Abstract
Blooms that are formed by cyanobacteria consist of toxic and nontoxic strains. The mechanisms that result in the occurrence of nontoxic strains are enigmatic. All the nontoxic strains of the filamentous cyanobacterium Planktothrix that were isolated from 9 European countries were found to have lost 90% of a large microcystin synthetase (mcy) gene cluster that encoded the synthesis of the toxic peptide microcystin (MC). Those strains still contain the flanking regions of the mcy gene cluster along with remnants of the transposable elements that are found in between. The majority of the strains still contain a gene coding for a distinct thioesterase type II (mcyT), which is putatively involved in MC synthesis. The insertional inactivation of mcyT in an MC-producing strain resulted in the reduction of MC synthesis by 94 ± 2% (1 standard deviation). Nontoxic strains that occur in shallow lakes throughout Europe form a monophyletic lineage. A second lineage consists of strains that contain the mcy gene cluster but differ in their photosynthetic pigment composition, which is due to the occurrence of strains that contain phycocyanin or large amounts of phycoerythrin in addition to phycocyanin. Strains containing phycoerythrin typically occur in deep-stratified lakes. The rare occurrence of gene cluster deletion, paired with the evolutionary diversification of the lineages of strains that lost or still contain the mcy gene cluster, needs to be invoked in order to explain the absence or dominance of toxic cyanobacteria in various habitats.
Keywords: toxicity, microcystin, mobile elements, gene loss, microevolution, ecotypes
Introduction
Harmful algal blooms that are formed by cyanobacteria in lakes and rivers frequently contain toxins, such as the hepatotoxic peptide microcystin (MC), and they pose a serious health risk to livestock and humans (World Health Organization 2004). It has been known for a long time that the blooms that are formed by the genera Microcystis sp., Planktothrix spp., and Anabaena spp. consist of hepatotoxic and nonhepatotoxic strains (Carmichael and Gorham 1981). A similar variation has been recorded among dinoflagellates that produce neurotoxins (Touzet et al. 2007) and fungi that produce mycotoxins, for example, Aspergillus sp. that produce aflatoxin (Cary and Ehrlich 2006). The factors that regulate the ecological success of nontoxic and toxic strains in cyanobacteria are intriguing. However, no clear answer has been obtained with regard to the biological function of the toxins or the factors favoring nontoxic over toxic strains and vice versa (Orr and Jones 1998; Schatz et al. 2005; Kardinaal et al. 2007).
For cyanobacteria, it has been found that hepatotoxic and nontoxic strains differ in their content of microcystin synthetase (mcy) genes that encode specific peptide synthetases, which have been shown to be involved in the production of the toxic heptapeptide MC (Meißner et al. 1996; Dittmann et al. 1997). The large gene cluster (∼55 kbp) consists of nonribosomal peptide synthetases (NRPS), polyketide synthases, and tailoring enzymes (Tillett et al. 2000). The gene clusters of species from 3 genera (Microcystis, Planktothrix, and Anabaena) have been sequenced, and comparisons revealed that the genes mcyA, mcyB, mcyC, mcyD, mcyE, mcyG, and mcyJ that are involved in MC synthesis are always present (Tillett et al. 2000; Christiansen et al. 2003; Rouhiainen et al. 2004), whereas the 3 genera can differ in the presence of tailoring enzymes. A phylogenetic tree that was calculated from 2 housekeeping genes, the 16S rDNA and rpoC1 genes, showed perfect congruency with the phylogeny that was calculated from mcyA, mcyD, and mcyE (Rantala et al. 2004). This implies that the mcy gene cluster is of monophyletic origin and that several lineages must have lost the mcy gene cluster during cyanobacterial evolution. On a species level on a much shorter timescale of evolution, most closely related strains are observed either containing or lacking the mcy gene cluster. In blooms that are formed by Microcystis, the percentage of the strains containing the mcy gene cluster is low (1–38%, Kurmayer and Kutzenberger 2003). In contrast, blooms that are formed by the red-pigmented (phycoerythrin-rich) populations of Planktothrix rubescens that occur in deep-stratified lakes in the Alps, Scandinavia and in reservoirs solely consist of strains that contain the mcy gene cluster (Kurmayer et al. 2004). Accessory photosynthetic pigments, such as the red-colored phycoerythrin, result in the ability to live under transparent light conditions in deep water layers. Under specific climatic conditions, red-colored blooms are observed, which are then called the “Burgundy-blood phenomenon” (Walsby et al. 2005). Green-pigmented (phycocyanin-rich) blooms of Planktothrix agardhii that flourish in more shallow and polymictic lakes (Davis and Walsby 2002) typically show a lower percentage of strains containing the mcy gene cluster (Kurmayer et al. 2004).
The process of the acquisition/loss of mcy genes among closely related strains is not understood. The sequencing of the mcy gene clusters and the closely related nodularin synthetase gene cluster has revealed the localization of open reading frames (ORFs) with a significant similarity to transposases at the downstream 3′ end of the gene clusters (Tillett et al. 2000; Moffitt and Neilan 2004). Recently, a type IV pilus system has been described in Microcystis and has been suggested to allow for the receiving of the mcy gene cluster via lateral transfer (Nakasugi et al. 2007). The phylogenetic congruence between the 2 housekeeping genes and the mcy genes rules out the possibility of the horizontal transfer of the mcy gene cluster between genera (Rantala et al. 2004). However, the frequency of the transfer of the mcy gene cluster among other closely related strains remains unclear. In order to find out whether the variation in toxicity among strains in Planktothrix resulted from gene loss, 25 nontoxic strains were analyzed for remnants of the mcy gene cluster. In all the nontoxic strains, mcy operon remnants were found that were derived from a major gene cluster deletion event that succeeded the insertion (IS) of mobile elements.
Materials and Methods
Organisms
The 62 strains that were used for this study were either isolated from European lakes (44 strains) or obtained from culture collections (table 1 and fig. 1). The strains originated from 9 countries and 28 different water bodies. All the strains were assigned to either P. rubescens or P. agardhii following the criteria published previously (Suda et al. 2002). All the strains were grown in BG11 medium (Rippka 1988) at 15 °C and under low light conditions (10 μmol m2 s−1, Osram L30W/77 Fluora).
Table 1.
