Abstract
3-D imaging and analysis techniques can be used to assess the organization of cells in biological tissues, providing key insights into the role of cell arrangement in growth, homeostasis, and degeneration. The objective of the present study was to use such methods to assess the growth-related changes in cell organization of articular cartilage from different sites in the bovine knee. Three-dimensional images of fetal, calf, and adult cartilage were obtained and processed to identify cell nuclei. The density of cells was lower with growth and with increasing depth from the articular surface. The cell organization, assessed by the angle to the nearest neighboring cell, also varied with growth, and reflected the classical organization of cells in adult tissue, with neighboring cells arranged horizontally in the superficial zone (average angle of 20°) and vertically in the deep zone (60°). In all other regions and growth stages of cartilage, the angle was ~32°, indicative of an isotropic organization. On the contrary, the nearest neighbor distance did not vary significantly with growth or depth. Together, these results indicate that cartilage growth is associated with distinctive 3-D arrangements of groups of chondrocytes.
Introduction
A variety of biomaterial scaffolds have been introduced in tissue engineering in an effort to induce repair or regeneration after damage or disease. Such materials have been employed to harbor cells and tissues and to guide tissue formation either in vitro or in vivo. The microstructure and biological activity of the scaffold dictate the formation of tissue by regulating cell infiltration and cell activities. Scaffold features, such as pore size, attachment sites, and fluid channels, may be of critical importance in organizing cells, and, subsequently, generating tissue with structural and functional properties effective for use as a replacement tissue.
The organization of cells into groups often dictates their behavior in functional subunits [1] within tissues. Such organization can affect cells through their regulation, via direct cell-cell contact, and paracrine and autocrine factors. Such organization can also affect the local solid and fluid mechanical environment in which the cells reside, when exposed to external mechanical or chemical stimuli. Thus, it would be useful to assess the precise 3-D organization of cells within native and engineered tissues, as well as biomaterial grafts after cellular infiltration after in vivo implantation. Such analysis of 3-D cell organization would also be relevant to study of cell organization in tissues during development, such as in genetically-manipulated animals, and during disease progression and treatment.
Biomedical imaging techniques are used frequently to characterize the changing microstructure of biological tissues at various stages of development, growth, aging, injury, and disease. Often, the constituent cells are of interest because of their role in mediating tissue formation, repair and degeneration. The changing function of a population of cells may be reflected by the changes in their organization. Thus, delineation of cell organization may lead to a tissue-scale understanding of cell-guided formation, responses to injury, and degradation. This paper provides a brief review of imaging techniques used to examine cells in a 3-D environment, and then describes an investigation of changes in cell organization in the context of articular cartilage development and growth by analysis of fetal, calf, and adult tissue.
Imaging of Cell Populations within Tissues
The traditional method of assessing the organization of groups of cells in tissue has been by analysis of thin sections with 2-D light microscopy. Using such 2-D images, stereological analysis has evolved to allow for unbiased estimation of cell morphology and number per volume [2–5]. In the case of quantifying cell number per volume, new cells are counted in sequential image sections and the number divided by the total volume of tissue inspected. These procedures have proven useful in tabulating depth-varying cellularity in articular cartilage [3,5]. However, evaluation of higher levels of cell organization is difficult because cells are not spatially localized in these procedures and sample thickness may not be large enough to identify entire clusters of cells.
Three-dimensional imaging and image processing modalities have been used to directly analyze cell organization within tissues. Laser-scanning confocal microscopy allows for capture of 3-D tissue features stained with specific fluorescent probes by using pinhole apertures to exclude the excitation beam and collect fluorescence from a thin (in the z direction) optical section [6] More recently, two photon excitation laser scanning confocal microscopy has been employed on a variety of tissues, and can detect endogenous fluorophores excited by absorption of two-photons of twice the wavelength (half the energy) that is normally needed for excitation [7]. Using this technique, cellular components can be visualized to a depth of up to several hundred microns. Another technique, Optical Coherence Tomography, uses a different strategy to collect a series of z data at a single x-y point in combination with scanning in the x-y plane to form a 3-D image [8] In this technique, broadband light is split and passed into the tissue sample and an adjustable path length reference mirror, and the magnitude of the interference between the light reflected from these two targets represents the image intensity. This technique is in contrast to a newer approach, Optical Projection Tomography, in which a series of images are collected from different angles, either of transmitted brightfield light, or by emitted fluorescence signal from excited molecules [9] Using this method, a 3-D image is formed by mathematical reconstruction of the series of views from 0 to 360 degrees, with algorithms similar to those used in micro-computed tomography. This method has the potential to resolve groups of stained molecules (either fluorescent or colorimetric) on the cellular level, up to a depth of 1 cm.
