA in neural cells. J Neurosci Methods. 2002;121:151–7. doi: 10.1016/s0165-0270(02)00230-3. [DOI] [PubMed] [Google Scholar]
- 30.Yi CE, Bekker JM, Miller G, Hill KL, Crosbie RH. Specific and potent RNA interference in terminally differentiated myotubes. J Biol Chem. 2003;278:934–9. doi: 10.1074/jbc.M205946200. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Tekle E, Oubrahim H, Mieyal JJ, Stadtman ER, Chock PB. Stable and controllable RNA interference: Investigating the physiological function of glutathionylated actin. Proc Natl Acad Sci U S A. 2003;100:5103–6. doi: 10.1073/pnas.0931345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svoboda P, Stein P, Anger M, Bernstein E, Hannon GJ, Schultz RM. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev Biol. 2004;269:276–85. doi: 10.1016/j.ydbio.2004.01.028. [DOI] [PubMed] [Google Scholar]
- 33.Paddison PJ, Caudy AA, Hannon GJ. Stable suppression of gene expression by RNAi in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:1443–8. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]


