Abstract
Gene content has been shown to contain a strong phylogenetic signal, yet its usage for phylogenetic questions is hampered by horizontal gene transfer and parallel gene loss and until now required completely sequenced genomes. Here, we introduce an approach that allows the phylogenetic signal in gene content to be applied to any set of sequences, using signature genes for phylogenetic classification. The hundreds of publicly available genomes allow us to identify signature genes at various taxonomic depths, and we show how the presence of signature genes in an unspecified sample can be used to characterize its taxonomic composition. We identify 8,362 signature genes specific for 112 prokaryotic taxa. We show that these signature genes can be used to address phylogenetic questions on the basis of gene content in cases where classic gene content or sequence analyses provide an ambiguous answer, such as for Nanoarchaeum equitans, and even in cases where complete genomes are not available, such as for metagenomics data. Cross-validation experiments leaving out up to 30% of the species show that ∼92% of the signature genes correctly place the species in a related clade. Analyses of metagenomics data sets with the signature gene approach are in good agreement with the previously reported species distributions based on phylogenetic analysis of marker genes. Summarizing, signature genes can complement traditional sequence-based methods in addressing taxonomic questions.
Keywords: signature genes, metagenomics, phylogenomics, gene content, slow–fast
Introduction
Gene content contains a strong phylogenetic signal (Snel et al. 1999; Tekaia et al. 1999) and has helped to clarify several taxonomic uncertainties (for review, see Snel et al. 2005). Classic gene content is based on the fraction of genes shared between 2 genomes and requires a data set of completely sequenced genomes to confirm not only the presence but also the absence of each gene. If a complete genome cannot be obtained, gene content can still be used to address taxonomical questions by means of signature genes. In the signature gene approach, we use the wealth of completely sequenced genomes to define cores of genes for every clade. A core is the set of all genes common to (ubiquitous among) all genomes in a phylogenetically coherent group (Charlebois and Doolittle 2004). For an unidentified, even incompletely sequenced organism, its relatives can be identified by finding the overlap between its gene repertoire and these cores. Using this idea, we found that the anaerobic ammonium-oxidizing bacterium Kuenenia stuttgartiensis is closely related to the Chlamydiae, supporting its phylogenetic classification based on a superalignment of 49 proteins (Strous et al. 2006).
When complete genomes are available, and when one wants to use a single method for phylogenomic inference, we have shown gene content to be less suitable than sequence similarity-based approaches, at least in the Fungi (Dutilh et al. 2007). However, gene content does contain a phylogenetic signal that can be exploited if the right genes are selected (Dutilh et al. 2004). Furthermore, sequence-based approaches are restricted to sequences with a wide phylogenetic distribution. The presence or absence of genes that are stable in evolution provides phylogenetic evidence that complements sequence-based information because 1) gene content evolves at a different level (whole genes instead of residues) and 2) signature genes specifically exploit those genes that do not have a very wide phylogenetic distribution.
Signature genes have been identified for several taxa on an ad hoc basis, using one or more reference genomes, sequence similarity searches and often manual inspection of the results (Martin et al. 2003; Kainth and Gupta 2005; Gao et al. 2006; Griffiths et al. 2006; Gao and Gupta 2007). The large variety of completely sequenced genomes that have become available in recent years, together with high-quality orthology definitions (Tatusov et al. 2000; von Mering, Jensen, et al. 2007) and superalignment-based species phylogenies of all sequenced genomes (Ciccarelli et al. 2006), enable us to take a more systematic approach, and find signature genes on a large scale for many clades throughout the tree of life. To do this, we introduce a simple, phylogeny-based definition: the signature genes of a clade are those genes that occur in every daughter lineage of that clade but nowhere outside it (fig. 1). The most parsimonious explanation for such a distribution is that the gene originated at the root of this clade and has been retained in all the descendant lineages because it has an important function for the species in this clade. With a predefined species tree as a guide (Ciccarelli et al. 2006), we use this definition to find cores of genes for clades of different ages at all levels in the tree. As our definition only requires that the gene is retained in at least one species per daughter of a clade, it allows for species-specific losses, for example, in the degenerated genomes of parasites (Fraser et al. 1995). Thus, it is broader than a definition that requires complete coverage of a clade. To quantify the presence of an orthologous group (OG) in a taxon, we introduce a coverage score that takes into account asymmetric taxon sampling.