Finally, methods have been developed to obtain a 3-D image by reconstruction of serial images acquired using sequential imaging and physical sectioning. In one such technique, brain tissue was imaged at a surface using two-photon laser scanning confocal microscopy, and then laser-ablated to expose the next deeper layer [10]. This process was repeated to form a 3-D image with high resolution achieved by two-photon confocal microscopy, and size sufficient to visualize 3-D structures on the millimeter length scale. In another commercial procedure, called digital volumetric imaging (DVI), 2-D images of a tissue surface are captured using conventional fluorescence microscopy, and thin sections are sequentially removed by a diamond knife microtome [11]. This procedure involves (A) fluorescence staining and fixation of tissue and embedding with an opacified polymer, (B) automated serial sectioning and imaging through a 3-D tissue block, and (C) post-processing for viewing the image data and extracting image features. In the plane (x-y) of sectioning, resolution and field of view are determined by the objective lens and imaging camera. In the depth (z-axis) direction, resolution is determined by section thickness (with opacity of the polymer matched to achieve a surface measure), and the number of z-sections is effectively unlimited. This technique was originally developed using specific fluorescent markers for cell nuclei (Acridine Orange) and extracellular matrix (Eosin-Y),although its capability may be extended to identifying cell subpopulations with distinct phenotypes by incorporation of more specific fluorescent markers such as tagged antibodies. Recent studies have demonstrated the accuracy of DVI imaging (versus confocal imaging) [12], and the efficacy in quantifying the depth-varying organization of cells in conjunction with validated image processing routines, with high sensitivity and specificity, in bovine articular cartilage at different stages of growth [13]. The current study uses these DVI methods to localize cells in a series of samples from animals at distinct growth stages, and to analyze cell organization by computing novel metrics of distance and angle from each cell to neighboring cells.
Cell Organization Changes in Articular Cartilage during Growth
Articular cartilage undergoes substantial growth and functional maturation between the time of joint cavitation and attainment of skeletal maturity [14]. The organization of chondrocytes in cartilage reflects, and may contribute to, articular cartilage dynamics during fetal development and post-natal growth, and at skeletal maturity [15]. In particular, the decrease in cell density during growth through the full tissue volume results from cells’ separation during accretion of extracellular matrix molecules. The relatively high density of evenly-spaced chondrocytes in immature tissue are arranged favorably to actively deposit and remodel nearby extracellular matrix, essential to tissue growth and maturation. The higher density of cells, and tight vertical packing of horizontally oriented clusters of cells, near the articular surface [16], may enhance growth in this region. On the other hand, in mature articular cartilage, the low density of cells together with their clustering leads to regions of tissue relatively far from the nearest cells. This structure may predispose tissue to ineffective remodeling and repair and make it highly susceptible to structural and functional deterioration. This effect may be exacerbated in the deeper tissue regions, where cells are in sparse groups arranged in vertical columns [17]. Analysis of the depth-variation in cell organization at different stages of growth and maturation allows further understanding of the role of the indwelling cells in articular cartilage growth and homeostasis.
In immature articular cartilage, the population of cells is situated to contribute to a highly anabolic state of tissue growth. From previous 3D analysis of cell locations in immature tissue, a large proportion of the tissue is within ~10 µm of the nearest cell [13], within the distance over which a cell can actively metabolize proteoglycan molecules [18]. This proximity to tissue results from both the high density of chondrocytes, as well as their arrangement. Near the articular surface, horizontal clusters of tightly packed, flattened chondrocytes are observed, whereas deeper in the tissue, cells are less dense but have a more homogeneous arrangement [17]. The way in which the cells reach this arrangement is not known; however, the increase in the distance between cells with growth suggests that accretion of extracellular matrix may be pushing cells apart, while the closely situated and horizontally-oriented cell groups at the articular surface may be being generated by cell proliferation. Recently, proliferating chondrocytes have been identified near the articular surface of immature tissue [19–23], as have progenitor or stem cells [24–27]. The organization of cells in immature tissue is likely to facilitate a high rate of tissue growth, and the position of cells relative to one another may provide insight into key features of growth, such as matrix accretion and cell proliferation.
Upon maturation, the cartilage attains a classical cell organization that is associated with maintenance of tissue homeostasis [17]. In particular, cell density decreases with depth from the articular surface [5, 28, 29], and cells are arranged into characteristic groups ranging from horizontal clusters at the surface to vertical columns in the deep regions [5, 16, 17]. Previous studies have also found that the cell density varies across the joint surface, and may depend on the magnitude of load which it bears [3, 30]. Thus, the intrinsic and extrinsic cellular cues may be different across a joint surface and lead to variable growth and evolution of distinct cell groups. The progression of articular cartilage cell organization to the final heterogeneous state may be better understood by quantifying the local organization of cells and cell groups as a function of depth and joint site at a sequence of developmental stages.
We recently described a 3-D histological method for determining the location of chondrocytes in cartilage in conjunction with DVI [13]. Although the study focused on determining cellularity in 3-D image datasets, these methodologies could be extended to other higher order metrics of chondrocyte organization, particularly the relative arrangements of small groups of cells. 3-D imaging and image processing could be applied to quantify features of cell organization, and assess the variation with growth.
The objective of the present study was to use DVI imaging and 3-D image processing methods in order to quantify the cellularity and local organization of cells in bovine articular cartilage, as a function of depth from the surface, at different stages of growth, both for the weight-bearing femoral condyle (FC) and the intermittently-loaded patellofemoral groove (PFG).
Materials and Methods
Sample Preparation
Articular cartilage was harvested from two sites of the bovine distal femur at different growth stages and prepared for 3-D histology. A total of 18 osteochondral blocks, 1 cm3 in volume, were harvested from the lateral femoral condyle and patellofemoral groove of 3 fetal (2nd trimester, 229±11 days gestation) [31], 3 calf (1–3 wk), and 3 adult (1–2 yr) bovine stifle joints, obtained from an abattoir. Samples were fixed in 4% paraformaldehyde in PBS at pH 7.4 and 4°C for 3 days, and then trimmed to pieces ~1.5 mm × ~3 mm in surface area and to a depth 4 of ~1.5 mm from the articular surface.