Planktothrix agardhii and Planktothrix rubescens Strains that Were Used in the Present Study and Grouped According to the Presence and Absence of the mcy Gene Cluster
| Strain Number | Species | North(°) | East(°) | Origin | Water Depth Zmean/Zmaxa |
| Strains not containing the mcy gene cluster | |||||
| 250I, 251I, 252IV, 253II, 254II, 255I, 256II, 257I | P. agb | 39°20 | 0°21 | Albufera Lagune, Valencia, Spain | 1/3 |
| PCC7811II | P. ag | 48°51 | 2°20 | Vert-le-Petit, France | 2/2 |
| 41II, 63II, 66II | P. ag | 48°49 | 15°16 | Jägerteich, Austria | 1/2 |
| 259II, 263II, 274II, 277II, 281II | P. ag | 52°31 | 13°20 | Wannsee, Germany | 6/9 |
| 299II, 307II, 320II | P. ag | 52°02 | 5°02 | Klinckenberger Plas, The Netherlands | -/30 |
| PCC7805I | P. ag | 52°21 | 4°52 | Veluwermeer, The Netherlands | 2/5 |
| SAG5.81III | P. ag | 51°32 | 9°57 | Kiessee, Göttingen, Germany | 1/2 |
| CCAP1459/15II | P. ag | 54°36 | 6°23 | Lough Neagh, North Ireland, United Kingdom | 9/34 |
| PH22II | P. ag | 55°40 | 12°34 | LakeBagsværd Sø, Copenhagen, Denmark | 2/3 |
| 2AII | P. ag | 60°14 | 19°55 | Lake Markusbölefjärden, Finland | -/9 |
| Strains containing the mcy gene cluster | |||||
| 31/1, 32, 39, 260 | P. ag | 52°31 | 13°20 | Wannsee, Germany | 6/9 |
| SAG6.89 | P. ag. | 54°10 | 10°23 | Plußsee, Germany | 9/30 |
| CCAP1459/11A | P. ag | 54°21 | 2°56 | Lake Windermere, United Kingdom | 21/64 |
| CCAP1459/21 | P. ag. | 54°21 | 2°58 | Esthwaite Water, United Kingdom | -/16 |
| CCAP1459/16, CCAP1459/17 | P. ag. | 54°40 | 2°98 | Blelham Tarn, United Kingdom | 7/15 |
| CCAP1459/31 | P. ag. | 53°25 | 7°56 | White Lough, United Kingdom | -/30 |
| 79 | P. ag | 55°43 | 12°34 | Lake Arresø, Denmark | 6/40 |
| CCAP1459/36* | P. ag | 59°47 | 10°47 | Lake Gjersjoen, Norway | 23/64 |
| CYA126/8 | P. ag | 60°15 | 19°55 | Lake Langsjön, Finland | -/18 |
| CCAP1460/5 | P. ag | 35°41 | 139°44 | Lake Kasumigaura, Japan | 4/10 |
| 64, 67* | P. rubc | 46°36 | 14°03 | Wörthersee, Austria | 42/86 |
| 139*, 145*, 161*, 166*, 169*, 170*, 178* | P. rub | 47°59 | 13°05 | Grabensee, Austria | 7/13 |
| 3, 40*, 91/1*, 97, 110*, 111 | P. rub | 47°48 | 13°22 | Mondsee, Austria | 37/68 |
| 108 | P. rub | 47°56 | 13°19 | Irrsee, Austria | 15/32 |
| 80 | P. rub | 47°45 | 13°30 | Schwarzensee, Austria | 27/54 |
| 82, 83/2 | P. rub | 47°16 | 11°04 | Ammersee, Germany | 47/81 |
| 21- | P. rub | 48°06 | 16°18 | Figur, Austria | 8/12 |
| CCAP1459/30 | P. rub | 54°08 | 10°25 | Plöner See, Germany | 16/60 |
| CCAP1459/14 | P. rub | 54°25 | 3°0 | Loughrigg Tarn, United Kingdom | 12/13 |
| PCC7821 | P. rub | 59°47 | 10°47 | Lake Gjersjoen, Norway | 23/64 |
NOTE.—International culture collections: SAG, culture collection of algae (Göttingen, Germany); PCC, Pasteur culture collection (Paris, France); and CCAP, culture collection of algae and protozoa (Windermere, United Kingdom). I,II,III,IVStrains showing mcy gene cluster deletions of types I, II, III, and IV, respectively. *Strains found inactivated by IS of transposable elements (Christiansen et al. 2006).
Zmean, mean water depth; Zmax, maximum water depth of the origin of isolation.
P. ag, P. agardhii, green-pigmented strains.
P. rub, P. rubescens, red-pigmented strains.
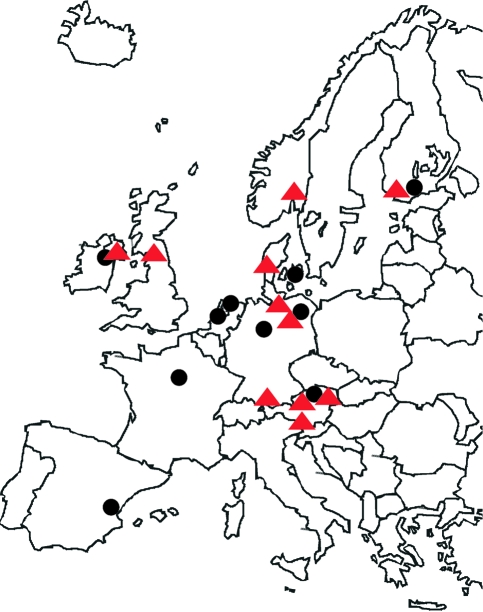
FIG. 1.—
Origin of the strains of the cyanobacterium Planktothrix spp. isolated from European freshwater sites. Red triangles, strains containing the mcy gene cluster encoding the synthesis of the toxic heptapeptide MC (37 strains) and black dots, nontoxic strains lacking the mcy gene cluster (25 strains).
Genetic Analysis of the Strains
In order to search for the remnants of the mcy gene cluster in nontoxic strains, primer pairs covering the whole mcy gene cluster were designed and used to amplify fragments of 500 bp without interruption (124 primer pairs, supplementary table S1, Supplementary Material online). Polymerase chain reaction (PCR) amplifications were performed in a volume of 20 μl, containing 2 μl of Qiagen PCR buffer (Qiagen, Vienna, Austria), 1.2 μl MgCl2 (25 mM, Qiagen), 0.6 μl deoxynucleotide triphosphates (10 μM each, MBI Fermentas, St Leon-Rot, Germany), 1 μl of each primer (10 pmol μl−1), 0.1 μl Taq DNA polymerase (5 units μl−1, Qiagen), 13.1 μl sterile millipore water, and 1.0 μl of DNA (50 ng μl−1). The PCR thermal cycling protocol included an initial denaturation at 94 °C for 3 min, followed by 35 cycles at 94 °C for 30 s, at 60 °C annealing temperature for 30 s, and elongation at 72 °C for 30 s. By the use of this technique, 8 strains (No41, 63, 66, PH22, 2A, SAG5.81, CCAP1459/15, PCC7805, and PCC7811) were found to contain mcyT. Seventeen additional nontoxic strains were used to test for the presence of mcyT by using the primers mcyTA-Z+: TAATTGATCCCCTGATCAATGATC and mcyTA-Z−: ATGCAAATAGACCAACTAAAGCC amplifying 791 bp. The strains containing mcyT were analyzed for the 5′ and the 3′ ends using a genome walking method (Siebert et al. 1995). The same method was used to sequence the 3′ end of the mcy gene cluster (AJ441056). The total DNA was digested with EcoRV and DraI at 37 °C overnight, and the DNA was extracted once by using phenol–chloroform–isoamyl alcohol (25:24:1, v/v/v) and once with chloroform and then washed with ethanol subsequent to glycogen-aided precipitation. A total of 100 ng μl−1 of DNA were then ligated to long suppression adapters at 4 °C overnight. One adapter contained an amino group at the 3′ end and ligated to any DNA fragment that was generated by restriction enzymes that yielded blunt ends. During primary PCR, this amino group blocked the extension of the lower adaptor strand unless a gene-specific primer extended to the opposite DNA strand. The primary PCR mixture was then diluted 1:100 and used as a template for a secondary or “nested” PCR with the nested adaptor-specific primer and a nested gene-specific primer. Primary PCR was performed with 2 Step Advantaq (Clontech, St. Germain-en-Lave, France) at 68 °C annealing (30 s) and 68 °C elongation (3 min). Nested PCR was performed by using Advantaq at 68 °C elongation (3 min).