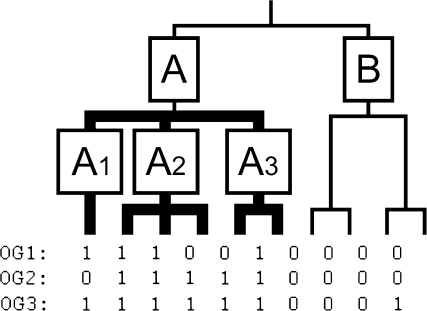
FIG. 1.—
Definition of signature genes based on a partially unresolved phylogeny. For every species, presence (1) or absence (0) of 3 genes (OGs) is indicated. In this example, only OG1 is a signature for clade A, as it is present in clade A1, clade A2 and clade A3, but not in clade B. Although OG2 and OG3 are present in more species within clade A, they are not a signature for clade A because OG2 is not present in clade A1, and OG3 is present outside of clade A.
Methods
Data
The reference phylogeny we used was based on a recent superalignment phylogeny of 31 universal protein families (Ciccarelli et al. 2006), including the 163 prokaryotic species that were also present in STRING 7.0 (von Mering, Jensen, et al. 2007). We excluded the Eukaryota because due to both the large sizes of the genomes and the highly asymmetrical taxon sampling, the eukaryotic signature genes would have obscured much of the statistical signal in the prokaryotic signature genes. To account for uncertainties in the Ciccarelli tree, we collapsed the nodes with a bootstrap value lower than 80%, resulting in a partly unresolved reference phylogeny (fig. 2). We chose a bootstrap cutoff of 80% as it is important that the phylogeny has a reasonable resolution, while weakly supported clades are collapsed. Provided that this cutoff is chosen in a reasonable range, we do not expect it to quantitatively influence our results.
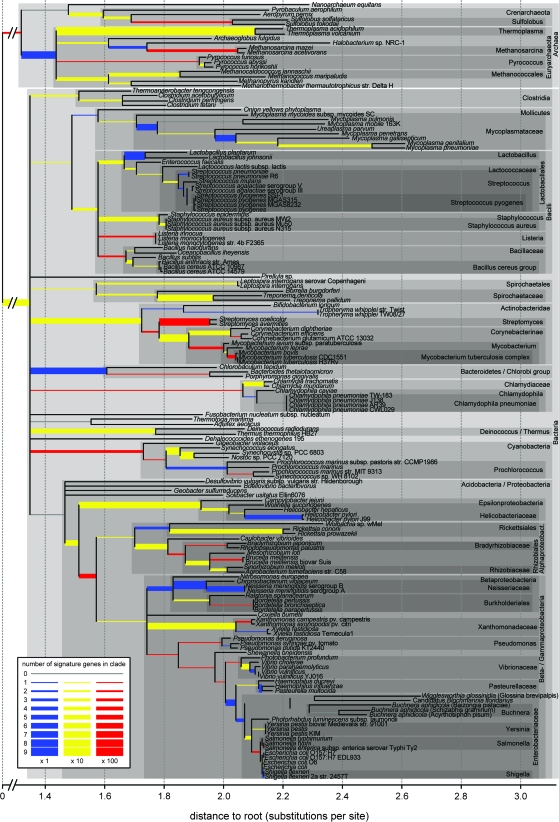
FIG. 2.—
Amounts of signature genes identified in prokaryotic taxa. The unresolved phylogeny is based on a superalignment tree (Ciccarelli et al. 2006) where we collapsed nodes with a bootstrap value lower than 80% and removed the Eukaryota. Several node names used in this paper are indicated with gray boxes. Branch widths and colors indicate the number of signature genes found for each node (see legend).