3-D Histology
Articular cartilage pieces were stained and imaged to localize cell nuclei within the 3-D cartilage block using DVI as described previously [12, 13]. Briefly, samples were stained using Eosin Y and Acridine Orange, sequentially dehydrated in graded alcohol, and held in xylene until processing. Samples were then embedded in Spurr resin with 50% opacity, so that the image depth effectively matched the thickness of material removed (1.77 µm), and 3-D images were acquired using DVI (MicroSciences Corp., Corte Madera, CA) at a voxel resolution of (1.77 µm)3.
3-D Image Processing
The cell organization in image datasets was analyzed qualitatively and quantitatively using Matlab® 7.1 software (The Mathworks, Inc., Natick, MA). Image subsets with a surface area of (0.2 mm)2 and a depth of ~1.0 mm were exported as a stack of 2-D images from RESView 3.1™ (MicroSciences Corp.). Image stacks were imported into Matlab® 7.1 to form a 3-D image matrix. Three-dimensional images were transformed so that the articular surface plane was parallel to the x-y plane and depth from the articular surface aligned with the z axis using a two step process. First, images were rotated in 3-D until the surface was approximately parallel to the x-y plane by inspection. Second, the articular surface was approximated by the best-fit plane through 9 reference surface points in a 3 by 3 grid, and the dataset transformed to a coordinate system where this best-fit plane was Z=0 everywhere [13].
Image datasets were processed to localize cell nuclei by their 3-D centroid location as described previously [13] with additional enhancement by 3-D spatial filtering. Briefly, the intensity was normalized as a function of depth, so that the average background intensity was 80 and the average nucleus intensity was 160. Next, image contrast of cell nuclei was enhanced relative to the background by 3-D convolution with a 5 × 5 × 5 Mexican Hat filter with σ equal to 1. In this way, objects of similar size and shape to the cell nuclei with a high intensity were enhanced, and high intensity, high frequency noise or low frequency background was attenuated. Images were again intensity normalized as a function of depth, as differently shaped nuclei through the tissue depth were enhanced to different degrees. Images were intensity segmented at a threshold of 130, determined by analysis of pixel histograms [13], into object and background voxels, and connected object voxels were grouped to form individual objects (cell nuclei). Accurate segmentation was confirmed by overlaying the segmented image on the original grayscale image, and estimating false positive and false negative objects relative to true positive results in different depth regions. Accuracy was found to be high (>95%) in a subset of 200 µm × 200 µm × 100 µm regions at varying depths in each of the samples analyzed, similar to that reported quantitatively previously [13]. Centroid locations were translated in the Z direction so that the most superficial cell nucleus was at Z=0. The number of cells localized for fetal, calf and adult samples were 7800±1200, 5100±600, and 2100±200, respectively.
A number of features of cell organization were assessed through the superficial, middle, and deep zones of adult cartilage, and corresponding depth regions of fetal and calf tissue. Cellularity was computed as a function of Z in successive 100 µm thick layers to a depth of 800 µm, excluding 5 µm at each x and y edge to eliminate bias from nuclei touching the image dataset boundary. Consistent with previous analyses using this method the intra-donor (within a donor) variability was small relative to the inter-donor (across donors) variability for the metric of cell density. Within a donor, the standard deviation had an average value over all depth layers of 15, 9, and 3 million cells per cm3 for fetal, calf, and adult samples, respectively, while across donors the standard deviation was much larger, being 53, 26, and 10 million cells per cm3. Thus the variability within the site sampled was small relative to that across donors. For each cell nucleus centroid, the nearest neighbor was identified as the closest cell nucleus centroid, and the distance and angle (relative to the x-y plane) to it were calculated, with an angle of 0° indicating a horizontal orientation and 90° indicating vertical. To eliminate edge effects, cells that were closer to the edge of the region of interest than to their nearest neighboring cell were not included. The average distance and angle was calculated as a function of depth in 100 µm thick layers to a depth of 800 µm. Finally, mutual neighbors were identified as those cells that were themselves the nearest neighbor of their nearest neighbor to quantify the distinct pairing of cells. The proportion of cells that were mutual neighbors was computed as a function of depth from the articular surface.
Simulations
For comparison of the above metrics of cell organization (angle to nearest neighboring cell and proportion of cells that were mutual neighbors), analyses were also performed on cells positioned randomly. For each image dataset, an equal sized image volume was created and cells were added randomly (using a random number generated for each of the x, y, and z axes) so cell density was equivalent in each tissue depth bin (200 µm × 200 µm × 100 µm), from 0–800 µm. The average nearest neighbor angle and proportion of mutual neighbors were calculated for each bin as described above. Statistical analyses (performed as described below) indicated that these 3-D metrics for such randomly organized cells did not vary significantly with depth, age, or site (i.e., with cell density, as described below). The average angle to the nearest neighboring object was 32.3±1.2°, and the average proportion of mutual neighbors was 0.588±0.017.
