Abstract
In Drosophila, the chromosomal region 75C1–2 contains at least three genes, reaper (rpr), head involution defective (hid), and grim, that have important functions in the activation of programmed cell death. To better understand how cells are killed by these genes, we have utilized a well defined set of embryonic central nervous system midline cells that normally exhibit a specific pattern of glial cell death. In this study we show that both rpr and hid are expressed in dying midline cells and that the normal pattern of midline cell death requires the function of multiple genes in the 75C1–2 interval. We also utilized the P[UAS]/P[Gal4] system to target expression of rpr and hid to midline cells. Targeted expression of rpr or hid alone was not sufficient to induce ectopic midline cell death. However, expression of both rpr and hid together rapidly induced ectopic midline cell death that resulted in axon scaffold defects characteristic of mutants with abnormal midline cell development. Midline-targeted expression of the baculovirus p35 protein, a caspase inhibitor, blocked both normal and ectopic rpr- and hid-induced cell death. Taken together, our results suggest that rpr and hid are expressed together and cooperate to induce programmed cell death during development of the central nervous system midline.
Keywords: apoptosis
Programmed cell death is a tightly regulated process that serves to eliminate unnecessary and/or deleterious cell types in a variety of developmental, physiological, and pathological contexts. The most well studied form of programmed cell death is apoptosis, which is characterized by distinctive morphological and molecular changes that include cell shrinkage, membrane blebbing, chromatin condensation, and the generation of nucleosomal ladders (1). Whereas a wide range of distinct signaling mechanisms can be used to elicit apoptosis, the cell death machinery itself appears to be highly conserved (reviewed in refs. 2–4). In Drosophila, genetic studies have shown that the 75C1,2 region of the third chromosome is essential for apoptosis, as deficiency strains that lack this region result in the blockade of essentially all embryonic cell deaths (5). Three different genes that map in this region, reaper (rpr), head involution defective (hid), and grim, are involved in regulation of apoptosis (5–7). These genes are all expressed in dying cells and can induce death in 75C1,2 deletion mutant embryos. In addition, ectopic expression of these genes results in stage-specific lethality and induces excess cell death in embryos and the adult eye (5–9). rpr, hid, and grim all function upstream of one or more caspases, as their ability to induce cell death is blocked by caspase inhibitors, including the baculovirus anti-apoptosis protein, p35, and N-benzyloxycarbonyl-Val-Ala-Asp fluoromethyl ketone (6–10). RPR is a small, 65-amino acid, protein which has homology to the cytoplasmic death domains of the vertebrate FAS and TNFR (tumor necrosis factor receptor) proteins, both of which play key roles in induction of apoptosis (5, 11, 12). Death domains are also found in several other proteins which dimerize with FAS and/or TNFR (13–15). Alternately, HID and GRIM are both novel proteins that do not exhibit extensive homology to other known proteins. RPR, HID, and GRIM do all share a similar 14-amino acid stretch at their amino termini (6, 7). The presence of these multiple closely linked cell death genes raises the question of whether they may functionally interact. For example, do these genes act combinatorially to regulate the elimination of individual cells, or do they each regulate the killing of distinct cells or cell types?
One cellular system that provides a useful model for addressing this issue is the central nervous system (CNS) midline of the Drosophila embryo. CNS midline cells derive from two stripes of mesectoderm in the blastoderm, which come together at the ventral midline during gastrulation and go on to generate a distinctive set of 6–8 CNS midline nerve cell precursor (CMP) cells per segment (16, 17). These CMP cells later differentiate into approximately 25 neurons and glia (17, 18). The midline cells play crucial roles in several distinct developmental events, including organization of the axon scaffold (16, 17), differentiation of the ventral epidermis (19), and migration of phagocytic macrophages (20). Previous studies have shown that most of the developing midline glia die and are quickly phagocytosed by migrating macrophages, whereas none of the ventral unpaired median (VUM) neurons die during embryogenesis (20, 21). 75C1,2 deficiencies, such as Df(3L)H99, that eliminate rpr, hid, and grim (5, 7), result in a blockade of all these deaths and an accumulation of ectopic glia (20, 21). The relative contributions of rpr, hid, and grim in midline cell death have not yet been defined.
