Abstract
Background
Multiple sclerosis (MS) is a complex trait in which alleles at or near the class II loci HLA-DRB1 and HLA-DQB1 contribute significantly to genetic risk. The MHC class II transactivator (MHC2TA) is the master controller of expression of class II genes, and methylation of the promoter of this gene has been previously been shown to alter its function. In this study we sought to assess whether or not methylation of the MHC2TA promoter pIV could contribute to MS disease aetiology.
Methods
In DNA from peripheral blood mononuclear cells from a sample of 50 monozygotic disease discordant MS twins the MHC2TA promoter IV was sequenced and analysed by methylation specific PCR.
Results
No methylation or sequence variation of the MHC2TA promoter pIV was found.
Conclusion
The results of this study cannot support the notion that methylation of the pIV promoter of MHC2TA contributes to MS disease risk, although tissue and timing specific epigenetic modifications cannot be ruled out.
Background
Genetic-epidemiological studies indicate unequivocally that there is a genetic influence on susceptibility to Multiple Sclerosis (MS) [1]. The only consistent genetic association with MS in Northern Europeans had been with extended MHC haplotypes especially those containing HLA-DRB1*1501 [1]. Recently, the interleukin 7 receptor (IL7R) and interleukin 2 receptor (IL2R) genes have been shown to be additional MS susceptibility loci [2,3]. However, any effect of IL7R or IL2R is small and it is clear that the MHC is the key MS susceptibility locus [4].
The MS MHC class II association has been fine mapped to the extended haplotype HLA-DQA1*0102-DQB1*0602-DRB1*1501-DRB5*0101 [5]. Intense linkage disequilibrium within the MHC has prevented the exact susceptibility locus from being conclusively identified. Analysis of the MHC region with a large number of markers as well as classical typing show evidence for the involvement of the class II region only [6,7]. However, the paradigm is more complex than one in which the HLA-DRB1*15 allele acts solely to increase MS risk. Our previous investigations have shown that HLA-DRB1*15 and HLA-DRB1*17 bearing haplotypes increase risk of MS, and HLA-DRB1*14 and HLA-DRB1*11 bearing haplotypes are protective [8,9]. Additionally, HLA-DRB1*10, DRB1*01 and DRB1*08 interact with HLA-DRB1*15 to influence disease risk [8,9].
Given the unequivocal MHC class II association with MS, the amount and cellular distribution of class II molecules may therefore be important factors in determining susceptibility to the disease. MHC class II molecule expression is regulated primarily through a transcriptional co-activator termed MHC2TA [10]. MHC2TA functions as a non-DNA-binding co-activator that coordinates multiple events that are required for the activation of transcription including the recruitment of transcription factors and phosphorylation of RNA Polymerase II [11]. The highly regulated pattern of expression of the gene encoding MHC2TA dictates where, when and to what level MHC class II genes are expressed [11]. Transcription of the gene encoding MHC2TA is controlled by a large regulatory region that contains three independent promoters (pI, pIII and pIV) [11]. The promoter pIV is essential for driving MHC2TA expression in cells that are sensitive to interferon-γ, and it has been shown that methylation of CpG dinucleotides in this promoter region can influence the expression of MHC2TA and thus MHC class II molecules [12].
Given a contentious association of MHC2TA polymorphisms with susceptibility to MS [13,14], we sought to assess whether or not methylation of the MHC2TA pIV promoter could contribute to MS aetiology using a cohort of monozygotic discordant twins, potentially ideal for entangling genetic and epigenetic contributions to disease susceptibility.
Methods
Subjects
All subjects used in the study were ascertained through the ongoing Canadian Collaborative Project on the Genetic Susceptibility to MS (CCPGSMS), for which the methodology has been previously described [15,16]. Each participating centre of the CCPGSMS obtained ethical approval (as set out in the Helsinki Declaration) from the relevant institutional review board, and the entire project was reviewed and approved by the University of British Columbia. Blood was obtained with appropriate consent.
Fifty pairs of monozygotic discordant twins (100 samples in total, 35 female and 15 male pairs) were chosen for analysis. The clinical data for the MS patients is shown in Table 1. The average age at blood sampling was 41.1 years (standard deviation = 3.7 years). 31 (62%) twin pairs were HLA-DRB1*15 positive.
Table 1.
Clinical details of MS patients
| Clinical/demographic details | |
| Sample Size (n) | 50 |
| Mean age of onset (years) | 31.1 |
| % Relapsing Remitting MS | 68 |
CpG Dinucleotide Prediction
The sequence of the pIV promoter from the NCBI Build 36.1 reference sequence was analysed to identify CpG islands that could be methylated. The methodology for this is described in [17].