Statistics
Data are presented as mean ± SEM. ANOVA was performed with significance level of 0.05 for the measures of cellularity, nearest neighbor distance, nearest neighbor angle, and mutual neighbor proportion, with growth stage as the main factor, and depth and joint site as the repeated measures. For the measures of cellularity and nearest neighbor distance, Tukey post-hoc pairwise comparisons were made between growth stages. For the measures of nearest neighbor angle and mutual neighbor proportion, the values from each growth stage were compared against those from computer simulations of randomly distributed cells, using a planned comparison with a Bonferroni adjustment of the p-value (×3). Where applicable, p values are presented after correction.
Results
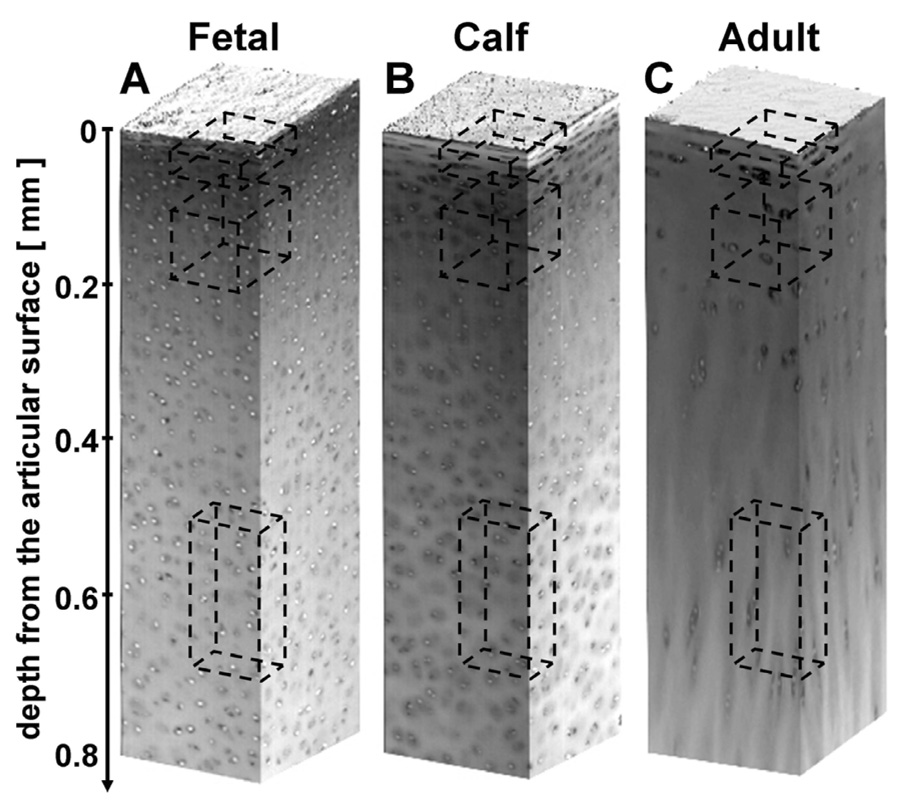
Qualitatively, the 3-D images clearly demonstrated cells and their nuclei, allowing for automated localization of cell nuclei (Figure 1A–C). The methods described for spatially normalizing nuclei and background intensities resulted in images that were accurately segmented using a consistent intensity threshold of 130. The sensitivity and specificity of cell nucleus localization was high (~95–100%) for all growth stages and tissue depths as assessed by inspection of raw image stacks overlaid with segmented images.