In addition, all the strains were sequenced for 16S rDNA and 3 intergenic spacer (IGS) regions: PC-IGS (the IGS between cpcB and cpcA genes), 16S ITS (the internal transcribed spacer located between the 16S and 23S rDNA), and PSA-IGS (the IGS between psaA and psaB genes). The forward primer (5′–3′) 16Sfwd581 “CAGTGGAAACTGGAAGACTAGAGTGTA” (63 °C) and the reverse primer 16Srev999 “GCACCTGTCTTCTGGTTCCTTAC” (62 °C) were used to sequence part of the 16S rDNA (418 bp). The primers that were used to amplify and sequence PC-IGS (271 bp) have been described previously (Kurmayer et al. 2004). For 16S ITS, the forward primer (5′–3′) 16SITSneu+ “GCTAGGAAAGAAAGGAACTTTCA” (61 °C) and the reverse primer untranslated leader region− “CCT CTG TGT GCC TAG GTA TC” (59 °C) (394 bp) were used (Janse et al. 2003). For PSA-IGS, the forward primer psafwd “GGGTGGTACTTGCCAAGTCTCT” (58 °C) and the reverse primer psarev “CGACGTGTTGTCGGGTCTT” (58 °C) (669 bp) were used. All the PCR products were cloned and sequenced according to the standard techniques. The sequences were submitted to GenBank under the following accession numbers: 16S rDNA (EU266118–EU266179), PC-IGS (EU266242–EU266303), 16S ITS (EU266180–EU266241), PSA-IGS (EU258202–EU258263), mcyT (EU266304–EU266364), mcyT 5′ flanking region (EU271829–EU271852), mcyT 3′ flanking region (EU271805–EU271828), mcyJ and flanking regions sequenced from strain No252 (EU271803), and the IS element sequenced from strain CYA126/8 (EU271804). The sequence of the IS element that was involved in the deletion of the mcy gene cluster was submitted to the IS finder database and denoted as ISPlag1 (Siguier et al. 2006).
Functional Analysis of the Thioesterase Type II mcyT
In order to confirm the involvement of mcyT in the biosynthesis of MC, the gene was insertionally inactivated in the transformable strain CYA126/8 by homologous recombination as described (Christiansen et al. 2003). To build the transformation construct, mcyT was amplified and cloned using the pDrive cloning system (Qiagen) and subsequently partially digested using HindIII according to the standard protocols (Sambrook et al. 1989). The 3′ overhangs were blunted using Klenow Polymerase (Fermentas, Hanover, MD) and ligated with the BsaAI fragment (1.9 kbp) from pACYC184 containing the chloramphenicol resistance cassette (CmR). The mcyTKOCmR construct contained 1.5 kbp of homologous sequence on both 5′ and 3′ ends and was introduced by electroporation as described (Christiansen et al. 2003). Transformed cyanobacterial cells were inoculated in BG11 medium. After 48 h, chloramphenicol was added to the culture medium (1 μg ml−1) and resistant clones emerged after 6 weeks. PCR using primers binding to the flanking regions of the inserted chloramphenicol resistance gene showed the stable integration of the construct mcyTKOCmR at the expected position as a result of a homologous crossover recombination event. In addition, the mcyT gene in the knockout mutant as well as in the wild-type strain was amplified by PCR using the mcyTA-Z primers. The calculated PCR product of 791 bp was obtained for the wild-type strain only, whereas the ΔmcyT mutant showed an amplicon of 2.7 kbp due to the IS of the CmR fragment (1.9 kbp). Consequently, the ΔmcyT mutant was fully segregated.
The wild-type strain CYA126/8 and the ΔmcyT mutant were compared in the MC production under semicontinuous culture conditions following the turbidostat principle (Kohl and Nicklisch 1988) at 20 °C and 40–60 μE m−2 s−1 (Philips LTD, 36W/965). The growth of the cells was monitored by measuring the absorbance at 880 nm (5-cm light path) every other day and each time optical density ≥ 0.1 the culture was diluted to optical density = 0.01 (corresponding to a biomass of 1.44 ± 0.17 [standard error] mm3 L−1 of biovolume). The cells were harvested at optical density = 0.1 onto glass fiber filters (BMC Ederol, Vienna, Austria) directly in the culture room using low-vacuum filtration (<−0.4 bar), in which the filters with the collected cells were flash frozen in liquid nitrogen for subsequent RNA isolation. Aliquots of the cells were filtered onto preweighed glass fiber filters and dried at room temperature in a vacuum centrifuge, reweighed, and stored frozen at −20 °C. The dissolved MCs were collected from the filtrate by using solid phase extraction via tC18 cartridges (Waters, Sep-Pak Vac 1cc [100 mg]) according to the standard techniques. For cell number determination, the cells were fixed in 2% formaldehyde and enumerated using 6-diamidino-2-phenylindole staining following the standard procedures.
Filters were extracted in 50% methanol (v/v), as described (Kurmayer et al. 2004), and the extracts were analyzed for MCs by using high-performance liquid chromatography coupled to diode array detection at 240 nm, using a linear gradient from 20% acetonitrile (0.05% trifluor acetic acid) to 50% acetonitrile on a LiChrosper 100, ODS, 5 μm, LiChroCART 250-4 cartridge system (Merck, Darmstadt, Germany) at 1 ml min−1. Aeruginosides were identified and quantified at 210 nm under the same conditions (Ishida et al. 2007). The quantification of MCs was achieved by using the calibrated standards that were obtained from Cyanobiotech GmbH (Berlin, Germany).
RNA was extracted following the standard protocols with Trizol reagent under liquid nitrogen as described (Kaebernick et al. 2000) and converted to cDNA by reverse transcription. The quantification of mcy transcripts was performed using real-time PCR by using the TaqMan assay (Kurmayer and Kutzenberger 2003). TaqMan Probes for PC-IGS and mcyB were used (Schober and Kurmayer 2006). All the growth experiments were repeated 4 times.