Sequencing of promoter pIV
Total genomic DNA was extracted from whole blood as part of the CCPGSMS. PCR was performed using the primers shown in Table 2 under standard conditions [18] with an annealing temperature of 60 degrees Celsius and using AmpliTAQ gold (Applied Biosystems), yielding a PCR amplicon 257 base pairs in size. Sequencing reactions were carried out using BigDye v3.1 after which the DNA was sequenced using an ABI 3700 automated sequencer.
Table 2.
Primer sequences used for sequencing
| Primers | |
| Forward Primer | GGTTGGACTGAGTTGGAGAGA |
| Reverse Primer | GGAGCAACCAAGCACCTACT |
Bisulfite treatment and Methylation Specific PCR
Genomic DNA was treated using methylSEQr Bisulfite Conversion Kit from Applied BioSystems, following the manufacturer's protocol. This converts unmethylated cytosines to uracils and leaves methylated cytosines unchanged. Methylation specific PCR [19], using methylated DNA and unmethylated DNA specific primer sets was performed on treated DNA to detect methylation of the CpG island in the MHC2TA promoter. PCR was performed using the primers shown in Table 3 under standard conditions [18] with an annealing temperature of 55.5 degrees Celsius. Each PCR was performed twice for each sample to ensure validity of results. Universal methylated DNA, universal unmethylated DNA (both from CpGenome™) and water was used as positive, negative and blank controls respectively. Amplified fragments were confirmed by a 2.0% agarose gel.
Table 3.
Primer sequences used for methylation specific PCR
| Primers | Sequence | Product Size |
| Methylated Forward | TGTTTGGTTGTTTTATAGTTTGGTTC | 60 bp |
| Methylated Reverse | CTACTAATAACCTCTCCCTCCCG | |
| Unmethylated Forward | TTGGTTGTTTTATAGTTTGGTTTGA | 157 bp |
| Unmethylated Reverse | CTACTAATAACCTCTCCCTCCCAC |
Results
In silico prediction of CpG islands in the pIV promoter uncovered 1 potential site (Figure 1) Sequencing of the region did not identify any polymorphisms in the pIV promoter sequence in any of the twin pairs.
Figure 1.
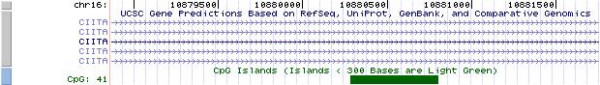
Predicted CpG island in the MHC2TA gene promoter IV.
Methylation specific PCR was able to distinguish between methylated and unmethylated control samples (Figure 2). All twin DNA samples produced amplicons only with the unmethylated DNA specific primers.
Figure 2.
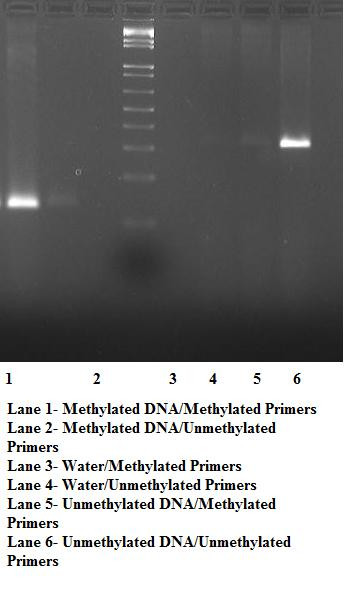
Gel electrophoresis of control and blank DNA amplicons after methylation specific PCR.
Discussion
Multiple sclerosis is unambiguously associated with the MHC class II region [6] and this locus exerts the strongest genetic effect on the risk of developing the disease [4]. MHC2TA is the master regulator of MHC class II gene expression and therefore variability at the MHC2TA gene could conceivably influence susceptibility to MS.
In this investigation we studied the sequence variability of the pIV promoter of the MHC2TA gene and found no variation. This is in agreement with previous studies and this conservation may be a result of the importance of this promoter to gene function.
