Abstract
Two mouse insulin genes, Ins1 and Ins2, were disrupted and lacZ was inserted at the Ins2 locus by gene targeting. Double nullizygous insulin-deficient pups were growth-retarded. They did not show any glycosuria at birth but soon after suckling developed diabetes mellitus with ketoacidosis and liver steatosis and died within 48 h. Interestingly, insulin deficiency did not preclude pancreas organogenesis and the appearance of the various cell types of the endocrine pancreas. The presence of lacZ expressing β cells and glucagon-positive α cells was demonstrated by cytochemistry and immunocytochemistry. Reverse transcription-coupled PCR analysis showed that somatostatin and pancreatic polypeptide mRNAs were present, although at reduced levels, accounting for the presence also of δ and pancreatic polypeptide cells, respectively. Morphometric analysis revealed enlarged islets of Langherans in the pancreas from insulin-deficient pups, suggesting that insulin might function as a negative regulator of islet cell growth. Whether insulin controls the growth of specific islet cell types and the molecular basis for this action remain to be elucidated.
Insulin is synthesized, stored, and secreted by the pancreatic islet β cells in a highly regulated manner and plays a vital role in glucose homeostasis. Insulin action also results in several other pleiotropic effects that are less well documented. Embryonic insulin synthesis begins early in gestation, but fetal glycemia closely follows maternal blood glucose levels. The question, therefore, arises as to what function embryonic insulin might fulfill during development. For instance, one might ask whether insulin plays an autocrine or paracrine role in pancreatic islet cell growth and differentiation, since insulin is synthesized with other hormones in developing islet cell types (1–3). Recently, this question has been addressed in a few transgenic studies. For instance, the gene encoding PDX-1 (4, 5), a homeodomain transcription factor synthesized in adult β cells and capable of transactivating insulin gene expression, has been inactivated by targeted disruption (6, 7). Agenesis of pancreas resulting from PDX-1 deficiency precluded from addressing the question of the possible role of insulin in islet cell growth and differentiation. Similarly, mice lacking the LIM homeodomain transcription factor ISL1, synthesized in all classes of islet cells in the adult, were arrested in development soon after embryonic day 9.5 (8). The requirement of ISL1 in pancreatic epithelium for the differentiation of all islet cell types was, however, demonstrated by in vitro culture of explants from ISL1-deficient embryonic day 9.5 embryos that gave rise to cells that were negative for glucagon, insulin, and somatostatin. In another study, transgenic mouse embryos expressing the gene encoding the diphteria toxin A chain under control of the rat Ins2 promoter were generated (9). The resulting genetic ablation of the insulin-producing cells did not appear to alter the development of the nontargeted islet cell types, but an incomplete penetrance of the toxigene was a limitation in this approach.
The present work describes the generation of mice carrying null mutations in the two nonallelic insulin genes, Ins1 and Ins2, and reports, to our knowledge, the first phenotypic description of double nullizygous insulin-deficient mice with particular emphasis on the effects of insulin deficiency on the endocrine pancreas.
MATERIALS AND METHODS
Engineering of the Targeting Vectors.
All bacterial, phage, and DNA manipulations were performed as described (10). Mouse strain 129 genomic DNA libraries in phage λ were screened with probes corresponding to mouse Ins1 or Ins2 cDNA. The restriction maps of the inserts in recombinant phages were established and several fragments were subcloned.
For Ins1, 8.7-kb HindIII–SmaI and 8.2-kb ApaI–XhoI fragments (5′ and 3′ sides of Ins1) were subcloned into pSK+ using HindIII/SmaI and ApaI/XhoI sites to produce p34 and p4, respectively. A 4-kb BamHI–HindIII fragment (5′ side of Ins1) was also subcloned into pSK+ using BamHI/HindIII sites to produce pR1. The vector used to engineer the targeting vector was pJorg (obtained from J. Hamm, European Molecular Biology Laboratory, Heidelberg), a pKS+ derivative containing neo at the BamHI site and tk at XhoI/HindIII sites. The 2.5-kb NotI–PvuII fragment from pR1 corresponding to the 5′ homology was cloned into pJorg at NotI/XbaI sites to produce pR2, in which the 6.5-kb HindIII fragment from p4 corresponding to the 3′ homology was cloned at the HindIII site leading to the targeting vector for Ins1. The probes 1 and 2 were synthesized with the BamHI fragment from p34 and the HindIII–XhoI fragment from p4, respectively.