Phylogenetic Analysis
Sequences of mcyT (751 bp) were aligned by using multiple sequence alignment (ClustalW 1.8). Maximum likelihood analysis was used in order to estimate the nucleotide substitution parameters under a general time-reversible nucleotide substitution model by estimating the gamma distribution for the variable rates among the sites. Ambiguous sites (wherein at least one sequence showed a gap) were removed (3 sites), and the discrete gamma algorithm was used to approximate a continuous gamma distribution using 5 categories of rates (ncatG = 5) in the program BASEML of the PAML software package (version 3.14, Yang 1997). Phylogenetic trees were also constructed using: 1) Neighbor-Joining from the nucleotide sequences distance matrix (calculated using Kimura's substitution model) and 2) maximum parsimony from nucleotide sequences by using the PHYLIP software package (Felsenstein 1993). The statistical significance of the branches was estimated by bootstrap analysis in turn generating 100 replicates of the original data set by using the PHYLIP software package. Finally, consensus trees following the 50% majority rule were computed.
For mcyT, the ratio of nonsynonymous (dN) and synonymous (dS) substitution rates per site was determined by using maximum likelihood estimates as implemented in PAML. For the nontoxic and toxic strains separately, the “one ratio” model, estimating the dN/dS ratio, and the “fixed-ratio” model, assuming a constant dN/dS ratio as observed for other mcy genes in the mcy gene cluster (dN/dS = 0.2), were statistically compared by constructing a likelihood ratio test (Yang 1998).
For multiple locus sequence typing (MLST), all the nontoxic (25) and toxic strains (37) were defined by the alleles (unique genotypes) that were present at 4 additional sequenced loci (the allelic profile): 16S rDNA, 16S–23S rDNA-ITS, PC-IGS, and PSA-IGS. Each unique allelic profile was assigned a sequence type (ST). Isolates with the same ST at all the loci were considered to be members of a single clone (Feil et al. 2004). The program eBurst (V3) was used via the MLST Web site (http://www.mlst.net/) in order to divide the 62 strains into clonal complexes. Each of the clonal complexes only contained strains sharing at least 3 of 4 identical alleles with at least one other strain in the group. The primary founder of a group was defined as the ST that differs from the largest number of other STs at only a single locus (i.e., the ST that has the greatest number of single locus variants [SLVs]). The level of confidence in the predicted founding ST was estimated by using the default settings, that is, 1,000 random data sets of the same size as the original data set were produced by resampling with replacement, and the bootstrap values, which were shown for each ST, are the percentage of times that the ST was predicted to be the primary founder of the group (Feil et al. 2004).
Results
Characterization of the Remnants of the mcy Gene Cluster
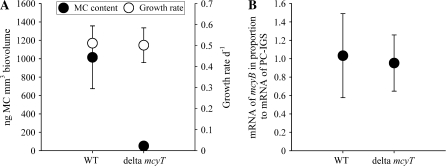
In 24 out of the 25 nontoxic strains, mcyT (751 bp), a gene coding for a distinct thioesterase type II (TeII), was amplified. A distinct TeII has been frequently associated with NRPS gene clusters and has been shown to positively influence the synthesis rate of corresponding NRPS (Mootz et al. 2001; Schwarzer et al. 2002). This is due to the regeneration of the misprimed 4′-phosphopantetheine (4′PP) cofactor of the peptidyl carrier protein because the 4′PP transferases are not able to distinguish between acetylated and free CoA (Quadri et al. 1998). The acetylation of 4′PP has the consequence that the biosynthesis is blocked until a TeII cleaves the acetyl residue from the 4′PP making the biosynthesis possible. In Planktothrix, the mcyT gene is located at the 5′ end of the mcy gene cluster but has not been found in the mcy gene cluster of other MC-producing cyanobacteria (Christiansen et al. 2003). To demonstrate the role of McyT in MC synthesis, the mcyT gene was inactivated by experimental mutagenesis in P. agardhii strain CYA126/8. The wild-type strain contained 964 ± 321 ng (±1 standard deviation [SD]) of MC-RR (68 ± 3%) and MC-LR (32 ± 3%) per mm3 of biovolume (n = 5). The insertional inactivation of mcyT resulted in a reduction of MC synthesis by 94 ± 2% (1 SD) compared with the wild type. In contrast, the proportion of MC variants, cellular growth rates, as well as the transcriptional rates of other mcy genes (mcyB) were not altered (fig. 2). The synthesis rate of the related, coproduced nonribosomally synthesized (NRS) peptides such as aeruginoside 126A, 126B (Ishida et al. 2007) was not significantly affected (37 ± 71% increase compared with the wild type). It is concluded that mcyT is directly involved in MC synthesis. In one nontoxic strain, we were unable to amplify mcyT but detected another mcy operon remnant at the other (3′) end of the mcy gene cluster, mcyJ, which was an O-methyltransferase that was previously shown to be involved in MC synthesis (Christiansen et al. 2003).
FIG. 2.—
Growth rate, MC content, and mcy transcripts of the wild-type Planktothrix strain CYA126/8 and its ΔmcyT mutant grown under semicontinuous culture conditions. (A) Mean (±1 SD) MC content (per mm3 of biovolume, black circles) and growth rates (μ d−1, white circles). (B) The mRNA contents of mcyB in proportion to PC-IGS (in percentage of mcyB) for the same experiment.
Characterization of the Sequence Breaking Points of the mcy Gene Cluster
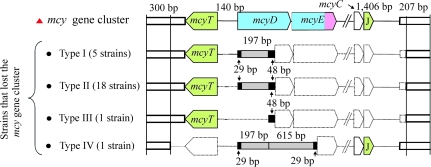
There were identical flanking sequences observed directly upstream and downstream of all the mcy operon remnants: at the 5′ end similarity was 99.0–100% (300 bp, the strain with the mcyJ remnant only had 219 bp) and at the 3′ end similarity was 98.7–100% (207 bp, the strain with the mcyJ remnant had 241 bp) (fig. 3, supplementary figs. S1 and S2, Supplementary Material online). Those flanking sequences that were found in the nontoxic strains also had the highest similarity to the corresponding region of the toxic strain CYA126/8: at the 5′ end similarity was 99.0–99.7% (300 bp, for AJ441056, Positions 149–449) and at the 3′ end similarity was 93.2–94.2% (207 bp, located 1,406 bp downstream of the stop codon of mcyJ AJ441056). In between the 2 regions that were originally flanking the mcy gene cluster, shorter fragments containing the inverted repeated sequences “CAGGACTTACGCAAGCACGCTATATATAG” (29 bp) occurred. These shorter fragments were found directly adjacent to the mcy operon remnants (mcyT and mcyJ) and were identified as terminal inverted repeats, part of the remnants of an IS element (197 bp) ISPlag1 that was sequenced from the toxic strain CYA126/8 (1,306 bp). ISPlag1 encoded a single ORF (337 aa) with similarity to the IS701 group that was originally a part of the heterogeneous IS4 family (Chandler and Mahillon 2002): ISMhu9 from Methanospirillum hungatei strain JF-1 (E value 5 × 10−43, YP_501731). Residues 257–270 (CAIRWKIEEFHREIK) corresponded to the [Y/L](2)R(3)[I/L/V]E(6)K signature characteristic of the C1 domain of bacterial transposases (Rezsöhazy et al. 1993). Notably, all the nontoxic strains contained remnants of the same mobile element that was located between the former flanking regions of the mcy gene cluster. According to the presence of the mcy operon remnants and the location of the remnants of the IS element 4 different types of mcy gene cluster deletion events were distinguished: types I (5 strains), II (18 strains), and III (1 strain), all of which contained mcyT but differed in the remaining intergenic promoter region between mcyT and mcyD (type I vs. type II) or in the sequence length of the remnant of the IS element (types I and II vs. type III). In contrast, type IV contained mcyJ instead of mcyT and 1 additional larger remnant of the same IS element (615 bp) that was inserted in opposite direction to the shorter remnant (197 bp) found also in types I and II. We conclude, therefore, that the remnants of the IS element that was found directly adjacent to the mcy operon remnants were derived from functional IS elements that were inserted into the mcy gene cluster.