The proteomes and orthology definitions were downloaded from STRING 7.0 (von Mering, Jensen, et al. 2007); only cluster of orthologous groups (COGs) and nonsupervised orthologous groups (NOGs) present in at least 2 prokaryotic species were included in this study. Our concept of signature genes identifies those genes that originated at the root of a clade and are present in all the descendant lineages. COGs and NOGs are based on pairwise sequence similarity. If, for some reason, an OG has undergone accelerated evolution in a certain clade, its homology to other genes outside the clade may not be detected by pairwise sequence comparison, and these genes may be erroneously assigned to a new OG. This could cause an overestimation of the number of signature genes for the accelerated clade or also an underestimation of the number of signature genes for the parent clade where the OG actually originated. To avoid this, we used a highly sensitive approach to identify homology between OGs by performing profile–profile searches. First, we aligned the sequences of each OG using MUSCLE (Edgar 2004). Hidden Markov models (HMMs) were created for each OG using HHmake (HHsearch 1.4 [Soding 2005]) and calibrated against a database comprising 1,250 random SCOP domain HMMs (Murzin et al. 1995). We then compared the HMM profiles all-against-all using HHsearch. For the homologous OG pairs (query and hit aligned over >50% of their sequence; score>90), we inspected their distribution in the species tree, and if the parent clade of the OG with the narrowest distribution did not contain the OG that was more widely distributed, they were considered mergeable. We then merged the mergeable OGs using CFinder (Palla et al. 2005) at the level of communities. Remaining OGs that were not included in these communities were merged as pairs. Thus, we merged 2,958 of the 18,611 OGs, obtaining a final total of 17,323 OGs. Note that the effect of this merging procedure is mainly qualitative, removing cases where homologous OGs may be a signature for different clades and obscuring the phylogenetic signal. Quantitatively, 268 of the 8,362 signature genes found (below) derive from merged OGs. The fact that we find a small fraction of merged signature OGs is robust with respect to the homology parameters. Varying the required aligned region from >50% to >90% reduced the number of merged signature OGs to 63; varying the required homology score from >90 to >50 increased the number of merged signature OGs to 322.
Signature Genes and Coverage Score
Signature genes were identified automatically based on the OGs and the reference phylogeny. Signature genes for a clade are those OGs that do not occur outside the clade and are represented by at least one copy in every one of its daughters (i.e., 2 for a resolved node and more than 2 for an unresolved node; e.g., OG1 for clade A in fig. 1). Using this approach, we identified 8,362 signature genes for 112 of the 128 clades (table 1, fig. 2 and supplementary table 1; Supplementary Material online), that is, an average of 64.8 signatures per clade. We found no correlation of the number of signature genes with the number of daughters (r = 0.07), the number of species (r = −0.06), the bootstrap value of the clade (r = 0.05), or the distance to the root (r = −0.01). The clade with the most signature genes was Streptomyces (796 signature genes). When we restricted our search to perfect signature genes (i.e., present in every species within the clade), we identified 4,342 signatures for 98 clades (table 1). Because for 2-species clades the daughters in which a gene is required are single species, all their signatures are perfect. A total of 2,972 perfect signature genes are a signature for 2-species clades, and 1,370 perfect signature genes are a signature for larger clades.
Table 1.
Statistics of All Signature Genes Identified, the Signature Genes with a Coverage Score Cutoff of 0.75 and Perfect Signature Genes (coverage = 1.00)
| Taxa with Signatures | Number of Signatures | Average Coverage Score | |
| Signatures | 112 | 8,362 | 0.80 |
| Signatures (coverage ≥0.75) | 106 | 6,177 | 0.94 |
| Perfect signatures | 98 | 4,342 | 1.00 |
The coverage score is calculated as a nested coverage, a method that takes into account potential asymmetrical taxon sampling. For terminal clades, the score is equal to the coverage, that is, the fraction of species containing the OG. For higher order clades, the score is the average of the scores in its daughter clades. This is best illustrated with an example (fig. 1). The coverage score of OG1 as a signature for clade A is 0.72:
Phylogenetic Signal in Gene Repertoires
To assess whether the number of signature genes found for a clade is significant, we composed 1,000 sets of randomized genomes. Bearing in mind that the size distribution of both the OGs and the genomes as well as the topology of the tree (e.g., the multifurcating branches) are important for the identification of signature genes, we kept the number of OGs per genome identical, as well as the number of genomes in which an OG is represented. This was done by randomly swapping every OG in every genome with one in another genome, taking care not to place the same OG twice in one genome. Because the tree topology was not randomized, we could calculate the expected number of signature genes for a clade as the average for that exact same clade, with the same distribution of species sizes, over the 1,000 randomized genome sets. In these randomized data sets, we found an average (standard deviation [SD]) of 1,667 (35.46) signature genes of which only 74 (8.50) had a coverage score ≥0.75 and 37 (6.13) were perfect. These small numbers contrast with the many signature genes found in the nonrandomized gene repertoires (see table 2), showing the strong phylogenetic signal, and therewith the relevance of signature genes.
Table 2.