In this study we examine the specific functions of the rpr, hid, and grim genes in regulating midline cell death. We show that rpr and hid are expressed in midline cells that normally die and that hid, as well as rpr and grim, appears to be required for normal patterns of midline cell death. Ectopic midline expression of rpr or hid alone did not efficiently induce ectopic midline cell death during embryogenesis. In contrast, coexpression of both rpr and hid induced very rapid midline cell death, indicating synergistic actions between these two death regulators.
MATERIALS AND METHODS
Fly Strains and Genetic Crosses.
The P[52A-Gal4] strain was isolated in the Nambu laboratory by means of an enhancer trap screen of approximately 400 independent P[GawB] (22) insertions. P[52A-Gal4] has a 2nd chromosome lethal insertion that drives strong embryonic Gal4 expression in the midline glia as well as most of the VUM neurons from stage 11 onward (stages defined in ref. 23). The P[UAS-rpr] and P[UAS-hid] strains were generated by cloning rpr cDNA clone 40KA or 13B2, and hid cDNA clone 5A1B, into the EcoRI site of the pUAST vector (22), and microinjecting these DNA constructs along with π25.7wc helper DNA (24) into w1118 or yw67c23 embryos. Two independent P[UAS-rpr] insertions were obtained that mapped to the X chromosome (using 40KA) and the 3rd chromosome (using 13B2), while one P[UAS-hid] insertion was obtained that mapped to the 3rd chromosome. Additional P[UAS-rpr] and P[UAS-hid] strains were generated by P element mobilization using the Δ2–3 source of transposase (25). Four strains—y, w,P[UAS-hid], y, w,P[UAS-rpr], w;;P[UAS-rpr]/TM3, and w;;P[UAS-hid]—were used in these experiments. Two P[UAS-lacZ] strains, one on the 2nd and one on the 3rd chromosome, were kindly provided by Andrea Brand (Univ. of Cambridge, Cambridge, U.K.). The P[1.0slit-lacZ] strain contains an X-linked insertion which drives strong expression of lacZ in developing midline glia from stage 11 onward (26). Two P[UAS-p35] strains with insertions on the 2nd and 3rd chromosomes were kindly provided by Bruce Hay (California Institute of Technology, Pasadena).
To test for cooperative functions of rpr and hid, six strains were generated:
(i) w, P[UAS-hid];;P[UAS-hid], P[UAS-lacZ];
(ii) w, P[UAS-rpr];;P[UAS-rpr], P[UAS-lacZ]/TM3;
(iii) w, P[UAS-hid];;P[UAS-rpr], P[UAS-lacZ]/TM3;
(iv) w, P[UAS-rpr];;P[UAS-hid], P[UAS-lacZ];
(v) w, P[UAS-hid], P[UAS-rpr];;P[UAS-lacZ]; and
(vi) w, P[UAS-lacZ];;P[UAS-hid], P[UAS-rpr]/TM3.
Embryos were collected from crosses between the P[52A-Gal4] strain and the above six strains. All crosses were performed at room temperature or 25°C.
Immunocytochemistry and in Situ Hybridization.
Embryos were collected and processed for immunocytochemistry and in situ hybridization as described previously (20). The monoclonal antibody BP102 (obtained from the Developmental Studies Hybridoma Bank) was used to visualize axon pathways and a mouse monoclonal anti-β-galactosidase antibody (Promega) was used to detect lacZ expression. cDNA clones 13B2 (rpr) and 5A1B (hid) were used to generate single-stranded digoxigenin-labeled cRNA probes using the DIG RNA Labeling Kit (Boehringer Mannheim).
Cell Counting and Statistical Analysis.
Analysis of P[1.0slit-lacZ]-expressing midline cells in stage 16 wild-type embryos indicated no significant difference among segments T1 to A8. lacZ-expressing cells in segments T2, A1, and A5 were counted for individual wild-type and mutant embryos, and the mean of lacZ-expressing cells per segment was calculated for each embryo. Twelve embryos were counted for each genotype. Cell counting data were then analyzed using the Statistical Analysis System software (version 6.11; SAS Institute, Cary, NC).