The only known epigenetic modification of DNA in mammals is methylation of cytosine at position C5 in CpG dinucleotides [20]. DNA methylation affects transcription directly, by influencing the binding of specific transcription factors, and indirectly, by recruiting methyl-CpG-binding proteins and their associated chromatin remodeling activities. It has been shown that methylation of the pIV promoter can influence MHC2TA expression. Monozygotic twins share a common genotype. However, genetically identical twin pairs exhibit differences in susceptibility to many diseases, including MS, where the monozygotic twin concordance rate at its highest does not exceed 30% [21]. There are several possible explanations for these observations, one of these being the existence of epigenetic differences. In this study, we used a cohort of monozygotic MS discordant twins to examine whether methylation differences of the MHC2TA promoter could explain differences in susceptibility to disease. We did not detect methylation of CpG dinucleotides in the pIV promoter in any of our samples, either MS affected or not. Although this study argues against a role of methylation of MHC2TA in MS disease pathogenesis, it must be remembered that whilst genomic information is uniform among the different cells of a complex organism, the epigenome varies from tissue to tissue, controlling the differential expression of genes and providing specific identity to each cell type. Hence, by looking solely at peripheral blood mononuclear cells we may have missed tissue specific methylation of the MHC2TA promoter. Furthermore, a recent study which compared global and locus specific methylation patterns in monozygotic twins, showed that although indistinguishable in early life, epigenetic profiles of monozygotic twins change with age [22] and hence for an adult onset disease with susceptibility determined early in life [23,24] timing of any epigenetic changes may be crucial, and our study may not have been able to detect methylation of MHC2TA at an early age that has since decayed. Additionally, we may have missed low level methylation patterns and it would be necessary to examine every CpG dinucleotide of MHC2TA to be confident that an association between methylation and disease had not been missed just because the wrong markers had been typed.
Conclusion
In summary, although our results do not completely rule out the possibility of an association between methylation of MHC2TA and MS we believe that our data is sufficient to exclude a major effect of methylation of this gene in MS pathology.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
GCE conceived and designed the experiments. SVR, DAD, BMH, GCD, MRL, SMO, LH, MJC and ADS performed the experiments. SVR and GCE analyzed the data and wrote the paper.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
This work was funded by the Multiple Sclerosis Society of the United Kingdom. SVR is funded by the Medical Research Council of the United Kingdom. The authors would like to thank all patients who generously participated in this study and physicians participating in the CCPGSMS. Experiments performed for this investigation comply with current guidelines and ethics. The sponsor of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and final responsibility for the decision to submit for publication.
Contributor Information
Sreeram V Ramagopalan, Email: sramagopalan@gmail.com.
David A Dyment, Email: ddyment@well.ox.ac.uk.
Katie M Morrison, Email: katiem@well.ox.ac.uk.
Blanca M Herrera, Email: blanca@well.ox.ac.uk.
Gabriele C DeLuca, Email: gcdeluca@gmail.com.
Matthew R Lincoln, Email: mlincoln@well.ox.ac.uk.
Sarah M Orton, Email: ortons@well.ox.ac.uk.
Lahiru Handunnetthi, Email: lahiru.handunnetthi@green.ox.ac.uk.
Michael J Chao, Email: michael.chao@well.ox.ac.uk.
A Dessa Sadovnick, Email: dessa.sadovnick@gmail.com.
George C Ebers, Email: george.ebers@clneuro.ox.ac.uk.
References
- Dyment DA, Ebers GC, Sadovnick AD. Genetics of multiple sclerosis. Lancet Neurol. 2004;3:104–110. doi: 10.1016/S1474-4422(03)00663-X. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Duvefelt K, Svensson F, Masterman T, Jonasdottir G, Salter H, Emahazion T, Hellgren D, Falk G, Olsson T, Hillert J, Anvret M. Two genes encoding immune-regulatory molecules (LAG3 and IL7R) confer susceptibility to multiple sclerosis. Genes Immun. 2005;6:145–152. doi: 10.1038/sj.gene.6364171. [DOI] [PubMed] [Google Scholar]
- Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, de Bakker PI, Gabriel SB, Mirel DB, Ivinson AJ, Pericak-Vance MA, Gregory SG, Rioux JD, McCauley JL, Haines JL, Barcellos LF, Cree B, Oksenberg JR, Hauser SL. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- Peltonen L. Old Suspects Found Guilty -- The First Genome Profile of Multiple Sclerosis. N Engl J Med. 2007 doi: 10.1056/NEJMe078147. [DOI] [PubMed] [Google Scholar]
- Fogdell A, Hillert J, Sachs C, Olerup O. The multiple sclerosis- and narcolepsy-associated HLA class II haplotype includes the DRB5*0101 allele. Tissue Antigens. 1995;46:333–336. doi: 10.1111/j.1399-0039.1995.tb02503.x. [DOI] [PubMed] [Google Scholar]
- Lincoln MR, Montpetit A, Cader MZ, Saarela J, Dyment DA, Tiislar M, Ferretti V, Tienari PJ, Sadovnick AD, Peltonen L, Ebers GC, Hudson TJ. A predominant role for the HLA class II region in the association of the MHC region with multiple sclerosis. Nat Genet. 2005;37:1108–1112. doi: 10.1038/ng1647. [DOI] [PubMed] [Google Scholar]
- Chao MJ, Barnardo MC, Bu GZ, Lincoln MR, Ramagopalan SV, Herrera BM, Dyment DA, Sadovnick AD, Ebers GC. Transmission of class I/II multi-locus MHC haplotypes and multiple sclerosis susceptibility: accounting for linkage disequilibrium. Hum Mol Genet. 2007 doi: 10.1093/hmg/ddm142. [DOI] [PubMed] [Google Scholar]
- Dyment DA, Herrera BM, Cader MZ, Willer CJ, Lincoln MR, Sadovnick AD, Risch N, Ebers GC. Complex interactions among MHC haplotypes in multiple sclerosis: susceptibility and resistance. Hum Mol Genet. 2005;14:2019–2026. doi: 10.1093/hmg/ddi206. [DOI] [PubMed] [Google Scholar]
- Ramagopalan SV, Morris AP, Dyment DA, Herrera BM, Deluca GC, Lincoln MR, Orton SM, Chao MJ, Sadovnick AD, Ebers GC. The Inheritance of Resistance Alleles in Multiple Sclerosis. PLoS Genet. 2007;3:e150. doi: 10.1371/journal.pgen.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109 Suppl:S21–33. doi: 10.1016/S0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- Reith W, LeibundGut-Landmann S, Waldburger JM. Regulation of MHC class II gene expression by the class II transactivator. Nat Rev Immunol. 2005;5:793–806. doi: 10.1038/nri1708. [DOI] [PubMed] [Google Scholar]
- Morris AC, Spangler WE, Boss JM. Methylation of class II trans-activator promoter IV: a novel mechanism of MHC class II gene control. J Immunol. 2000;164:4143–4149. doi: 10.4049/jimmunol.164.8.4143. [DOI] [PubMed] [Google Scholar]
- Swanberg M, Lidman O, Padyukov L, Eriksson P, Akesson E, Jagodic M, Lobell A, Khademi M, Borjesson O, Lindgren CM, Lundman P, Brookes AJ, Kere J, Luthman H, Alfredsson L, Hillert J, Klareskog L, Hamsten A, Piehl F, Olsson T. MHC2TA is associated with differential MHC molecule expression and susceptibility to rheumatoid arthritis, multiple sclerosis and myocardial infarction. Nat Genet. 2005;37:486–494. doi: 10.1038/ng1544. [DOI] [PubMed] [Google Scholar]
- Akkad DA, Jagiello P, Szyld P, Goedde R, Wieczorek S, Gross WL, Epplen JT. Promoter polymorphism rs3087456 in the MHC class II transactivator gene is not associated with susceptibility for selected autoimmune diseases in German patient groups. Int J Immunogenet. 2006;33:59–61. doi: 10.1111/j.1744-313X.2006.00568.x. [DOI] [PubMed] [Google Scholar]
- Sadovnick AD, Risch NJ, Ebers GC. Canadian collaborative project on genetic susceptibility to MS, phase 2: rationale and method. Canadian Collaborative Study Group. Can J Neurol Sci. 1998;25:216–221. doi: 10.1017/s0317167100034041. [DOI] [PubMed] [Google Scholar]
- Ramagopalan SV, Dyment DA, Valdar W, Herrera BM, Criscuoli M, Yee IM, Sadovnick AD, Ebers GC. Autoimmune disease in families with multiple sclerosis: a population-based study. Lancet Neurol. 2007;6:604–610. doi: 10.1016/S1474-4422(07)70132-1. [DOI] [PubMed] [Google Scholar]
- Bock C, Walter J, Paulsen M, Lengauer T. CpG island mapping by epigenome prediction. PLoS Comput Biol. 2007;3:e110. doi: 10.1371/journal.pcbi.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning : a laboratory manual. 3rd. Cold Spring Harbor, New York , Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Dyment DA, Risch NJ, Sadovnick AD, Ebers GC. Twin concordance and sibling recurrence rates in multiple sclerosis. Proc Natl Acad Sci U S A. 2003;100:12877–12882. doi: 10.1073/pnas.1932604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Dyment DA, Sadovnick AD, Rothwell PM, Murray TJ, Ebers GC. Timing of birth and risk of multiple sclerosis: population based study. Bmj. 2005;330:120. doi: 10.1136/bmj.38301.686030.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean G, Elian M. Age at immigration to England of Asian and Caribbean immigrants and the risk of developing multiple sclerosis. J Neurol Neurosurg Psychiatry. 1997;63:565–568. doi: 10.1136/jnnp.63.5.565. [DOI] [PMC free article] [PubMed] [Google Scholar]