FIG. 3.—
Schematic view of the mcy operon remnants and flanking regions in strains that lost the mcy gene cluster. The 4 types (I–IV) of gene cluster deletion events are shown. Vertical straight lines enclose the identical 5′ and 3′ ends. The gray gene regions represent the remnants of the IS elements (197 bp) containing terminal inverted repeats (29 bp or 48 bp) (black boxes). The dotted areas indicate the deletions. Red triangle, strains containing the mcy gene cluster and black dots, nontoxic strains lacking the mcy gene cluster.
In silico Characterization of the Remnants of the mcy Gene Cluster
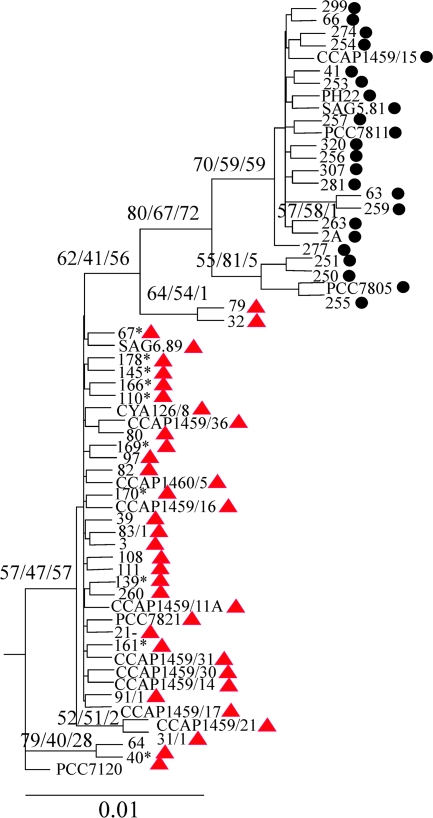
To determine whether mcyT, as a remnant of the mcy gene cluster, is still a functional gene that is involved, for example, in the biosynthesis of other NRS peptides occurring frequently in Planktothrix (Fujii et al. 2000; Ishida et al. 2007), it was sequenced (751 bp) from all the strains that contained (37 strains) or that had lost the mcy gene cluster (24 strains). The nontoxic strains formed a phylogenetic lineage that was distinct from the lineage that was formed by strains containing the mcy gene cluster (fig. 4). The ratio of nonsynonymous (dN) and synonymous (dS) substitution rates was used to differentiate between purifying selection and relaxation of selective constraints (Yang 1998). Typically, the gene regions that show a high degree of purifying selection have dN values < dS values (negative purifying selection), whereas dN values ≈ dS values are indicative of a relaxation of selective constraints (Yang 2005). For various genes of the mcy gene cluster, a dN/dS ratio = 0.2 was calculated (Tanabe et al. 2004; Kurmayer and Gumpenberger 2006). A likelihood ratio test was performed in order to test the hypothesis of the relaxation of selective constraints against the purifying selection for mcyT of both lineages. Only for the strains that had lost the mcy gene cluster (dN/dS = 1.45), the likelihood was significantly improved (degree of freedom = 1, P < 0.0001), whereas for the strains still containing the mcy gene cluster (dN/dS = 0.46), the estimated likelihood did not differ significantly from the null model (dN/dS = 0.2). Thus, it is concluded that purifying selection for mcyT was lost subsequent to its separation from the mcy gene cluster.
FIG. 4.—
Maximum likelihood tree of Planktothrix strains containing mcyT as part of the mcy gene cluster (red triangles) and as a remnant of the mcy gene cluster (black dots). The numbers at the nodes indicate the percent bootstrap frequency (100 replicates) that was obtained from maximum likelihood–Neighbor-Joining–maximum parsimony calculated using the PHYLIP package. The tree was rooted using the Nostoc sp. strain PCC7120 as an outgroup. Only the bootstrap values of >50% are shown. Asterisks indicate strains that were inactivated by transposable elements but still contained the whole mcy gene cluster (Christiansen et al. 2006).
Phylogenetic Origin of the Deletion of the mcy Gene Cluster
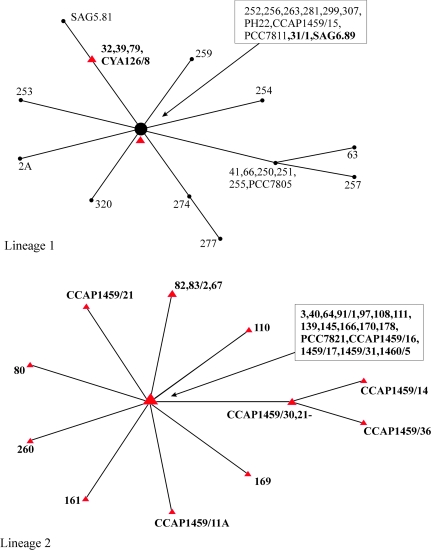
The phylogenetic origin of the mcy gene cluster deletion was identified by sequencing all the strains for gene regions that are representative of essential housekeeping genes, that is, the 16S rDNA region (302 bp), the IGS between cpcB and cpcA, PC-IGS (211 bp), the ITS that is located between the 16S and 23S rDNA, 16S rDNA-ITS (317 bp), and the IGS between psaA and psaB, PSA-IGS (615 bp). All the strains shared nearly identical 16S rDNA sequences, in contrast to the number of polymorphic sites was higher at the other more variable spacer regions (table 2). In total, 25 unique genotypes were detected with 19 genotypes occurring only once. By using MLST (Feil et al. 2004), 2 clonal complexes were observed: 1) lineage 1 was comprised of both the strains that lost and that contained the mcy gene cluster and 2) lineage 2 was comprised of the strains containing the mcy gene cluster including all the red-pigmented strains that are assigned to P. rubescens (fig. 5). In lineage 1, the primary founder (11 strains), which is defined as the ST with the greatest number of SLVs, contained the 2 most divergent types of mcy gene cluster deletion (types II and IV). The other types of deletions (I and III) were detected as SLVs or double locus variants. Therefore, nontoxic strains are all derived from a single ancestral strain that became inactive due to IS element-mediated IS followed by mcy gene cluster deletions and divergence.