For analysis of ectopic rpr and hid killing of midline cells, the number of lacZ-expressing cells was monitored in embryos bearing P[52A-Gal4], P[UAS-lacZ], P[UAS-rpr], and/or P[UAS-hid]. For analysis of midline glia, the number of lacZ-expressing glia was determined in segments T2 and A5, while for analysis of the VUM neurons, cells in segments T1 to A8 were monitored. Because of the high number and close proximity of the labeled VUM neurons, calculations on the effects of ectopic rpr and hid expression were assessed by determining the number of segments in each embryo that had fewer than three lacZ-expressing VUM neurons. No differences were observed among segments T1 to A8 in the elimination of midline glia or neurons (n = 10 embryos for each cross).
RESULTS
Both rpr and hid Are Expressed in Dying Midline Cells.
It was previously shown (20, 21) that extensive cell death occurs among developing midline glia and that these deaths are blocked by 75C1,2 deficiencies that eliminate rpr, hid, and grim. These three genes each exhibit dynamic and complex patterns of expression in both dying and nondying cells throughout the embryo (5–7). In this study we specifically analyzed the pattern of rpr and hid expression in CNS midline cells. This was accomplished by in situ hybridization of rpr and hid cRNA probes, derived from rpr and hid cDNA clones, to wild-type embryos.
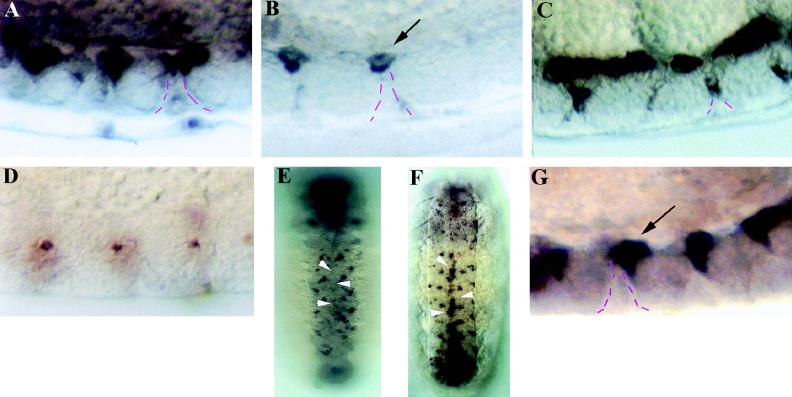
The hid gene exhibited expression in many midline cells from stages 12–14 (Fig. 1A). Interestingly, hid expression was largely restricted to midline cells that reside adjacent to a series of basement membrane midline pores through which phagocytic macrophages migrate and engulf dying midline cells. hid-expressing midline cells decrease in number after stage 14, and in stage 16 embryos few hid-expressing midline cells are detected (Fig. 1B). This reduction in hid-expressing midline cells correlates well with the timing of normal midline cell death. In addition, use of P[52A-Gal4] and P[UAS-p35] strains (see Materials and Methods) to drive ectopic midline expression of p35, an anti-apoptotic protein (27, 28), prevented the death of midline cells, resulting in ectopic hid-expressing cells along the dorsal surface of the nerve cord and alongside the midline pores (Fig. 1C). This further suggests that hid is normally expressed in dying midline cells.
Figure 1.
Both hid and rpr are expressed in dying midline cells. hid is expressed in several dorsally placed midline cells in stage 13 wild-type embryos (A). Most of this expression was eliminated by stage 16 (B), and residual hid expression above midline pores was inside macrophages (arrow). The broken lines indicate the interface between midline cells and the midline pore, a basement membrane structure separating segmentally reiterated clusters of midline cells. In stage 16 P[52A-Gal4]/P[UAS-p35] embryos there were numerous ectopic hid-expressing midline cells localized along the dorsal surface of the nerve cord and alongside of the midline pore (C). rpr expression was detected in only 1–2 midline cells per segment at stage 12 wild-type embryos (D) and was not detected in midline at later stages (arrowheads, E). In P[52A-Gal4]/P[UAS-p35] embryos (F), there were many ectopic rpr-expressing cells in the midline (arrowheads). In Df(3L)X25 mutant embryos (G) there were also greater than normal numbers of rpr-expressing midline cells (arrow). A–D and G are sagittal views with anterior to left. E and F are ventral views with anterior up. (A–D and G, ×200; E and F, ×100.)