For Ins2, 5-kb and 7-kb EcoRI fragments (5′ and 3′ sides of Ins2) were subcloned into pSK+ at the EcoRI site to produce p11 and p12, respectively. A NsiI site present in the 7-kb insert in p12 was destroyed to produce p12ΔNsiI. A 5-kb XbaI fragment containing Ins2 was also subcloned into pSK+ at XbaI site to produce p13. Ins2 region (positions −950 to +20) was PCR-amplified with the primers 5′-CGCTCTAGACCCTCCTCTTGCATTTCAAA-3′ and 5′-CGCATGCATGTAGCGGATCACTTAGGGT-3′ that provide XbaI and NsiI sites and cloned upstream of lacZ coding sequence in pGN (obtained from P. Brûlet; ref. 11) at XbaI/NsiI sites; the NsiI site was destroyed and this resulted in p15. The XbaI/SfiI fragment in p15 was replaced by a 2.5-kb XbaI–SfiI fragment from p13 to produce p16. To engineer the targeting vector, pJorg was first modified by inserting a NsiI linker at HindIII site to produce p10. The XbaI–XhoI fragment from p16 containing the 5′ homology (2.7 kb) and lacZ were cloned into p10 using XbaI/SpeI sites to produce p17, in which the 5.5-kb SmaI–EcoRI fragment from p12ΔNsiI corresponding to the 3′ homology was cloned at the NsiI site using NsiI linkers to produce the targeting vector for Ins2. The probe 3 was synthesized with the XhoI–EcoRI fragment from p12 and the probe 4 was synthesized with the BamHI fragment generated from pJorg.
Generation of Mutant Mice.
All embryonic stem (ES) cell cultures and mouse embryo manipulations were performed as described (12–14). The targeting vector DNAs were linearized with NotI prior to electroporation into D3 ES cells (obtained from P. Chambon, Laboratorie de Genetique Moleculaire des Eucaryotes, Illkirch, France). After G418 and ganciclovir (a gift from Syntex, Palo Alto, CA) selection, 3 and 12 recombinant clones of Ins1 and Ins2, respectively, were identified. Recombinant cells (10–15 cells) were microinjected into B6 blastocysts that were transferred into the oviduct of pseudopregnant females. Several male germ-line chimeras were obtained that were crossed with B6 females. The various crosses that resulted in double homozygous mice are described in the text. At least two clones were used for each construct. Southern blot analyses using cellular or mouse-tail DNAs were performed as described (12) with the probes indicated.
Reverse Transcription-Coupled PCR (RT–PCR) Analysis.
Total pancreatic RNA (1 μg), prepared as described (15), was subjected to a combined RT–PCR using a Promega kit under the conditions specified. Ins1 and Ins2 expression was analyzed by using the strategy previously described (16). Briefly, a unique primer pair (5′-GGCTTCTTCTACACACCCA-3′/5′-CAGTAGTTCTCCAGCTGGTA-3′) was used for the PCR amplification step (40 cycles). The MspI digestion of PCR products analyzed by Southern blot using a single 5′-32P-labeled oligonucleotide (5′-ACAATGCCACGCTTCTG-3′) revealed a fragment of 71 and 112 bp for Ins1 and Ins2, respectively. Glucagon mRNA was amplified with the primers (5′-GGTGCAAGGCAGCTGGCAGC-3′/5′-CACGGCGGGAGTCGAGGTAT-3′) and the probe was 5′-[32P]-ACAGGGCACATTCACCAGCG-3′. For somatostatin mRNA the primers were (5′-CAGAAGTCTCTGGCGGCTGC-3′/5′-GGCAGACCTCTGCAGCTCCA-3′) and the probe was 5′-[32P]-CAGAGCTGCTGTCCGAGCCC-3′. For pancreatic polypeptide mRNA the primers were (5′-TGCTGCCTCTCCCTGTTTCT-3′/5′-TCTGCGGAGCTGAGTTTCAT-3′) and the probe was 5′-[32P]-TGCTGCTGCAGCCCCTGCAG-3′. β-actin mRNA was amplified with the primer pair (5′-CGTGGGCCGCCCTAGGCACCA-3′/5′-TTGGCCTTAGGGTTCAGGGGGG-3′) and a 5′-32P-labeled oligonucleotide (5′-AAGGACTCCTATGTGGGTGACG-3′) as probe.