Sensitivity, Specificity, Precision, and Accuracy of the Signature Gene Method
| Number of Species Left Out | 1 | 16 (10%) | 32 (20%) | 48 (30%) |
| Sensitivity (tp/(tp + fn)) | 77.6 | 76.3 | 74.1 | 71.5 |
| Specificity (tn/(tn + fp)) | 98.9 | 98.9 | 98.7 | 98.7 |
| Precision (tp/(tp + fp)) | 93.1 | 92.9 | 92.0 | 91.7 |
| Accuracy (true/all) | 95.6 | 95.3 | 94.7 | 94.1 |
NOTE.—Results are based on several cross-validation analyses, leaving out 1 or 10%, 20%, or 30% of the species (averages of 100 experiments) from the data set and identifying signature genes in the removed genomes.
Because our randomization procedure retained the structure of the phylogeny as well as the size distribution of the genomes, we could calculate an observed over expected ratio (o/e ratio) for each individual clade, based on the number of signature genes found in the original data set and in the random gene repertoires. Out of the 129 clades in the phylogeny, 103 contained more and 24 contained less signature genes than expected (see supplementary table 1, Supplementary Material online). For the Chlamydophila pneumoniae clade and the Acidobacteria/Proteobacteria clade, no signature genes were found or expected based on the 1,000 randomized gene sets, and for the Mycoplasma genitalium/pneumoniae clade, 29 signature genes were found but none expected. For the remaining 126 taxa, the average o/e ratio was as high as 1,321, which is indicative of the strong phylogenetic signal in the gene repertoires. If we applied a coverage score cutoff of 0.75, 104 clades contained more signature genes than expected, and for 41 clades, no signature genes were expected at all. Twelve clades contained less signature genes than expected, and for 13 clades no signature genes were found or expected.
Results
Using the definition of signature genes and the method outlined above (fig. 1), we have identified 8,362 sets of signature genes (OGs) for 112 clades throughout the prokaryotic tree of life (see fig. 2, Methods and supplementary table 1, Supplementary Material online) using a partly unresolved reference phylogeny (Ciccarelli et al. 2006) and a predefined set of OGs (von Mering, Jensen, et al. 2007). Homologous OGs that had largely complementary phylogenetic distributions were merged to prevent high rates of sequence evolution from causing an overestimation of the number of signature OGs (see Methods). Subsequently, signatures for a given clade were defined as those OGs that are specific for the corresponding node and occur in every daughter lineage (fig. 1). The many signature genes we found underline the phylogenetic signal that exists in gene content. Conversely, the results justify the suspicion of clades that are completely void of signature genes. Figure 2 shows the number of signature genes identified for each branch that defines a taxon (see also supplementary table 1, Supplementary Material online). Most taxa are confirmed by the signature genes. For example, even the Bacteroidetes/Chlorobi group, which is a difficult bacterial division to retrieve in gene content trees (see supplementary fig. 1, Supplementary Material online), is supported by 7 signature genes with an average coverage score of 0.86 (see supplementary table 1, Supplementary Material online). In contrast, the controversial grouping of the hyperthermophilic bacteria Thermotoga maritima and Aquifex aeolicus is not supported by any signature genes.
Reliability of the Signature Genes for Identifying Related Clades
To assess the reliability of the signature genes method for assigning an unknown species to a taxonomic clade, we use a cross-validation procedure, and do a leave-one-out analysis for each of the 163 prokaryotic species (supplementary table 2, Supplementary Material online), as well as a leave-n-out analysis, removing 10%, 20%, and 30% of the genomes randomly. Note that leaving out even more species would make this analysis trivial as the reference tree should contain enough species to provide a meaningful taxonomic resolution. These analyses mimic the situation in which an unidentified sample has to be taxonomically characterized by identifying the genes from the removed genomes as signature genes in the adjusted tree of life. In this analysis, the OGs in the removed genomes could be a signature for one of the ancestral nodes of the removed species (true positive, tp), a signature for another node (false positive, fp), or not a signature. In that case, the OG could have been a signature in the situation where no species were excluded (false negative, fn) or not (true negative, tn). Using these values, we computed sensitivity (tp/(tp + fn)), specificity (tn/(tn + fp)), precision (tp/(tp + fp)), and accuracy (true/all) of the method (table 2).