The midline expression of rpr was more restricted and difficult to detect. One or two rpr-expressing midline cells per segment were detected in stage 12 embryos, and we were unable to detect rpr expression in midline cells at later stages (Fig. 1 D and E). This might suggest a more transient function for rpr than hid in dying midline cells; however, no data are currently available regarding the translation pattern of the corresponding proteins. Strong midline rpr expression was detected in P[52A-Gal4]/P[UAS-p35] embryos (Fig. 1F), providing additional evidence that rpr, like hid, is normally expressed in dying midline cells. These ectopic rpr-expressing cells were located at the same positions as the ectopic hid-expressing cells, suggesting that at least some of these rescued cells express both genes. We also examined rpr expression in Df(3L)X25 embryos, where hid and grim but not rpr are removed (7). These embryos exhibited blockade of many, though not all, midline cell deaths (see below). In Df(3L)X25 mutant embryos there was an accumulation of ectopic rpr-expressing midline cells (Fig. 1 D and G). However, there were still fewer rpr-expressing midline cells than in P[52A-Gal4]/P[UAS-p35] embryos. This result indicates that rpr alone was not sufficient for the normal pattern of midline cell death and suggests that hid and/or grim functions are also required.
Normal Patterns of Midline Glial Death Require hid.
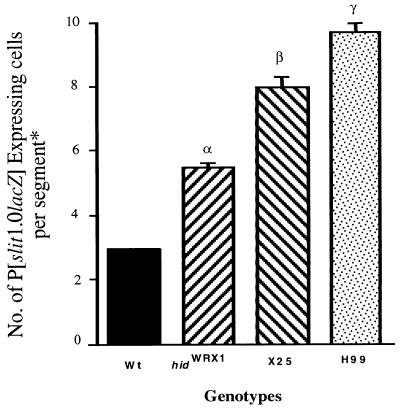
To further address whether rpr, hid, and grim all function in midline cell death, we utilized a P[1.0slit-lacZ] marker chromosome (26) to examine development of the midline glia in a series of cell death mutations, including the following: Df(3L)H99, which eliminates rpr, hid, and grim (5–7); hidWR+X1 and hidWR+E6 null mutants (6); and Df(3L)X25 (see above). In stage 15 Df(3L)H99 mutant embryos there was a 3-fold increase in the normal number of lacZ-expressing midline glia measured in segments T2 and A5 (Fig. 2 and refs. 20 and 21). In both hidWR+X1 and hidWR+E6 mutant embryos there was a 2-fold increase in the number of lacZ-expressing cells, indicating that hid is essential for many, but not all, midline cell deaths. Df(3L)X25 mutant embryos exhibited a slight increase in the number of lacZ-expressing midline cells compared with hid mutants, though there were still fewer lacZ-expressing cells than in Df(3L)H99 mutants (Fig. 2). This suggests that rpr and grim are also essential for some midline cell deaths. Thus, the normal pattern of midline cell deaths may require the functions of all three of these death genes.
Figure 2.
Numbers of P[1.0slit-lacZ]-expressing cells in stage 16 wild-type, hidWR+X1, Df(3L)X25, and Df(3L)H99 embryos. ANOVA testing placed the phenotypes of three mutants at different statistical rankings (α, β, and γ) using the wild-type value as the control group.
* See Materials and Methods for cell counting and statistical analysis.
rpr and hid Cooperate to Kill Midline Cells.