Histological, Immunocytochemical, Histochemical, and Skeletal Analyses.
Liver and pancreas samples from Ins1−/− Ins2−/− and normal littermates were fixed by immersion in 10% formaldehyde. After dehydration and embedding in paraffin, 5-μm sections were performed and stained with hematoxylin and eosin for direct microscopic examination. Immunocytochemistry was performed on paraffin sections with primary rabbit antibodies against C-peptide 1 and C-peptide 2 (provided by O. D. Madsen, ref. 16) reacting with proinsulin 1 and proinsulin 2, respectively, and glucagon (Euro-diagnostica, Malmö, Sweden). For C-peptide 1 and C-peptide 2, goat anti-rabbit serum coupled to peroxidase (Immunotech, Westbrook, ME) was used as secondary antibody. For glucagon immunostaining, incubation with swine anti-rabbit serum (Dako, Glostrup, Denmark) was followed by incubation with rabbit peroxidase-anti-peroxidase (Dako). The reactions were revealed with diaminobenzidine and slides were counterstained with hematoxylin. For 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) staining, pancreas were fixed for 30 min in 4% paraformaldehyde in phosphate-buffered saline at 4°C before incubation with X-Gal solution overnight. Specimens were then post-fixed in 4% paraformaldehyde, embedded into paraffin, sectioned, and counterstained with nuclear red.
For skeleton analysis, embryos were dissected, eviscerated, and fixed overnight in 95% ethanol at room temperature. After a 1-week incubation in alcian blue at 37°C, skeletons were incubated with 2% KOH for 24 h and then with alizarin red for 12 h. Tissues were then cleared in 20% glycerol/1% KOH for 1 week and skeletons were transferred to 50% glycerol/50% ethanol at room temperature for storage as described (17) and photographed.
RESULTS AND DISCUSSION
Generation of Insulin-Deficient Mice.
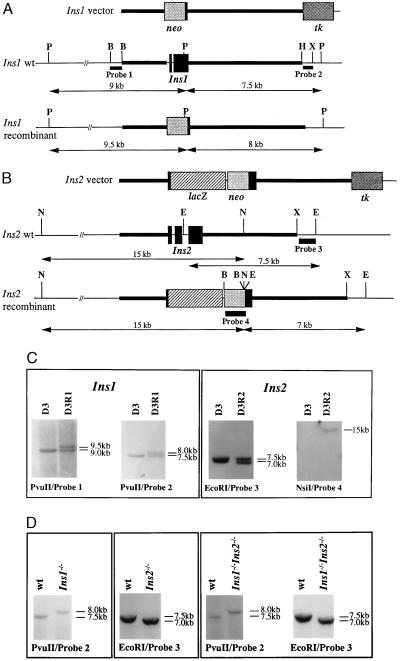
The targeting vectors that we used to inactivate Ins1 and Ins2 are presented Fig. 1 A and B. These vectors were designed to delete most of the Ins1 and Ins2 coding sequences. Moreover, the Ins2 vector was engineered to put lacZ under the control of the Ins2 promoter. Both vectors contained neo and tk, which allowed positive and negative selection, respectively, of transfected ES cells. After electroporation of vector DNAs into D3 ES cells, G418- and ganciclovir-resistant clones were screened by Southern blot analysis using the restriction enzymes and probes indicated in Fig. 1 A and B and recombinant ES cell clones for each mutation were identified (Fig. 1C). Such cells were used to generate male germ-line chimeras for each mutation. Heterozygous animals were identified and crossed to obtain homozygous Ins1−/− or Ins2−/− animals (Fig. 1D). These single homozygous mutants were viable and fertile. The intercross of Ins1−/− and Ins2−/− animals generated double heterozygotes that were then mated, and the offspring were genotyped. Double homozygous mutant pups were identified as illustrated in Fig. 1D and were present among the progeny in a ratio of approximately 1:16 (15 double homozygotes among 235 pups born). Absence of Ins1 and Ins2 transcripts in total RNA from the pancreas of double homozygous pups as evidenced by RT–PCR analysis (Fig. 2A) confirmed that these animals carry a null mutation for both Ins1 and Ins2. Thus, the lack of embryonic insulin did not result in any embryonic lethality of double nullizygous embryos.
Figure 1.