The species with the lowest numbers of correctly assigned signature genes in this cross-validation, that is, the one that would be most often incorrectly placed back into the phylogeny, is Solibacter usitatus: only ∼40% of the signature genes correctly link this species to an ancestral clade, depending on which sets of other species were left out in these experiments (results not shown). Bdellovibrio bacteriovorus and Pirellula sp. (∼50%), Desulfovibrio vulgaris and Geobacter sulfurreducens (∼60%), and Chromobacterium violaceum and Gloeobacter violaceus (∼65%) also violate their taxonomic recognition by signature genes. In the phylogeny (fig. 2), the species that cannot robustly be placed in the tree using signature genes are present in particularly ill-resolved parts of the reference phylogeny. Apparently, the unresolved taxonomic position of these species in the sequence-based reference tree is reflected in the fact that they share many signature genes with taxa that are unrelated in the reference phylogeny. The 2 exceptions are C. violaceum and, strikingly, G. violaceus that share 323 signature genes with the other Cyanobacteria. This observation suggests that G. violaceus may be more derived than its position in the reference phylogeny at the root of the Cyanobacteria. As it shares 78 signature genes with Synechococcus/Synechocystis/Nostoc and only 6 signature genes with the Prochlorococci (see supplementary table 2, Supplementary Material online), it may actually cluster with that Cyanobacterial subclade.
These cross-validation experiments also allowed us to assess the stability of the set of signature genes when up to 30% of the genomes are removed. Leaving out random subsets of 10%, 20%, or 30% of the genomes yielded subsets of 89.6% (5.1%), 79.6% (6.1%), and 68.8% (6.8%) of the original set of 8,362 signature genes, respectively (averages [SD] of 100 samples). Conversely, the restricted species sets contained only very few new signature genes: 2.0% (1.0%), 5.1% (1.3%), and 5.9% (1.4%) for the 100 random subsets of 10%, 20%, or 30% of the species, respectively. Owing to the fact that we do not require complete coverage of a clade (the average coverage score of the remaining signature genes remained the same: ∼0.80) and that we include signature genes for all clades in the tree of life, the total number of signature genes will grow rather than shrink as the number of species increases (Charlebois and Doolittle 2004). Addressing this issue in another light, we performed a historical reconstruction (fig. 3), showing that with the inclusion of more completely sequenced genomes, the number of signatures grows, rather than shrinks, and the number of signature genes per taxon remains quite stable. This is the result of, on the one hand, the sampling of more daughters per taxon, which increases the coverage requirement for a signature gene, and on the other hand the sampling of more species per daughter, which increases the species sampling, leading to more imperfect signatures.
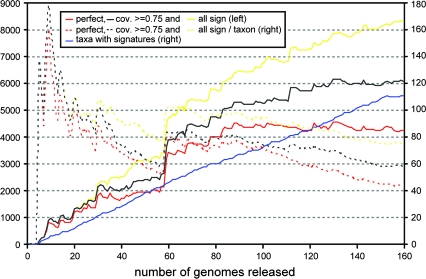
FIG. 3.—
The number of signature genes, perfect signature genes (coverage score 1), and signature genes with a coverage score cutoff of 0.75 found with increasing numbers of completely sequenced genomes. The genomes are added one by one, in order of appearance (according to www.ncbi.nlm.nih.gov/genomes). Initially, the number of signature genes increases almost linearly with the appearance of more genomes. The 60th genome, that of Streptomyces avermitilis, completes the signature-rich Streptomyces clade (Streptomyces coelicolor was the fourth genome), and causes a great jump in the number of both perfect and normal signature genes.
Difficult Taxonomic Questions Addressed with Signature Genes
As an independent method to address taxonomic questions, the signature gene procedure also allows us to investigate in detail the taxonomic position of some early branching prokaryotic species, for which the phylogenetic signal in the sequences may have been lost. As in the cross-validation experiments (see “Reliability of the Signature Genes for Identifying Related Clades” above), we removed the species A. aeolicus, Fusobacterium nucleatum, Halobacterium sp., Nanoarchaeum equitans, and T. maritima from the data set one by one and reidentified signature genes in the remaining 162 species. Table 3 shows which genes from the removed genomes were found as signature genes in the corresponding restricted data set (for the leave-one-out analysis of all species, see supplementary table 2, Supplementary Material online). Thus, these signature genes can classify the removed genomes in terms of their taxonomic relatives.
Table 3.