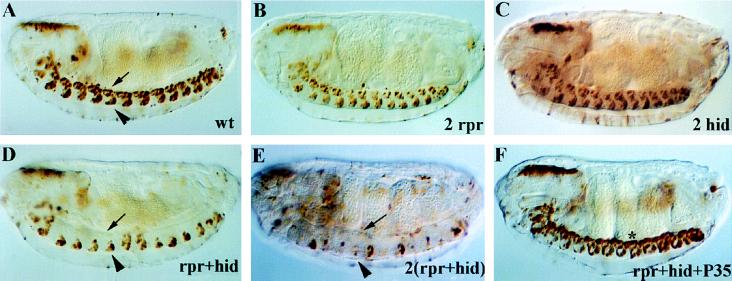
We then analyzed the ability of ectopic rpr and hid expression to kill midline cells that normally survive. Targeted expression of rpr and hid was achieved by crossing the P[52A-Gal4] strain to P[UAS-rpr] and P[UAS-hid] strains. To visualize the developing midline neurons and glia, a P[UAS-lacZ] reporter was introduced into the strains containing P[UAS-rpr] and/or P[UAS-hid] (see Materials and Methods). Embryos were collected from several different crosses and stained by anti-β-galactosidase immunocytochemistry. In embryos where two copies of either rpr or hid alone were targeted to the midline, most or all of the midline cells appeared normal (Fig. 3A–C). [In some embryos bearing two copies of P[UAS-rpr] there was a slight reduction of midline glia in a few segments (Fig. 3B, Table 1).] This analysis indicated that prolonged expression of rpr or hid alone was not sufficient to induce substantial apoptosis in embryonic CNS midline cells. In contrast, when one copy each of both P[UAS-rpr] and P[UAS-hid] were driven together by P[52A-Gal4], a striking loss of midline glia was detected (Fig. 3D). Loss of midline glia was apparent by stage 12, 1–2 hr after ectopic expression of rpr and hid was first detected, and by stage 14 most of the midline glia were eliminated. Interestingly, the VUM neurons exhibited a greatly reduced sensitivity to the effects of rpr and hid coexpression, as these cells appeared normal in position, number, and morphology in most or all segments (Fig. 3D, Table 1).
Figure 3.
rpr and hid act cooperatively to kill CNS midline cells. Shown are anti-β-galactosidase staining of stage 16 embryos bearing only P[52A-Gal4] and P[UAS-lacZ] (A), or additionally bearing two copies of P[UAS-rpr] (B), two copies of P[UAS-hid] (C), one copy of P[UAS-rpr] and one copy of P[UAS-hid] (D), two copies of P[UAS-rpr] and two copies of P[UAS-hid] (E), or one copy each of P[UAS-rpr], P[UAS-hid], and P[UAS-p35] (F). (A) P[52A-Gal4]/P[UAS-lacZ] embryos exhibited β-galactosidase expression in 2–3 midline glia (arrow) and the VUM neurons (arrowhead) in each segment. In embryos containing two copies of P[UAS-rpr] (B) or two copies of P[UAS-hid] (C), essentially normal numbers of midline glia and VUM neurons were present, indicating that these genes were not individually sufficient to kill midline cells. In embryos containing one copy each of P[UAS-rpr] and P[UAS-hid] (D), there was a dramatic loss of midline glia, although most of the VUM neurons remained present. In embryos containing two copies of P[UAS-rpr] and P[UAS-hid] (E), there was more severe loss of midline glia, as well as elimination of most of the VUM neurons. In embryos containing one copy of P[UAS-rpr] and P[UAS-hid] as well as one copy of P[UAS-p35], both ectopic and normal midline cell deaths were blocked. Note that many of the ectopic midline cells aggregated at the dorsal surface of the nerve cord (star). All views sagittal with anterior to left. (×100.)
Table 1.
Quantitation of rpr- and hid-induced midline cell deaths
| Ectopically expressed cell death genes | No. of midline glia in segments T2 and A5 | No. of segments with fewer than 3 VUM neuron* |
|---|---|---|
| 0 copies of rpr or hid | 2.30 ± 0.15 | 0.00 ± 0.00 |
| 2 copies of hid | 2.00 ± 0.21 | 0.10 ± 0.10 |
| 2 copies of rpr | 1.80 ± 0.25 | 0.00 ± 0.00 |
| 1 copy of rpr and hid | 0.30 ± 0.21 | 1.10 ± 0.35 |
| 2 copies of rpr and hid | 0.00 ± 0.00 | 7.80 ± 0.47 |
Numbers of lacZ-expressing midline cells in embryos where P[52A-Gal4] is used to drive P[UAS-lacZ] along with 0, 1, or 2 copies of rpr and/or hid. n = 10 for each group. All numbers are mean ± SE.
Segments T1 to A8 scored.
These results suggest that rpr and hid can cooperate to induce ectopic midline cell death, since the simultaneous targeted expression of both genes produced a much stronger effect than expression of two copies of either gene alone. The onset of ectopic midline glial cell death is similar to the timing of normal midline glial cell death, and the weak effect on the VUM neurons is also consistent with the lack of death that normally occurs in these cells. To examine whether the effects of targeted rpr and hid expression in the midline cells is dosage sensitive, crosses were performed to generate embryos where two copies of P[UAS-rpr] and P[UAS-hid] were driven by P[52A-Gal4]. These embryos displayed a complete elimination of the midline glia and dramatic loss of VUM neurons (Fig. 3E; Table 1). This result indicates that all midline cells are capable of undergoing cell death, and it suggests that the midline neurons and glia may have different sensitivities to rpr and hid expression.