Figure 1.
The depth-varying organization of cells depicted by 3-D image renders of bovine articular cartilage at different growth stages. Raw images of fetal (A), calf (B), and adult (C) tissue, with dashed boxes indicating regions depicted in Figure 2.
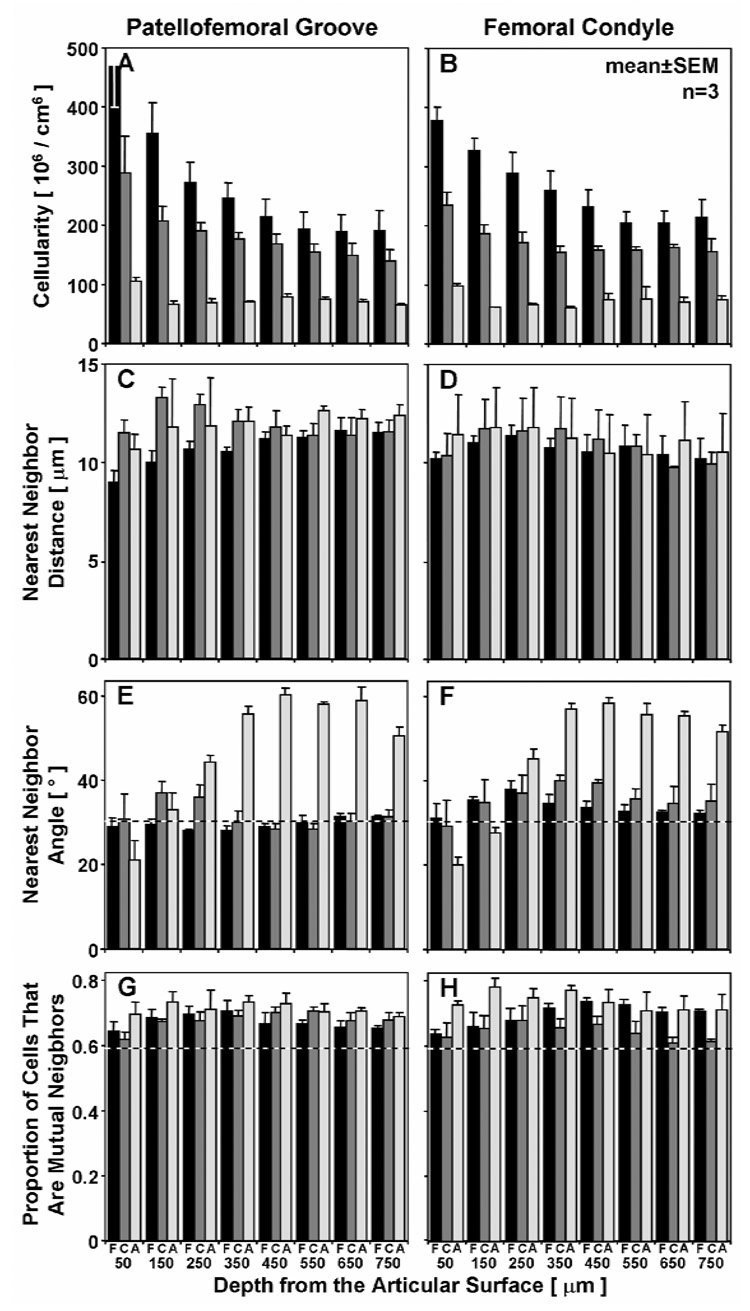
The density of cells in a given tissue volume depended on the growth stage and the depth from the articular surface (Figure 1, Figure 2, Figure 3A,B). The cellularity of articular cartilage decreased markedly with maturation (p<0.001), with reductions of 30% (fetal to calf) and 60% (calf to adult) in the tissue volume analyzed. The cellularity also decreased with depth from the articular surface (p<0.001). This depth-variation in cellularity was more marked for more immature tissue (depth-growth stage interaction, p<0.001), decreasing from 420±50 million cells/cm3 at 0–100 µm to 200±30 at 700–800 µm in fetal tissue, 260±20 to 150±5 in calf, and 100±6 to 70±4 in adult. The depth-associated variation in cellularity was also distinct in the PFG and FC (p<0.001), especially relative to the superficial (0–100 µm) region.
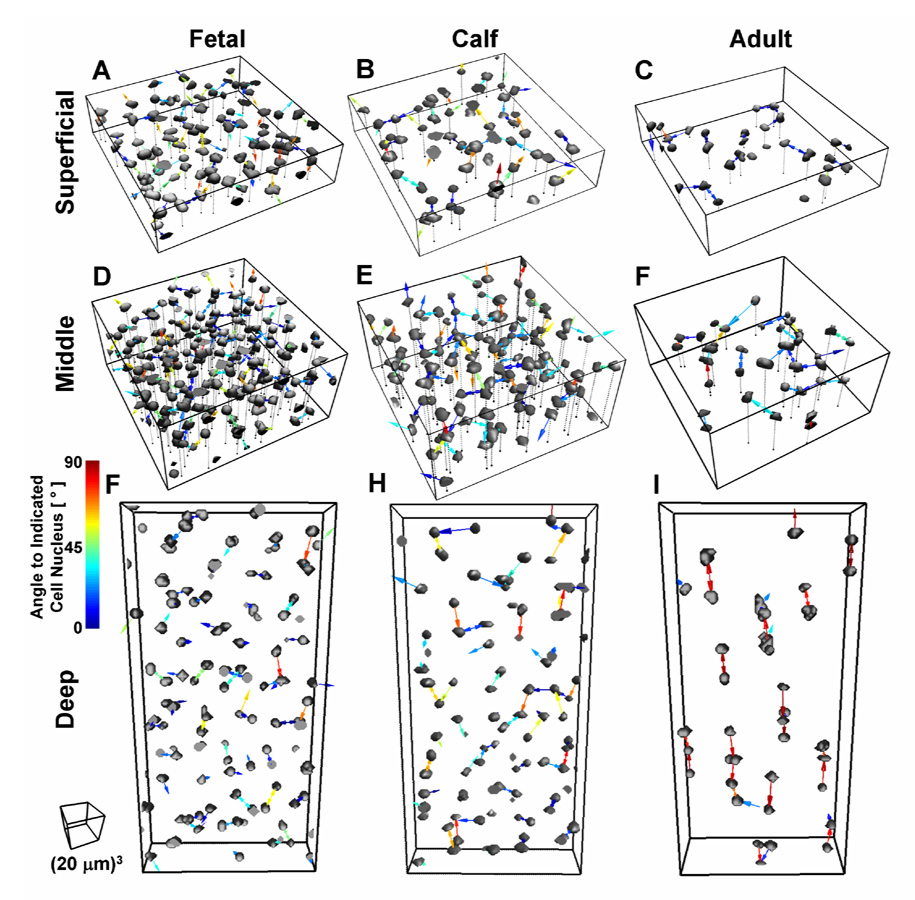
Figure 2.