Targeted disruption of Ins1 and Ins2. Structures of the targeting vectors as well as of the wild-type (wt) and recombinant alleles are shown in A for Ins1 and in B for Ins2. The restriction enzymes and probes used for genotyping of cellular and mouse DNAs are indicated. Autoradiograms of Southern blot analyses confirming the presence of ES cells (D3) carrying recombined alleles for Ins1 (D3R1) or Ins2 (D3R2) and shown in C and of Ins1−/−, Ins2−/− and double homozygous mutant mice are shown in D. neo, Neomycin phosphotransferase gene; tk, thymidine kinase gene from herpes simplex virus type 1; B, BamHI; E, EcoRI; H, HindIII; N, NsiI; P, PvuII; X, XbaI.
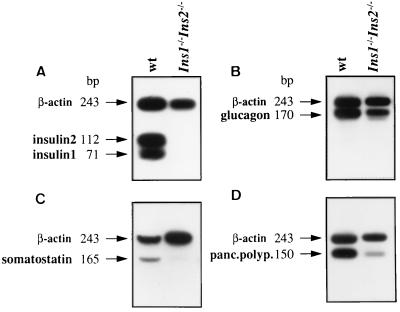
Figure 2.
RT–PCR amplification of mRNAs for insulin 1/insulin 2 (A), glucagon (B), somatostatin (C), and pancreatic polypeptide (D) in the pancreas of wild-type (wt) mice and double homozygous mutants. β-actin mRNA was amplified as control. pBR322/HpaII was used as size marker.
Growth Retardation and Metabolic Disorders in Insulin-Deficient Mice.
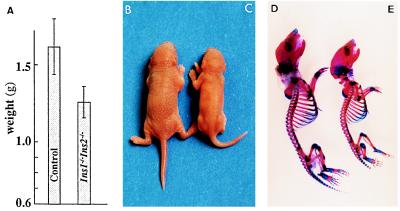
Double homozygous mutant pups appeared morphologically normal except that they were smaller and their mean body weight at birth or within a few hours of birth was 22% less than that of their siblings (Fig. 3). The growth difference between insulin-deficient and normal pups was also obvious when comparing their skeletons (Fig. 3). The growth retardation occurred during fetal life, since the few insulin-deficient embryos examined after cesarean around day 18 of gestation weighed 15–20% less than the other embryos. At that time, the liver appeared normal. Examination of the time course of the embryonic growth retardation to determine how it correlates with insulin accumulation requires the sacrifice of a large number of pregnant females and must await the generation of a larger colony of these mutant mice.
Figure 3.
Growth retardation of double nullizygotes. (A) Weights of double nullizygous (n = 11) neonates compared with control animals, which include wild-type mice and various heterozygotes (n = 69). Size and skeletons of control (B and D) and insulin-deficient (C and E) pups.
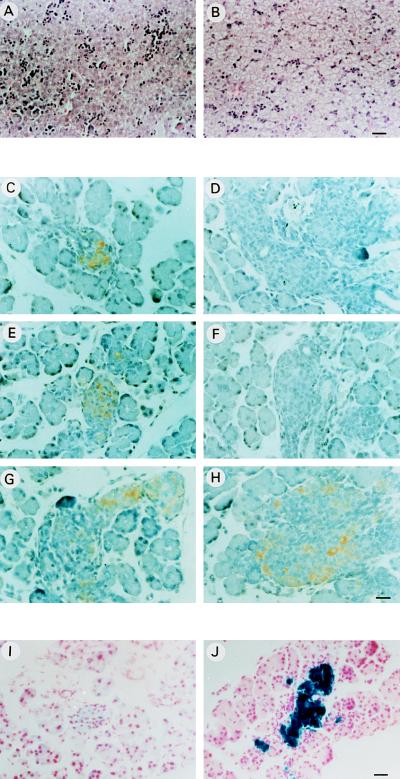
Insulin deficiency resulted in the rapid development of diabetes mellitus and ketoacidosis. Glycosuria was detected as soon as the mutant pups had suckled, and the appearance of ketone bodies followed within a day. The triglyceride levels in plasma were judged to be high from its milky appearance. Anatomical examination of the mutant pups revealed a marked hepatomegaly with liver steatosis (Fig. 4 A and B). Finally, insulin-deficient mice showed early neonatal lethality since the mutant pups died on average within 48 h.
Figure 4.