To determine whether the ectopic midline cell deaths occur by a caspase-dependent mechanism, we tested whether they could be blocked by p35. Crosses were carried out to generate embryos where P[52A-Gal4] drove the simultaneous midline expression of rpr, hid, lacZ, and p35. These embryos exhibited an absence of ectopic cell deaths (Fig. 3F), demonstrating that midline cell deaths induced by ectopic rpr and hid require the functions of one or more caspases. Interestingly, the Drosophila caspase gene, DCP-1, exhibits strong CNS midline expression (29). Expression of p35 was able to block both rpr- and hid-induced as well as normal midline cell death, as stage 16 embryos exhibited greater than normal numbers of midline cells and no engulfed midline cells were detected in phagocytic macrophages.
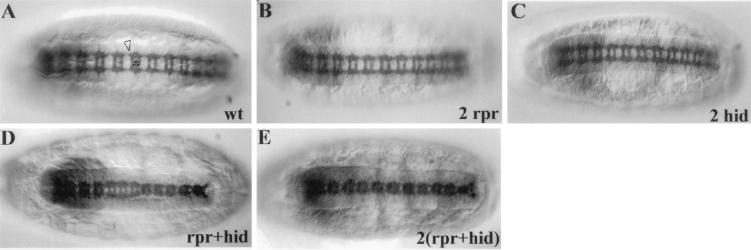
To analyze the developmental consequences of the rpr- and hid-induced ectopic midline cell death, we monitored ventral nerve cord organization by means of immunostaining with the monoclonal antibody BP102, which labels all CNS axons (30). Midline expression of one or two copies of rpr or hid alone did not result in detectable axon scaffold defects (Fig. 4A–C), suggesting that the midline cells were not only surviving but were capable of carrying out normal axon guidance functions. In contrast, in embryos where both rpr and hid were expressed, significant defects in axon scaffold organization were detected. In several segments, the anterior and posterior commissures exhibited partial fusion and the two longitudinal connectives were narrowed (Fig. 4D). Embryos bearing two copies of both rpr and hid exhibited more severe disruption of axon scaffold organization, including fusion of the commissures in every segment and severe narrowing of the longitudinals (Fig. 4E). This CNS phenotype is similar to that of single-minded, slit, and spitz class mutants, where some or all midline cells fail to develop or maintain their normal positions (see ref. 17). These axonal defects imply that the death of midline cells by means of targeted expression of rpr and hid was quite rapid and occurred over a time course comparable to that seen with targeted neuronal expression of metabolic toxins, such as ricin (31).
Figure 4.
Ectopic midline cell deaths result in axon scaffold defects. (A) Stage 16 wild-type embryo stained with monoclonal antibody BP102 to visualize the CNS axon scaffold (star indicates commissural axon tracts, arrowhead indicates longitudinal connectives). (B and C) Similarly stained stage 16 embryos in which two copies of P[UAS-rpr] (B) or P[UAS-hid] (C) were driven in the CNS midline cells by P[52A-Gal4]. Note normal CNS organization. (D) Stage 16 embryo in which one copy each of both rpr and hid were driven by P[52A-Gal4]. Note that several segments exhibited fusion of commissures and narrowing of longitudinal connectives. (E) Stage 16 embryo in which two copies each of both rpr and hid were driven by P[52A-Gal4]. Note severe commissure fusions and narrowing of longitudinals in all segments. All views ventral with anterior to left. (×100.)