Table 2.
Number of Unique Genotypes (Alleles) and Variable Sites of Sequences at the Gene Loci that Were Analyzed in the Planktothrix Strains that Lack, or Contain, the mcy Gene Cluster
| Locus | Bpa | Maximum Dissimilarity (%) | Nb | Number of Variable Sites | Number of Alleles |
| Strains lacking the mcy gene cluster (n = 25) | |||||
| mcyT | 751 | 0.93 | 24 | 36 | 22 |
| 16S ITS | 316 | 2.23 | 25 | 10 | 5 |
| PC-IGS | 211 | 6.16 | 25 | 13 | 2 |
| PSA-IGS | 615 | 0.49 | 25 | 5 | 6 |
| 16S | 302 | 1.32 | 25 | 4 | 3 |
| Strains containing the mcy gene cluster (n = 37) | |||||
| mcyT | 751 | 0.67 | 37 | 14 | 11 |
| 16S ITS | 317 | 2.23 | 37 | 10 | 2 |
| PC-IGS | 211 | 7.11 | 37 | 17 | 7 |
| PSA-IGS | 615 | 3.74 | 37 | 24 | 6 |
| 16S | 302 | 0 | 37 | 0 | 1 |
| All strains (n = 62) | |||||
| mcyT | 751 | 1.74 | 61 | 48 | 33 |
| 16S ITS | 317 | 3.82 | 62 | 16 | 6 |
| PC-IGS | 211 | 7.11 | 62 | 17 | 7 |
| PSA-IGS | 615 | 3.9 | 62 | 28 | 10 |
| 16S | 302 | 1.32 | 62 | 4 | 3 |
Bp, base pair number sequenced.
N, number of sequences.
FIG. 5.—
MLST diagrams showing 2 clonal complexes of strains that lost (lineage 1) or still contained the mcy gene cluster (lineages 1 and 2). Red triangles and bold letters: strains containing the mcy gene cluster encoding the synthesis of the toxic heptapeptide MC (37 strains) and black dots and normal letters: nontoxic strains that lost the mcy gene cluster (25 strains). Lineage 1, central founder (11 strains), bootstrap confidence 85% and lineage 2, central founder (17 strains), bootstrap confidence 92%. The connected STs differ at only 1 sequenced locus. The size of the symbols is proportional to the number of strains representing each ST.
Discussion
IS elements are generally recognized as important mutagenic agents (Doolittle and Sapienza 1980; Chandler and Mahillon 2002). Little is known, however, about the relative importance of the IS- or excision-induced variation in natural populations of bacteria as opposed to other mechanisms, for example, lateral gene transfer. In the present study, all the strains that lost the mcy gene cluster still contained inverted terminal repeated sequences between the 5′ and 3′ end mcy operon remnants that were probably derived from more than one copy of a putative IS element. The most likely mechanism that caused the mcy gene cluster deletion in all the strains was conservative site-specific recombination (Craig 1988), which resulted from the IS of 2 copies of the IS element at both ends of the mcy gene cluster. Indeed, in the vast majority of the strains that lost the mcy gene cluster, the inverted repeated sequences showed direct orientation when flanking the mcy operon remnants at the 5′ and the 3′ ends. It is, therefore, concluded that the loss of the mcy genes is the result of a 2-step process, that is, the inactivation of the mcy gene cluster via the peripheral IS of 2 copies of an IS element that finally led to its deletion.
It is anticipated that the genotypes that become inactivated in MC synthesis by an IS element must be able to persist under natural conditions until the inactive mcy gene cluster is lost due to site-specific recombination. We previously described the inactivation of the mcy gene cluster by another IS element among the strains of the second lineage that always contain the mcy gene cluster (Christiansen et al. 2006). Those mcy genotypes that became inactivated by an IS element cannot be distinguished from active mcy genotypes via MLST (figs. 4 and 5) and have been observed to occur in deep-stratified lakes for at least 5 years. Although the inactive mcy genotypes make up the minor part of the population only (e.g., 21% in Lake Mondsee, Kurmayer et al. 2004), it is obvious that the inactivation of the mcy gene cluster through IS elements does not imply a selective disadvantage to the individual. On the other hand, the insertional inactivation of MC synthesis in Microcystis aeruginosa did not increase growth under different light conditions (4–110 μmol m−2 s−1) compared with the MC-producing wild type (Hesse et al. 2001). This implies that the metabolic costs that are attributable to MC synthesis are not necessarily of relevance for the cell division rate. In the present study, only those strains that lost more than 90% of the mcy gene cluster were spread throughout Europe. One explanation for this could be that only larger gene deletion events may provide sufficient selective advantage for a specific genotype due to the energy requirements that are involved in the translational machinery (Mira et al. 2001). Under laboratory conditions, a sudden appearance of mutants that lost a larger part of the mcy gene cluster has in fact been reported (Schatz et al. 2005). Because the strain CYA126/8 is the only toxin-producing cyanobacterium that has been repeatedly transformed in several laboratories (Christiansen et al. 2003; Ishida et al. 2007), the experimental deletion of parts or the full mcy gene cluster of this strain is proposed as a tool for differentiating between the costs that are attributable to MC synthesis (e.g., ΔmcyT) and the costs of the replication/translation of the mcy gene cluster (e.g., constructing a mutant by deleting larger parts of the mcy gene cluster) compared with the wild type.