Renders of isolated cell nuclei depicting cell organization highlighted by nearest neighbor vectors. The angle of vector in the z direction relative to the x/y plane is indicated by the color legend from 0–90°. Data presented for fetal, calf, and adult tissue in subsets of the superficial (0–100 µm), middle (100–200 µm) and deep (400–600 µm) tissue regions.
Figure 3.
Variation in cell organization with depth in fetal (▪), calf ( ), and adult (□) bovine articular cartilage. Cell density (A,B), nearest neighbor distance (C,D), nearest neighbor angle (E,F), and proportion of mutual neighbors shown for the patellofemoral groove (A,C,E,G) and femoral condyle (B,D,F,H) from the articular surface to 800 µm in depth.
), and adult (□) bovine articular cartilage. Cell density (A,B), nearest neighbor distance (C,D), nearest neighbor angle (E,F), and proportion of mutual neighbors shown for the patellofemoral groove (A,C,E,G) and femoral condyle (B,D,F,H) from the articular surface to 800 µm in depth.
Cell organization also varied with growth and tissue depth (Figure 1, Figure 2, Figure 3C–H). Adult cartilage demonstrated horizontal clusters in the superficial zone, and oblique and vertical columns in the middle and deep zones, respectively (Figure 1, Figure 2). Quantitatively, the 3-D metric of angle to the nearest neighbor was sensitive to such organization, being significantly different across growth stages (p<0.001) and through the depth of the tissue (p<0.001) with an interactive effect (p<0.001, Figure 3E,F). In mature tissue, this angle increased from 20° in the superficial zone (0–100 µm) to 60° in the deep zone (>500 µm), while in fetal and calf tissue the angle was ~32° except in certain regions (100–600 µm, especially in the FC), where the angle was intermediate to that in adult deep tissue (~40°). In contrast, the distance to nearest neighbor (Figure 3C,D) did not vary with age (p=0.60) or site (p=0.21), and showed little and insignificant variation (average distances of 8–13 µm) in depth (p=0.06), with all average distances only varying from 8–13 µm. The proportion of neighbors that were mutual also did not change significantly with growth stage, from 68±3% in fetal, 66±3% in calf, and 72±3% in adult, but did vary with depth (p<0.001). The proportion was higher than the average of ~59% calculated for a 3-D volume with randomly distributed objects (Figure 3G,H), achieving 1 significance (p<0.01) at all depths of adult tissue, and most depths of the immature fetal and calf (p<0.05).
Scatter plots and averages of angle versus distance to the 1st, 2nd, and 4th nearest neighboring cells further highlighted the distinct populations of cells with preferentially oriented neighbors in adult tissue and the isotropic organization in fetal and calf (Figure 4, Figure 5). In the surface-most bin, the angles to the nearest neighboring cells were distributed from 0 to 90° in fetal and calf, but showed a high concentration near 0° in the superficial zone of adult tissue, and near 90° in the deep zone. 3-D renders of cell nuclei with the vectors to the nearest neighboring cell nuclei (Figure 2) further illustrate the horizontal orientation of neighboring cells in the superficial zone and the vertical orientation in the deep zone in the adult, and the isotropic arrangement in fetal tissue. These trends continued for the 2nd nearest cell where the average angle was 26° in the superficial and 43° in the deep, and a smaller proportion of the cells had horizontal (in the superficial) or vertical (in the deep) orientations. By the 4th neighbor, the average angle approached that for isotropic tissue (~32°) in all growth stages and depths. The scatter plots also illustrate the consistent distance of nearest neighboring cells across depth and growth stage, despite substantial changes in cellularity. On the other hand, the trends become more consistent with cellularity in the 2nd and 4th nearest neighbors, where distances increased with decreasing cellularity, and the distance to the 4th neighbor varied from ~15 µm in the middle regions of fetal tissue, to 18 µm in calf, and 24 µm in adult (Figure 5).
Figure 4.
Depth- and growth-associated variation in vectors to neighboring cells. The angle and distance to the A) 1st, B) 2nd, and C) 4th nearest neighboring cell nuclei are plotted for 1000 cell nuclei along with the mean±SD in the superficial (0–100 µm), middle (100–200 µm), and deep (400–600 µm) regions of fetal( ), calf (
), calf ( ), and adult (
), and adult ( ) tissue.
) tissue.
Figure 5.
Depth-and growth-associated variation in average vector to neighboring cells. The average angle and distance to the 1st, 2nd, and 4th nearest neighboring cell nuclei are plotted for the superficial (S), middle (M) and deep (D) regions of fetal (A), calf (B), and adult (C) tissue. Error bars were not included for simplicity, since Figure 3 shows 1st neighbor SEM, and 2nd and 4th neighbor SEMs are similar.
Discussion
This study investigated the variations in the organization of cells in the superficial-most 800 µm of articular cartilage on the lateral patellofemoral groove and femoral condyle in three stages of growth in bovine stifle joints. Using previously described 3-D imaging and analysis methods, and novel metrics of the distance and angle to neighboring cells, the organization of cells was characterized in the various tissue regions. The cell density decreased monotonically with growth from the fetal to calf to adult stage, and with depth from the articular surface in a way that varied with joint site (Figure 1, Figure 2, Figure 3A,B). Despite the variations in cell density, the distance to the nearest neighboring cell did not vary significantly (Figure 3C,D). On the other hand, the angle to the nearest neighboring cell was sensitive to changes in cell organization by the adult stage of growth, being particularly low (20°) in the superficial zone and high (60°) in the deep zone (Figure 2, Figure 3E,F, Figure 4 and Figure 5). Further, cells tended to associate as small groups, with a higher incidence of mutual neighbors (68% for fetal, 66% for calf, 72% for adult) than expected by chance (59%) (Figure 2, Figure 3G,H). The growth-associated changes in cell organization highlighted in this study may offer further insight into the role of populations of cells in articular cartilage growth, and the evolution of characteristic groupings of cells witnessed in mature tissue.