Analysis of the liver and pancreas of wild-type and double nullizygous mice. Histological analysis of liver sections from wild-type (A) and insulin-deficient mice (B). Immunocytochemical staining of pancreas sections of wild-type (C, E, and G) and insulin-deficient mice (D, F, and H) with antibodies against C-peptide 1 (C and D), C-peptide 2 (E and F), and glucagon (G and H). lacZ expression in pancreatic sections of insulin-deficient mice visualized by X-Gal staining (J) and background staining obtained with sections from wt animals (I) are shown. [Bars = 10 μm (A and B) and 8 μm (C–J).]
Insulin action is mediated through the insulin receptor, a receptor with tyrosine kinase activity widely distributed in the body. Insulin and its receptor are structurally very similar respectively to insulin-like growth factors (IGF-1 and IGF-2) and the IGF-1 receptor. Insulin and IGFs lead to some common and some specific effects on cellular metabolism, growth, and differentiation. These different ligands can bind to and activate the heterologous receptor. The role that IGFs play during embryonic development has been investigated by targeted disruption of the genes encoding IGF-1, IGF-2, or the IGF-1 receptor (18–20). The phenotype of insulin-deficient mice described herein was comparable to that of the insulin-receptor-deficient mice that have been obtained recently (21, 22), except that the disorders resulting from the lack of insulin were more severe and appeared more rapidly. This finding suggests that in the insulin-receptor-deficient mice that become hyperinsulinemic, some effects of insulin might be achieved through its interaction with the IGF-1 receptor. More recently, it has been reported that growth retardation in embryos lacking both the insulin and IGF-1 receptors was more pronounced than in IGF-1-receptor-deficient embryos, in agreement with a role of the insulin receptor in embryonic development (23).
Pancreatic Islet Cell Growth and Differentiation in the Absence of Insulin.
In our analysis of insulin-deficient animals, we have focused our efforts on examining the pancreas, which was clearly visible by anatomical observation. Histological analysis indicated that both endocrine and exocrine components were present. As expected, immunocytochemical staining of pancreatic sections using anti-C-peptide 1 and anti-C-peptide 2 antibodies specific for proinsulin 1 and proinsulin 2, respectively, was negative (Fig. 4 D and F). Since lacZ was inserted at the Ins2 locus under the control of the Ins2 promoter, the pancreas was assayed for cytochemical detection of β-galactosidase activity. As shown in Fig. 4J, most of the cells in the islets were stained blue when X-Gal was used as substrate and, therefore, represent β cells. Immunocytochemical staining of pancreatic sections using antibodies against glucagon revealed the presence of α cells in the islets (Fig. 4 G and H). The glucagon mRNA was readily detected by RT–PCR analysis (Fig. 2B). Somatostatin and pancreatic polypeptide mRNAs were also detected by RT–PCR, accounting for the presence also of δ and pancreatic polypeptide cells. These mRNAs, however, were present at very reduced levels (Fig. 2 C and D) and we do not yet know whether this is due to a down-regulation of the transcription of these two genes or whether the number of δ and pancreatic polypeptide cells in the islets is reduced.
Thus, embryonic insulin appears to be dispensable for pancreas organogenesis and the appearance of various islet cell types. It can be concluded from the present study that the agenesis of pancreas observed in PDX-1-deficient mice (6, 7) cannot be accounted for by the absence of insulin gene activity. Rather, it came as a surprise in the present study that the absence of embryonic insulin could result in hyperplastic islets as observed in microscopic examination (Fig. 4, compare C, E, and G with D, F, and H). In line with this observation, the surface occupied by the islets in pancreatic serial sections appeared to be ∼1.48 times larger in diabetic nullizygous (n = 5) than in normal littermates (n = 4) in preliminary morphometric analysis. An interesting question raised by this observation is whether insulin functions as a negative regulator of islet cell growth in pancreas organogenesis, and it will be interesting to further explore the underlying molecular mechanisms for this action of insulin.
Finally, insulin-deficient mice represent interesting tools for addressing many other basic issues related to insulin action and for testing different therapeutic strategies (i.e., injection of insulin analogues, transplantation of insulin producing cells, or gene therapy) for insulin-dependent diabetes. In this respect, first attempts of injecting insulin into insulin-deficient pups showed that glycosuria, ketoacidosis, and liver steatosis can be corrected within 1 day. This opens the possibility of obtaining fertile insulin-deficient adult mice by conventional insulin therapy, which would be useful for many other studies.