In contrast to observations in the laboratory (Schatz et al. 2005), the results of this study point to a much slower replacement of toxic strains by nontoxic strains that lost the mcy gene cluster under natural conditions. Assuming a random spontaneous mutation rate of 4.1 × 10−10 (Drake 1991) along with an annual mean growth rate of 0.14 per day (Davis and Walsby 2002), the time necessary to create 5 random spontaneous mutations (the minimum dissimilarity between mcyT [791 bp] of nontoxic strains compared with toxic strains, fig. 4) was calculated to be 3.6 Myr. It could be argued that, for most of the time, cells do not grow exponentially but rather linearly, and nontoxic strains may competitively exclude toxic strains under nearly exponential growth conditions during prebloom periods in eutrophic lakes (e.g., Kardinaal et al. 2007). Indeed, Planktothrix occurring in deep-stratified lakes can dominate phytoplankton for years without pronounced periods of exponential growth/decline, whereas Planktothrix occurring in more shallow lakes may show an exponential increase/decline during the season (Salmaso and Padisak 2007). However, even a 1,000-fold underestimation of a random spontaneous mutation rate for mcyT would imply that shifts between toxic strains and nontoxic strains as observed during seasonal succession in lakes cannot be directly of relevance to the evolution of the mcy gene cluster. Due to the extremely rare event of the mcy gene cluster deletion, the sole occurrence of toxic genotypes in P. rubescens populations in deep-stratified lakes (Kurmayer et al. 2004) can be explained by the evolutionary diversification of a genotype that still contains the mcy gene cluster. It has been suggested that P. rubescens diversified from P. agardhii due to its pigment adaptation to an underwater light climate relatively recently (Suda et al. 2002). We anticipate that evolutionary diversification that is driven by selective factors other than an underwater light climate, for example cyanophages or hydrostatic pressure, will lead to a clonal dependence of MC production also in other taxa. In a recent study, numerous cyanophages active against Anabaena, Planktothrix, and Microcystis were reported (Deng and Hayes 2008). Cyanophage dynamics may further select phage-resistant genotypes and indirectly affect the shifts between MC-producing and non–MC-producing genotypes in Microcystis (Yoshida et al. 2008). Correspondingly, Tanabe et al. (2007) by using MLST found a clonal population structure of Microcystis sp., suggesting that each phylogenetic cluster might represent a “cryptic” ecotype (Cohan 2002). It is noteworthy that the occurrence of mcyG showed a clonal dependence, that is, all the strains containing mcyG were part of 2 lineages (groups A and B), whereas the other groups (C, D, and E) consisted of strains lacking the mcyG gene. Consequently, the frequently observed co-occurrence of toxic and nontoxic strains (Carmichael and Gorham 1981; Vezie et al. 1998) can only be understood if it is interpreted in terms of evolutionary diversification that is driven by selective factors including those not directly linked to MC production. Future experiments that aim at a cost–benefit analysis of MC production need to consider the fact that even closely related strains that differ in MC production might also differ in other ecological traits, reflecting the process of adaptation to their respective environment. For example, it is well known that the natural isolates of bacteria (Mikkola and Kurland 1992) and cyanobacteria (Van Liere and Mur 1980) usually differ widely in their maximum specific growth rate under standardized laboratory conditions. This implies that the measured growth rates of nontoxic and toxic strains under experimental conditions cannot be used to relate the fitness of a particular strain to the presence/absence of MC production.
Supplementary Material
Supplementary figures S1 and S2 and table S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We are most grateful to our colleagues for sending water samples to us or who delivered strains (in alphabetical order): Antonio Camacho, Peter Henriksen, Kaarina Sivonen, and Linda Tonk. Johanna Schmidt provided rather valuable assistance in the laboratory. We very much appreciate the comments of 3 anonymous reviewers. This study was financed by a grant from the Austrian Science Fund P18185 “Microevolution of toxin synthesis in cyanobacteria.”
References
- Carmichael WW, Gorham PR. The mosaic nature of toxic blooms of cyanobacteria. In: Carmichael WW, editor. The water environment: algal toxins and health. New York: Plenum Press; 1981. pp. 161–172. [Google Scholar]
- Cary J, Ehrlich K. Aflatoxigenicity in Aspergillus: molecular genetics, phylogenetic relationships and evolutionary implications. Mycopathologia. 2006;162:167–177. doi: 10.1007/s11046-006-0051-8. [DOI] [PubMed] [Google Scholar]
- Chandler M, Mahillon J. Insertion sequences revisited. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington (DC): American Society for Microbiology; 2002. pp. 305–366. [Google Scholar]
- Christiansen G, Fastner J, Erhard M, Börner T, Dittmann E. Microcystin biosynthesis in Planktothrix: genes, evolution, and manipulation. J Bacteriol. 2003;185:564–572. doi: 10.1128/JB.185.2.564-572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen G, Kurmayer R, Liu Q, Börner T. Transposons inactivate biosynthesis of the nonribosomal peptide microcystin in naturally occurring Planktothrix spp. Appl Environ Microbiol. 2006;72:117–123. doi: 10.1128/AEM.72.1.117-123.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan FM. What are bacterial species? Annu Rev Microbiol. 2002;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- Craig NL. The mechanism of conservative site-specific recombination. Ann Rev Genet. 1988;22:77–105. doi: 10.1146/annurev.ge.22.120188.000453. [DOI] [PubMed] [Google Scholar]
- Davis PA, Walsby AE. Comparison of measured growth rates with those calculated from rates of photosynthesis in Planktothrix spp. isolated from Blelham Tarn, English Lake District. New Phytol. 2002;156:225–239. doi: 10.1046/j.1469-8137.2002.00495.x. [DOI] [PubMed] [Google Scholar]
- Deng L, Hayes PK. Evidence for cyanophages active against bloom-forming freshwater cyanobacteria. Freshwater Biology. 2008;53:1240–1252. [Google Scholar]
- Dittmann E, Neilan BA, Erhard M, von Döhren H, Börner T. Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Mol Microbiol. 1997;26:779–787. doi: 10.1046/j.1365-2958.1997.6131982.x. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- Drake J. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. Eburst: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (phylogeny inference package). Version 3.5c. Seattle (WA): Department of Genetics, University of Washington; 1993. [Google Scholar]
- Fujii K, Sivonen K, Naganawa E, Harada K. Non-toxic peptides from toxic cyanobacteria, Oscillatoria agardhii. Tetrahedron. 2000;56:725–733. [Google Scholar]
- Hesse K, Dittmann E, Börner T. Consequences of impaired microcystin production for light-dependent growth and pigmentation of Microcystis aeruginosa PCC 7806. FEMS Microbiol Ecol. 2001;37:39–43. [Google Scholar]