The study focused on a region of articular cartilage at the articular surface, including the superficial, middle, and deep zones of adult tissue, and corresponding depths of fetal and calf tissue. This region, of ~1 mm thickness, extends through only a small proportion (~10%) of the total thickness of fetal and calf tissue (>1 cm). The depth zones of cartilage are traditionally defined by the morphology of cells and cell groups, the structure of the extracellular matrix, or the phenotype (secretion of zones-specific molecules such as superficial zone protein). In terms of cell morphology, articular cartilage is characterized by flattened cells in the superficial zone, oblique groups of cells in the transitional (middle) zone, and vertical columns of cells in the radial (deep) zone (16). In the immature tissue analyzed here, there is a thin layer (<50 µm) of flattened cells near the superficial zone of cartilage (Figure 1), followed by spheroidal cells in small groups, typical of the middle zone; the classical deep zone may not be present until later in the development process (22, 31). Thus, the regions of tissue analyzed here may not provide a direct comparison of different zones of cartilage, as defined classically. On the other hand, cellularity decreases with depth from the articular surface (Figure 3A,B), stabilizing at 500–800 µm in depth in the fetal, calf, and adult tissues, studied here. Finally, much of the cartilage present in the immature joint is destined for calcification by expansion of the underlying secondary center of ossification, whereas the most superficial tissue gives rise to the articular cartilage layer of the mature joint. The organization of adult cartilage may also be specialized at and near the calcified cartilage region, but was not investigated in this study.
The 3-D image datasets generated using digital volumetric imaging were analyzed using similar methods to the previous study, but improved by application of 3-D spatial filtering. The image datasets showed staining consistent with the previous study [13], with intense bright nuclei and non-specific staining of background extracellular matrix. In some sample regions, the intensity of this background staining exceeded that of nuclei. This would have led to classification of matrix inappropriately as cells, i.e. false positive error (>20%). To prevent such errors, 3-D convolution using a Mexican Hat filter was employed prior to segmentation in all images to enhance the cell nuclei and attenuate the background. The robustness of the previously developed methods in conjunction with 3-D spatial filtering was demonstrated by its efficacy in the heterogeneous fetal, calf, and adult tissue, from the articular surface to a depth of 800 µm.
The cellularity quantified here from 3-D imaging shows variation with age, depth, and joint site consistent with other biochemical and histological studies. From biochemical analysis of DNA content in developing and growing bovine tissue, the cellularity decreased with growth stage [14, 32] and depth from the articular surface [33], as in the current study. The reduction in cell density with depth from the articular surface of adult articular cartilage has been identified in numerous 2-D and 3-D histological studies in a variety of animals [2–5, 28, 29] The effect of joint site on cell density seen in other studies [3, 30] was apparent at certain depths from the articular surface, but not when overall tissue thickness was considered. The more subtle differences observed in the current work could be due to the location of the source tissue. In this study, joint sites with very different loading regimes, such as the weight-bearing region (central femoral condyle) and non weight-bearing region (central patellofemoral groove) were examined, as opposed to the medial vs. lateral [30] or central vs. peripheral [3] femoral condyle. Absolute estimates of cell density were consistent with other studies [13, 33]. The cell density in the fetal tissue was high in the present study compared to our previous study (~450 compared to 290 million cells per cm3 in the superficial zone) [13], likely due to sampling at an earlier stage of growth (2nd trimester compared to 3rd trimester).
The analysis of cell organization revealed a relatively constant distance to the nearest neighboring cell, despite the substantial decrease in cellularity with depth and growth stage. These results suggest that small groups of 2–3 cells are closely situated, whereas the gap between these groups increases from fetal to calf to adult as illustrated by an increase in the 4th nearest neighbor with growth. Further, nearest neighboring cells were often mutual (Figure 2, Figure 3G,H), suggesting the presence of pairs of cells with some sort of affinity, either through cell-cell adhesion, or through a shared pericellular matrix. These groups seem to remain during growth through the calf to the adult stage, as the proportion of cells in the tissue decreases dramatically but the distance to nearest cell does not change, and cells are mutual neighbors with a high incidence. Such a phenomenon may also arise from active cell fate processes of proliferation and death, where cells throughout the tissue are constantly being removed by apoptosis or necrosis, and regenerated by proliferation as the tissue expands, leading to closely situated daughter cells. Previous studies [21, 23] have investigated the role of cell death in developing articular cartilage, and identified only very localized bands of cell death near the subchondral bone and proliferation either near the bone or the articular surface [19–23]. A more detailed analysis of cell fates and organization during growth may uncover the role of cell-cell interactions and cell kinetics.