Signature Genes Shared by Several Species and Potential Sister Clades
| Species | Clade | o/e ratio | Shared Signature Genes |
| Aquifex aeolicus | Bacteria | 60/0 | 60 COGs |
| Acidobacteria/Proteobacteria | 1/0 | COG3034 | |
| Alpha-/Beta-/Gamma-/Epsilonproteobacteria | 4.36 | COG3302, NOG13261, NOG09591–NOG17096 | |
| Alpha-/Beta-/Gammaproteobacteria | 0.56 | COG4618, COG5611 | |
| Helicobacteraceae (Epsilonproteobacteria) | 500 | NOG18902 | |
| Rickettsiales (Alphaproteobacteria) | 1/0 | NOG07928 | |
| Beta-/Gammaproteobacteria | 1,000 | COG4969 | |
| Archaea | 368.42 | COG1423, COG1458, COG1503, COG1517, COG1730, COG2112, COG4831 | |
| Crenarchaeota | 1/0 | COG4353 | |
| Sulfolobus (Crenarchaeota) | 1,000 | NOG18904 | |
| Methanosarcina (Euryarchaeota) | 500 | NOG09683 | |
| Fusobacterium nucleatum | Bacteria | 33,500 | 67 COGs |
| Lactobacillales (Firmicutes) | 83.33 | NOG17664 | |
| Mycoplasmataceae ex. Mycoplasma mycoides (Firmicutes) | 1/0 | NOG19254–NOG36375 | |
| Treponema (Spirochaetales) | 1/0 | NOG17678 | |
| Alpha-/Beta-/Gamma-/Epsilonproteobacteria | 3.51 | COG2992, COG3713, NOG11181 | |
| Alpha-/Beta-/Gammaproteobacteria | 0.47 | COG4797, NOG18514 | |
| Pasteurellaceae ex. Haemophilus ducreyi (Gammaproteobacteria) | 1,000 | NOG09881 | |
| Vibrionaceae/Pasteurellaceae/Enterobacteriaceae (Gammaproteobacteria) | 1.20 | COG2926 | |
| Methanosarcina (Euryarchaeota) | 1,000 | NOG22419 | |
| Halobacterium sp. | Archaea | 9,000 | 114 COGs, COG1591–NOG14885, COG3353–NOG29648, COG4023–NOG17603, NOG39364–NOG10118 |
| Euryarchaeota | 5/0 | COG1422, COG1777, COG2150, COG3390, COG1711–NOG33052 | |
| Archaeoglobus/Methanosarcina (Euryarchaeota) | 1,500 | COG4749, COG4885, COG5427 | |
| Methanosarcina (Euryarchaeota) | 1,000 | NOG06067, NOG17658, NOG15033 | |
| Methanococcales/Methanopyrus kandleri/Methanothermobacter thermoautotrophicus (Euryarchaeota) | 1/0 | COG3363 | |
| Pyrococcus ex. Pyrococcus furiosus (Euryarchaeota) | 1/0 | NOG24228 | |
| Leptospira (Spirochaetaceae) | 1/0 | NOG15034 | |
| Actinobacteridae | 76.92 | COG5282 | |
| Mycobacterium (Actinobacteridae) | 166.67 | NOG20057 | |
| Streptomyces (Actinobacteridae) | 83.33 | NOG36090, NOG15774 | |
| Cyanobacteria | 181.82 | COG4250, COG5524 | |
| Alpha-/Beta-/Gammaproteobacteria | 0.56 | COG3205, COG4538 | |
| Caulobacter vibrioides/Rhizobiales (Alphaproteobacteria) | 43.48 | COG3743 | |
| Nanoarchaeum equitans | Archaea | 67/0 | 66 COGs, NOG21880 |
| Euryarchaeota | 2/0 | COG1311, COG1933 | |
| Methanosarcina (Euryarchaeota) | 1/0 | NOG11162 | |
| Pyrococcus (Euryarchaeota) | 1/0 | NOG17563 | |
| Thermotoga maritima | Bacteria | 60/0 | 60 COGs |
| Clostridia (Firmicutes) | 200 | NOG22606 | |
| Archaea | 352.94 | COG1031, COG1184, COG1635, COG1992, COG3374, COG5014 | |
| Pyrococcus (Euryarchaeota) | 1/0 | NOG13536 | |
| Pyrococcus ex. P. furiosus (Euryarchaeota) | 1/0 | NOG23777 |
NOTE.—In some cases, no shared signature genes were found in the 1,000 randomized genome sets (e.g., o/e ratio 1/0). OGs that are linked with a hyphen were merged because they are homologous and have a nonoverlapping taxon distribution (see Methods). For the species names and clades see fig. 2.