- Ishida K, Christiansen G, Yoshida WY, Kurmayer R, Welker M, Bonjoch J, Hertweck C, Börner T, Hemscheidt T, Dittmann E. Biosynthetic pathway and structure analysis of aeruginoside 126A and B, cyanobacterial peptide glycosides bearing an unusual 2-carboxy-6-hydroxyoctahydroindole moiety. Chem Biol. 2007;14:565–576. doi: 10.1016/j.chembiol.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janse I, Meima M, Kardinaal WEA, Zwart G. High-resolution differentiation of cyanobacteria by using rRNA-internal transcribed spacer denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2003;69:6634–6643. doi: 10.1128/AEM.69.11.6634-6643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaebernick M, Neilan BA, Börner T, Dittmann E. Light and the transcriptional response of the microcystin biosynthesis gene cluster. Appl Environ Microbiol. 2000;66:3387–3392. doi: 10.1128/aem.66.8.3387-3392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardinaal W, Tonk L, Janse I, Hol S, Slot P, Huisman J, Visser P. Competition for light between toxic and nontoxic strains of the harmful cyanobacterium Microcystis. Appl Environ Microbiol. 2007;73:2939–2946. doi: 10.1128/AEM.02892-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl J, Nicklisch A. Ökophysiologie der Algen. Wachstum und Ressourcennutzung. Jena (Germany): Urban & Fischer; 1988. [Google Scholar]
- Kurmayer R, Christiansen G, Fastner J, Börner T. Abundance of active and inactive microcystin genotypes in populations of the toxic cyanobacterium Planktothrix spp. Environ Microbiol. 2004;6:831–841. doi: 10.1111/j.1462-2920.2004.00626.x. [DOI] [PubMed] [Google Scholar]
- Kurmayer R, Gumpenberger M. Diversity of microcystin genotypes among populations of the filamentous cyanobacteria Planktothrix rubescens and Planktothrix agardhii. Mol Ecol. 2006;15:3849–3861. doi: 10.1111/j.1365-294X.2006.03044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurmayer R, Kutzenberger T. Application of real-time PCR for quantification of microcystin genotypes in a population of the toxic cyanobacterium Microcystis sp. Appl Environ Microbiol. 2003;69:6723–6730. doi: 10.1128/AEM.69.11.6723-6730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meißner K, Dittmann E, Börner T. Toxic and non-toxic strains of the cyanobacterium Microcystis aeruginosa contain sequences homologous to peptide synthetase genes. FEMS Microbiol Lett. 1996;135:295–303. doi: 10.1111/j.1574-6968.1996.tb08004.x. [DOI] [PubMed] [Google Scholar]
- Mikkola R, Kurland CG. Selection of laboratory wild-type phenotype from natural isolates of Escherichia coli in chemostats. Mol Biol Evol. 1992;9:394–402. doi: 10.1093/oxfordjournals.molbev.a040731. [DOI] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- Moffitt MC, Neilan BA. Characterization of the nodularin synthetase gene cluster and proposed theory of the evolution of cyanobacterial hepatotoxins. Appl Environ Microbiol. 2004;70:6353–6362. doi: 10.1128/AEM.70.11.6353-6362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz HD, Finking R, Marahiel MA. 4 ‘-phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. J Biol Chem. 2001;276:37289–37298. doi: 10.1074/jbc.M103556200. [DOI] [PubMed] [Google Scholar]
- Nakasugi K, Alexova R, Svenson CJ, Neilan BA. Functional analysis of PilT from the toxic cyanobacterium Microcystis aeruginosa PCC 7806. J Bacteriol. 2007;189:1689–1697. doi: 10.1128/JB.01640-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr PT, Jones GJ. Relationship between microcystin production and cell division rates in nitrogen-limited Microcystis aeruginosa cultures. Limnol Oceanogr. 1998;43:1604–1614. [Google Scholar]
- Quadri LEN, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- Rantala A, Fewer DP, Hisbergues M, Rouhiainen L, Vaitomaa J, Börner T, Sivonen K. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc Natl Acad Sci USA. 2004;101:568–573. doi: 10.1073/pnas.0304489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezsöhazy R, Hallet B, Delcour J, Mahillon J. The IS4 family of insertion sequences—evidence for a conserved transposase motif. Mol Microbiol. 1993;9:1283–1295. doi: 10.1111/j.1365-2958.1993.tb01258.x. [DOI] [PubMed] [Google Scholar]
- Rippka R. Isolation and purification of cyanobacteria. Meth Enzymol. 1988;167:3–27. doi: 10.1016/0076-6879(88)67004-2. [DOI] [PubMed] [Google Scholar]
- Rouhiainen L, Vakkilainen T, Siemer BL, Buikema W, Haselkorn R, Sivonen K. Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaena strain 90. Appl Environ Microbiol. 2004;70:686–692. doi: 10.1128/AEM.70.2.686-692.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmaso N, Padisak J. Morpho-functional groups and phytoplankton development in two deep lakes (Lake Garda, Italy and Lake Stechlin, Germany) Hydrobiologia. 2007;578:97–112. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning—a laboratory manual. 2nd ed. Vol. 1–3. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schatz D, Keren Y, Hadas O, Carmeli S, Sukenik A, Kaplan A. Ecological implications of the emergence of non-toxic subcultures from toxic Microcystis strains. Environ Microbiol. 2005;7:798–805. doi: 10.1111/j.1462-2920.2005.00752.x. [DOI] [PubMed] [Google Scholar]
- Schober E, Kurmayer R. Evaluation of different DNA sampling techniques for the application of the real-time PCR method for the quantification of cyanobacteria in water. Lett Appl Microbiol. 2006;42:412–417. doi: 10.1111/j.1472-765X.2006.01857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer D, Mootz HD, Linne U, Marahiel MA. Regeneration of misprimed nonribosomal peptide synthetases by type II thioesterases. Proc Natl Acad Sci USA. 2002;22:14083–14088. doi: 10.1073/pnas.212382199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda S, Watanabe MM, Otsuka NS, Mahakahant A, Yongmanitchai W, Nopartnaraporn, Liu Y, Day JG. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int J Syst Evol Microbiol. 2002;52:1577–1595. doi: 10.1099/00207713-52-5-1577. [DOI] [PubMed] [Google Scholar]
- Tanabe Y, Kasai F, Watanabe M. Multilocus sequence typing (MLST) reveals high genetic diversity and clonal population structure of the toxic cyanobacterium Microcystis aeruginosa. Microbiology. 2007;153:3695–3703. doi: 10.1099/mic.0.2007/010645-0. [DOI] [PubMed] [Google Scholar]
- Tanabe Y, Kaya K, Watanabe MM. Evidence for recombination in the microcystin synthetase (mcy) genes of toxic cyanobacteria Microcystis spp. J Mol Evol. 2004;58:633–641. doi: 10.1007/s00239-004-2583-1. [DOI] [PubMed] [Google Scholar]
- Tillett D, Dittmann E, Erhard M, von Döhren H, Börner T, Neilan BA. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chem Biol. 2000;7:753–764. doi: 10.1016/s1074-5521(00)00021-1. [DOI] [PubMed] [Google Scholar]
- Touzet N, Franco JM, Raine R. Characterization of nontoxic and toxin-producing strains of Alexandrium minutum (Dinophyceae) in Irish coastal waters. Appl Environ Microbiol. 2007;73:3333–3342. doi: 10.1128/AEM.02161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Liere L, Mur LR. Occurrence of Oscillatoria agardhii and some related species, a survey. Dev Hydrobiol. 1980;2:67–77. [Google Scholar]
- Vezie C, Brient L, Sivonen K, Bertru G, Lefeuvre J-C, Salkinoja-Salonen M. Variation of microcystin content of cyanobacterial blooms and isolated strains in Lake Grand-Lieu (France) Microb Ecol. 1998;35:126–135. doi: 10.1007/s002489900067. [DOI] [PubMed] [Google Scholar]
- Walsby A, Schanz F, Schmid M. The Burgundy-blood phenomenon: a model of buoyancy change explains autumnal waterblooms by Planktothrix rubescens in Lake Zürich. New Phytol. 2005;169:109–122. doi: 10.1111/j.1469-8137.2005.01567.x. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Guidelines for drinking-water quality, Vol. 1: recommendations. 3rd ed. Geneva (Switzerland): World Health Organization; 2004. [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z. The power of phylogenetic comparison in revealing protein function. Proc Natl Acad Sci USA. 2005;102:3179–3180. doi: 10.1073/pnas.0500371102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Yoshida T, Kashima A, Takashima Y, Hosoda N, Nagasaki K, Hiroishi S. Ecological dynamics of the toxic bloom-forming Microcystis aeruginosa and its cyanophages in freshwater. Appl Environ Microbiol. 2008;74:3269–3273. doi: 10.1128/AEM.02240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.