DISCUSSION
In this study we have analyzed the cell death functions of the rpr, hid, and grim genes in the embryonic CNS midline cells. Both rpr and hid are expressed in dying midline cells, and it will be of interest to identify the mechanisms used to regulate midline transcription of these genes. Analyses of hid null and Df(3L)XL25 mutant embryos suggested both that multiple 75C1,2 genes are required for the normal pattern of midline cell death, and that rpr, hid, and grim may all be essential. Definitive resolution of this issue will likely require isolation of specific rpr and grim mutants. The mechanism through which these genes may interact to regulate midline cell death is not yet clear. While rpr and hid were able to synergistically induce midline cell death, it is not known whether RPR and HID proteins interact directly or simply function in converging pathways. In vertebrates, the FAS and TNFR transmembrane proteins form oligomers and their cytoplasmic death domains interact with death domains present on adapter proteins that, in turn, are linked to a caspase cascade (32, 33). The RPR protein, which possesses some similarity to death domains, is capable of forming multimers in vitro (Cerinda Carboy-Newcomb, Chia-Lin Wei, and H.S., unpublished results), suggesting that it may interact with related adapter proteins in Drosophila. However it should be noted that two recent studies suggest that the homology between RPR and vertebrate death domain proteins may not be indicative of conserved functions (34, 35). Neither HID nor GRIM possesses death domain homology; however, they, along with RPR, do possess a related 14-amino acid stretch at their amino termini (6, 7). Perhaps this shared region of the three proteins allows for homotypic or heterotypic interactions among themselves, or with common intermediaries. While in vitro biochemical studies will clearly facilitate analysis of rpr and hid interactions, the requirement of both rpr and hid to kill CNS midline cells may provide an in vivo system for mapping which regions of the RPR and HID proteins are essential for their cooperative functions.
Our results confirm and extend previous findings (5–7) that multiple genes in the 75C1,2 region are required for the proper patterns of cell death during embryogenesis. By focusing on a discrete subset of CNS midline cells, we have uncovered synergistic actions of the rpr and hid genes and also found that these genes may exhibit different efficiencies in killing midline neurons and glia. Our findings raise interesting possibilities, given that (i) it was previously shown that expression of rpr, hid, or grim alone was sufficient to induce cell death in wild-type or Df(3L)H99 embryos (5–7, 9), and (ii) synergistic functions of rpr and hid were not detected in targeted expression in the adult eye (6). Collectively, these data suggest that different cell types may have distinct requirements for 75C1,2 cell death genes during normal development and distinct sensitivities to ectopic rpr, hid, and grim expression. Thus, in previous cell killing experiments that analyzed the effects of ubiquitous embryonic expression of these genes (5–7), it was not determined whether the same cells were killed by each gene, or whether each gene participated in the death of a distinct subset of cells. Additionally, while rpr and hid can act synergistically to induce CNS midline cell deaths, their ability to do so may depend upon specific cellular contexts. One attractive hypothesis is that these three genes may act in a combinatorial fashion to regulate cell death in different tissues.
It will also be of interest to determine the effectiveness of ectopic grim expression in killing midline cells and to analyze its potential functional interactions with rpr and hid. This is a particularly interesting question, given that ectopic grim is capable of inducing cell death at earlier embryonic stages than either rpr or hid (7). Ultimately, additional studies of the functional interactions between these key death proteins will likely have relevance not only for the regulation of cell death during normal development and homeostasis but also for how ectopic cell death occurs during disease and injury.
Acknowledgments
We are grateful to Bruce Hay for providing unpublished P[UAS-p35] strains, Andrea Brand for providng P[UAS-lacZ] strains, and Barbara Osborne and John Wing for comments on the manuscript. This work was supported by National Institutes of Health Grant NS32251 and a March of Dimes Basic Research Grant to J.R.N., National Institutes of Health Grant GM40458 to L.M.S., and National Institutes of Health Grant AG55118 to L.M.S and J.R.N. A.S. was supported by a Howard Hughes Medical Institute-sponsored Junior Fellows Undergraduate Award from the University of Massachusetts. H.S. is an Associate Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- CNS