The angle to the nearest neighboring cell showed small variations from an isotropic organization in the femoral condyle of immature tissue. This result may suggest an earlier accelerated maturation process occurring in weight-bearing regions relative to intermittently loaded regions, consistent with the previous growth related studies showing larger increases in collagen content [14, 34], DNA, GAG, and collagen precursors [34], and tensile properties [32] hallmarks of maturation, from birth to early immature stages of growth. Further, horizontal clusters have been qualitatively shown in immature tissue from en face views in the superficial zone [13]. However, because of the relatively dense packing of cells in immature tissue, the nearest neighboring cell may often reside in a different cell cluster. Further development of image processing methods could allow characterization of cell clusters within densely packed groups of cells.
The change in angle of neighboring cells from horizontal to vertical from the superficial to deep zones of adult articular cartilage may be physiologically important, although this remains to be elucidated. Some authors have proposed models of articular cartilage growth which mimic that of the growth plate, where cells in progenitor and proliferating pools progress through to the hypertrophic zone and become calcified as the bone increases in length. In such a model, the groups of cells that arise in the superficial zone, possibly from proliferation of a progenitor-like cell residing in the region [19–27], advance through to the deep regions, becoming aligned vertically. Also possible is a relationship between the orientation of groups of cells within a pericellular region and the local collagen fibers. In the superficial zone, the collagen is oriented parallel to the surface, arcading downward through the middle zone, eventually becoming vertical in the deep zone [35]. Thus in the superficial zone, horizontal clusters of cells may reside in between sheets of collagen, where in the deep regions, the vertical columns of cells may inter-digitate with columns of collagen fibers.
Analysis of the distance and angle to neighboring cells beyond the 1st is consistent with the presence of multi-cell units in adult cartilage (Figure 4, Figure 5). In the superficial zone, the angle to 2nd nearest neighboring cell is 26°, suggesting that this neighbor still has a slight tendency toward a horizontal orientation to the original cell and clusters of 3 or greater cells may be present. In the deep zone, the 2nd nearest neighboring cell has an average angle of 43 degrees, reflecting columns of at least 3 cells. The scatter plots confirm this result, showing a small but distinct population of cells with a 2nd nearest neighbor still at an angle near 0° in the superficial and near 90° in the deep zone.
These quantitative metrics may be used to identify and characterize local subpopulations with distinct arrangements, as well as to describe global variations in the organization of cells in a tissue volume. The dynamics of biological tissues such as cartilage may be dependent on the activity of small distinct subpopulations of cells. During growth, the articular surface contains progenitor cells, and may also contain an adjacent small population of ‘transit amplifying’ cells that are undergoing proliferation. These events may be rare, especially in more mature tissue, yet still contribute substantially to the expansion of the cell population governing tissue growth. Such rare events could potentially be identified by appropriately validated metrics, such as proximity of neighboring cell nuclei (<5µm), or orientation of mutual neighbors. Further, focal sites of pathogenic cell death could be localized by identifying cells or tissue regions relatively far from the nearest cell, amidst a population of normally arranged chondrocytes, and may be associated with tissue undergoing early yet treatable pathogenic degeneration. The identification of anomalous cell groups within a large tissue sample may reveal the rare events critical for normal growth, and those detrimental to maintenance of joint homeostasis.
This paper describes the application of 3-D imaging and analysis to deduce the changes in cell organization that occur during in vivo growth of a biological tissue, namely articular cartilage. This cell organization changes dramatically during growth, with the expanding articular cartilage undergoing a decrease in cell density and ultimately attaining the classical zonal organization described previously. The novel metric, determined from 3-D images, of angle to nearest neighboring cell may be particularly useful in gauging the extent and mechanisms of tissue maturation. Further investigation of chondrocyte populations in cartilage during growth, aging, injury, and degradation, may uncover the physiological significance of the cell arrangements described here. Additionally, investigation of cells at this scale within other native and engineered biological tissues may uncover tissue-specific patterns of growth, and the role of cell populations and sub-populations in development of complex tissue geometries and microstructure as well as the propagation of tissue degeneration and disease. Finally, these metrics may be used to assess the organization of cells in biomaterial scaffolds engineered for implantation, or in biomaterial grafts after infiltration of cells and tissue in vivo. Such measures may highlight the behavior of cells and cell groups in such scaffolds, and offer insight into more effective design of scaffolds for use as a support mechanism for regeneration of native tissues.
Acknowledgments
This work was supported by NIH and NSF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kyle D. Jadin, Department of Bioengineering, University of California-San Diego, 9500 Gilman Drive, MC 0412, La Jolla, CA 92093-0412, Tel: (858) 534-0821, Fax: (858) 822-1614
Won C. Bae, Department of Bioengineering, University of California-San Diego, 9500 Gilman Drive, MC 0412, La Jolla, CA 92093-0412, Tel: (858) 534-0821, Fax: (858) 822-1614
Barbara L. Schumacher, Department of Bioengineering, University of California-San Diego, 9500 Gilman Drive, MC 0412, La Jolla, CA 92093-0412, Tel: (858) 534-0821, Fax: (858) 822-1614
Robert L. Sah, Department of Bioengineering, University of California-San Diego, 9500 Gilman Drive, MC 0412, La Jolla, CA 92093-0412, Tel: (858) 534-0821, Fax: (858) 822-1614, Email: rsah@ucsd.edu