A difficult case in classic gene content trees is Halobacterium sp. (Dutilh et al. 2004). Due to horizontal gene transfers (HGTs) with the Bacteria (Kennedy et al. 2001), this euryarchaeon is often found at the root of the Archaea in gene content trees (see also supplementary fig. 1, Supplementary Material online). However, our alternative application of gene content shows that many more signature genes than expected are shared with several Euryarchaeota clades (table 3), supporting the sequence-based taxonomic positioning of Halobacterium sp. in the Euryarchaeota.
Nanoarchaeum equitans is a tiny thermophilic archaeal parasite that was originally assigned to a novel, anciently branching archaeal phylum on the basis of an unpolished superalignment approach (Huber et al. 2002; Waters et al. 2003). Because of the split structure of many of its genes, the position that N. equitans is a living fossil still receives support (Di Giulio 2006), but the argument in that paper leans heavily on the tRNA molecule, which is usually codified in a single gene, but in N. equitans comprises 2 separate genes that are not contiguous in the genome. However, evidence for other affiliations can also be found. A BlastP-based survey of the phylogenetic pattern of all N. equitans open reading frames finds a strong link with the Euryarchaeota (Brochier et al. 2005), more specifically the Thermococcales. We also find that N. equitans clusters with the Pyrococci in a classic gene content tree (supplementary fig. 1, Supplementary Material online). Conversely, in the curated superalignment phylogeny we used as a reference (Ciccarelli et al. 2006), N. equitans clusters with the Crenarchaeota with high-bootstrap value (cf., fig. 2). However, not one signature is found for this N. equitans/Crenarchaeota clade (fig. 2). If we re-identify signature genes for all clades in the phylogeny after removing N. equitans, we find that several Euryarchaeota, among which Pyrococcus, share many more signature genes with N. equitans than expected, whereas no links to any Crenarchaeota clade are observed (table 3). Therefore, our results support the position of N. equitans as a derived Euryarchaeote, possibly related to Pyrococcus (Brochier et al. 2005).
Assessing Species Distribution in Metagenomics Samples
To show that the signature gene application can be applied to incomplete genomes, we have mapped the taxonomic distribution of signature genes identified in 3 metagenomics samples from the Sargasso sea (Venter et al. 2004), agricultural soil, and 3 deep-sea “whale fall” carcasses that have been assigned to OGs (Tringe et al. 2005). Beside the phylogenetic analyses in the papers that introduced these data sets, these environmental samples have recently been included in another phylogenetic analysis based on 31 universal marker genes (von Mering, Hugenholtz, et al. 2007), which provides insightful additional reference material to compare our signature genes approach with sequence-based approaches.
In the sequence-based approaches, the soil and seawater samples were shown to contain the largest species diversity. The soil sample mainly consisted of Chloroflexi (not in our data set of complete genomes), Alphaproteobacteria, and Bacteroidetes but also many Betaproteobacteria, Gammaproteobacteria, Gemmatimonadetes (not in our data set), Deltaproteobacteria and Acidobacteria (both not a clade in the reference tree, see fig. 2), and Actinobacteria (fig. 4g, supplementary fig. S2B in Tringe et al. 2005 and supplementary fig. S1A in von Mering, Hugenholtz, et al. 2007). In the original analysis of the Sargasso sea sample that was based on 6 phylogenetic markers (16S rRNA, RecA, EF-Tu, EF-G, HSP70, and RNA polymerase B) and in the later analysis based on 31 universal marker genes, the phylotypes were shown to be dominated by Alpha- and Gammaproteobacteria, but they were also shown to contain many Cyanobacteria, Bacteroidetes, and Betaproteobacteria (fig. 4h and fig. 6 in Venter et al. 2004 and supplementary fig. S1B in von Mering, Hugenholtz, et al. 2007). Finally, the whale fall samples were primarily mapped to Alphaproteobacteria, Bacteroidetes, Epsilon-, and Gammaproteobacteria (fig. 4i, supplementary fig. S4A in Tringe et al. 2005 and supplementary fig. S1C in von Mering et al. 2007). As figure 4a–f show, the previously reported species distributions based on the phylogenetic analyses of marker genes show a surprisingly good correspondence with the clades for which we find signature genes in these metagenomic samples, although in some cases, the precise proportions vary. Clearly, signature genes provide an independent tool that can be used to phylogenetically map unidentified, even incomplete genomes, or metagenomics data sets, allowing the exploitation of a complementary fraction of the data contained in these sequence samples.