central nervous system
- VUM
ventral unpaired median
References
- 1.Kerr J F R, Wyllie A, Currie A R. Brit J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hengartner M O, Horvitz H R. Curr Opin Genet Dev. 1994;4:581–586. doi: 10.1016/0959-437x(94)90076-f. [DOI] [PubMed] [Google Scholar]
- 3.Steller H. Science. 1995;267:1445–1449. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 4.White E. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 5.White K, Grether M E, Abrams J M, Young L, Farrell K, Steller H. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 6.Grether M E, Abrams J M, Agapite J, White K, Steller H. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- 7.Chen P, Nordstrom W, Gish B, Abrams J M. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- 8.Hay B A, Wassarman D A, Rubin G M. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 9.White K, Tahaoglu E, Steller H. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- 10.Pronk G J, Ramer K, Amiri P, Williams L T. Science. 1996;271:808–810. doi: 10.1126/science.271.5250.808. [DOI] [PubMed] [Google Scholar]
- 11.Tartaglia L A, Ayres T M, Wong G H W, Goeddel D V. Cell. 1993;74:845–853. doi: 10.1016/0092-8674(93)90464-2. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein P, Marguet D, Depraetere V. Cell. 1995;81:185–186. doi: 10.1016/0092-8674(95)90327-5. [DOI] [PubMed] [Google Scholar]
- 13.Hsu H, Xiong J, Goeddel D V. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 14.Chinnaiyan A M, O’Rourke K, Tewari M, Dixit V M. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 15.Stanger B Z, Leder P, Lee T-H, Kim E, Seed B. Cell. 1995;81:513–524. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- 16.Thomas J B, Crews S T, Goodman C S. Cell. 1988;52:133–141. doi: 10.1016/0092-8674(88)90537-5. [DOI] [PubMed] [Google Scholar]
- 17.Klämbt C, Jacobs J R, Goodman C S. Cell. 1991;64:801–815. doi: 10.1016/0092-8674(91)90509-w. [DOI] [PubMed] [Google Scholar]
- 18.Bossing T, Technau G M. Development (Cambridge UK) 1994;120:1895–1906. doi: 10.1242/dev.120.7.1895. [DOI] [PubMed] [Google Scholar]
- 19.Kim S H, Crews S T. Development (Cambridge UK) 1993;118:893–901. doi: 10.1242/dev.118.3.893. [DOI] [PubMed] [Google Scholar]
- 20.Zhou L, Hashimi H, Schwartz L M, Nambu J R. Curr Biol. 1995;5:784–490. doi: 10.1016/s0960-9822(95)00155-2. [DOI] [PubMed] [Google Scholar]
- 21.Sonnenfeld M G, Jacobs J R. Development (Cambridge UK) 1995;121:569–578. doi: 10.1242/dev.121.2.569. [DOI] [PubMed] [Google Scholar]
- 22.Brand A, Perrimon N. Development (Cambridge UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 23.Campos-Ortega J A, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Berlin: Springer; 1985. [Google Scholar]
- 24.Karess R E, Rubin G M. Cell. 1984;38:135–146. doi: 10.1016/0092-8674(84)90534-8. [DOI] [PubMed] [Google Scholar]
- 25.Robertson H M, Preston C R, Phillis R W, Johnson-Schlitz D M, Benz W K, Engels W R. Genetics. 1988;118:461–471. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nambu J R, Lewis J O, Wharton K A, Crews S T. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- 27.Xue D, Horvitz H R. Nature (London) 1995;377:248–251. doi: 10.1038/377248a0. [DOI] [PubMed] [Google Scholar]
- 28.Bump N J, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, Licari P, Mankovich J, Shi L, Greenberg A H, Miller L K, Wong W W. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 29.Song Z, McCall K, Steller H. Science. 1997;275:536–540. doi: 10.1126/science.275.5299.536. [DOI] [PubMed] [Google Scholar]
- 30.Elkins T, Zinn K, McAllister L, Hoffman F M, Goodman C S. Cell. 1990;60:565–575. doi: 10.1016/0092-8674(90)90660-7. [DOI] [PubMed] [Google Scholar]
- 31.Hidalgo A, Urban J, Brand A H. Development (Cambridge UK) 1995;121:3703–3712. doi: 10.1242/dev.121.11.3703. [DOI] [PubMed] [Google Scholar]
- 32.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 33.Muzio M, Chinnaiyan A M, Kischkel F C, O’Rourke K, Shevchenko A, Scaffidi C, Bretz J D, Zhang M, Ni J, Gentz R, Mann M, Krammer P H, Peter M E, Dixit V M. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 34.Chen P, Lee L, Otto L, Abrams J. J Biol Chem. 1996;271:25735–25737. doi: 10.1074/jbc.271.42.25735. [DOI] [PubMed] [Google Scholar]
- 35.Vucic D, Seshagiri S, Miller L K. Mol Cell Biol. 1997;17:667–676. doi: 10.1128/mcb.17.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]