Acknowledgments
We thank L. Lamotte for excellent assistance in genotyping mouse mutants, J. M. Chirgwin for kindly providing us with cDNAs corresponding to Ins1 and Ins2, O. D. Madsen for anti-C-peptide antibodies, P. Chambon for the D3 ES cells, P. Brûlet for pGN, J. Hamm for pJorg, and Syntex Laboratory for ganciclovir. B.D. is recipient of a fellowship from the Ministère de l’Enseignement Supérieur et de la Recherche. This work was partly supported by grants from the Association pour la Recherche sur le Cancer and the Ligue Nationale contre le Cancer.
ABBREVIATIONS
- ES
embryonic stem
- RT–PCR
reverse transcription-coupled PCR
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
- IGF
insulin-like growth factor
References
- 1.Teitelman G. Tumor Biol. 1993;14:167–173. doi: 10.1159/000217832. [DOI] [PubMed] [Google Scholar]
- 2.Slack J M W. Development (Cambridge, UK) 1995;121:1569–1580. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- 3.Madsen O D, Jensen J, Blume N, Petersen H V, Lund K, Karlson C, Andersen F G, Jensen P B, Larsson L I, Serup P. Eur J Biochem. 1996;242:435–445. doi: 10.1111/j.1432-1033.1996.435rr.x. [DOI] [PubMed] [Google Scholar]
- 4.Ohlsson H, Karlsson K, Edlund T. EMBO J. 1993;12:4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guz Y, Montminy M R, Stein R, Leonard J, Gamer L W, Wright C V E, Teitelman G. Development (Cambridge, UK) 1995;121:11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- 6.Jonsson J, Carlsson L, Edlund T, Edlund H. Nature (London) 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 7.Offield M F, Jetton T L, Labosky P A, Ray M, Stein R W, Magnuson M A, Hogan B L M, Wright C V E. Development (Cambridge, UK) 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 8.Ahlgren U, Pfaaf S L, Jessell T M, Edlund T, Edlund H. Nature (London) 1997;385:257–260. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- 9.Herrera P L, Huarte J, Zyfferey R, Nichols A, Mermillod B, Philippe J, Muniesa P, Sanvito F, Orci L, Vassalli J D. Proc Natl Acad Sci USA. 1994;91:12999–13003. doi: 10.1073/pnas.91.26.12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 11.Le Mouellic H, Lallemand Y, Brûlet P. Proc Natl Acad Sci USA. 1990;87:4712–4716. doi: 10.1073/pnas.87.12.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradley A, Evans M, Kaufman M H, Robertson E J. Nature (London) 1984;309:255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- 13.Mansour S L, Thomas K R, Capecchi M R. Nature (London) 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 14.Joyner A L, editor. Gene Targeting: A Practical Approach. Oxford: IRL; 1993. [Google Scholar]
- 15.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 16.Deltour L, Leduque P, Blume N, Madsen O, Dubois P, Jami J, Bucchini D. Proc Natl Acad Sci USA. 1993;90:527–531. doi: 10.1073/pnas.90.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLeod M J. Teratology. 1980;22:299–301. doi: 10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- 18.DeChiara T M, Efstratiadis A, Robertson E J. Nature (London) 1990;345:78–80. doi: 10.1038/345078a0. [DOI] [PubMed] [Google Scholar]
- 19.Liu J P, Baker J, Perkins A S, Robertson E J, Efstratiadis A. Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- 20.Powell-Braxton L, Hollingshead P, Warburton C, Dowd M, Pitts-Meek S, Dalton D, Gillett N, Stewart T A. Genes Dev. 1993;7:2609–2617. doi: 10.1101/gad.7.12b.2609. [DOI] [PubMed] [Google Scholar]
- 21.Accili D, Drago J, Lee E J, Johnson M D, Cool M H, Salvatore P, Asico D, José P A, Taylor S I, Westphal H. Nat Genet. 1996;12:106–109. doi: 10.1038/ng0196-106. [DOI] [PubMed] [Google Scholar]
- 22.Joshi R L, Lamothe B, Cordonnier N, Mesbah K, Monthioux E, Jami J, Bucchini D. EMBO J. 1996;15:1542–1547. [PMC free article] [PubMed] [Google Scholar]
- 23.Efstratiadis, A. (1996) Exp. Clin. Endocrinol. Diabetes 104, Suppl. 2, 4–6.