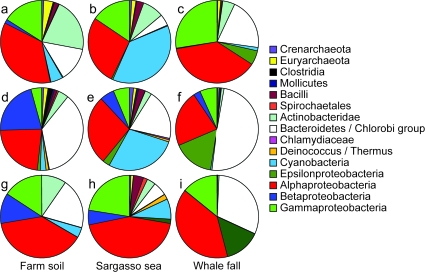
FIG. 4.—
Phylogenetic distribution of 3 metagenomics data sets (Venter et al. 2004; Tringe et al. 2005). Pies (a–c) are the total numbers of signature genes found for each clade (including subclades); pies (d–f) are the percentages of the total number signature genes that exist for each clade; pies (g–i) are the percentages of sequences found with several phylogenetic markers in the original publications (averages of all measurements; taxa that were not in the reference tree are not shown). According to the phylogenetic marker-based analyses, all 3 metagenomics data sets were highly dominated by bacterial signature genes (farm soil: 72%; sea: 78%; and whale fall: 70%), archaeal signature genes were present in much lower percentages (farm soil: 0.05%; sea: 0.6%; and whale fall: 0.1%). These phylogenetically less informative clades are not shown in the charts. This analysis is based on STRING 6.3 OGs as the mapping of the metagenomics data sets was only available for that version (kindly provided by C. von Mering).
A question that can be asked is what would be the extent of genome incompleteness that can be tolerated for accurate locating its position in the reference phylogeny? Or, how many signature genes should the incomplete genome have for accurate locating? This is a very valid question that does not have a straightforward answer. In principle, a single signature gene already pinpoints the taxonomic relatives of a new, incomplete genome with ∼92% precision (see table 2). The o/e ratio (table 3, supplementary table 1, Supplementary Material online) indicates the significance of finding a signature OG for a certain clade. In a web tool we have developed for the taxonomic characterization of a sequence sample using the signature gene approach (Dutilh et al. 2008), we take an additional step, summing the number of signature genes present in all the ancestral nodes for every species. This heuristic gives a good indication as to which species are most closely related to the origin of a sequence sample.
Discussion
One of the weaknesses of classic gene content trees is that they require completely sequenced genomes (Snel et al. 1999; Tekaia et al. 1999), which may not always be available (Tringe et al. 2005). Here, we solve this problem by introducing signature genes to employ gene content for phylogenetic analysis. The wealth of complete genomes allows us to identify signature genes for a range of taxa, and the presence of signature genes in an unidentified sample can help to detect the taxonomic composition of the query. However, the comprehensive overview of the gene repertoires of a diversity of species has also uncovered a great plasticity in gene content, with examples of extensive gene loss (e.g., in parasitic genomes [Fraser et al. 1995]), and HGT in prokaryotes (Doolittle 1999) as well as in Eukaryotes (Andersson 2005). Thus, a strict search for signature genes, which requires complete coverage of all genomes within the taxon, will only yield limited results (Charlebois and Doolittle 2004). To overcome this, we develop an intuitive definition that defines as signatures of a clade those genes that occur in every daughter of that clade, but complete coverage is not required. A coverage score indicates how well the signature gene has been retained in the descendant lineages.
We identified 8,362 signature genes for 112 clades throughout the prokaryotic tree of life (fig. 2), underlining the phylogenetic signal that exists in gene content. Based on a historical reconstruction (fig. 3) and on several cross-validation experiments, we expect that with the inclusion of more sequenced genomes, the number of signatures will grow, rather than shrink, and using a reasonable coverage score cutoff, the number of signature genes per taxon will remain quite stable (fig. 3). Theoretically, the number of signature genes may decrease due to their identification in species from other clades or increase due to a more complete sampling of the taxon. So far, the Global Ocean Sampling project, the largest environmental sequencing project published, identified almost 4,000 protein families in 7.7 million sequences (Rusch et al. 2007; Yooseph et al. 2007). Nevertheless, this abundance of data has hardly reduced the number of signatures for very ancient taxa (Bacteria and Archaea). Within the prokaryota, the authors find one Pfam domain that was thought to be Bacteria specific to be present in the Archaea and 4 Archaea specific Pfam domains in the Bacteria (Yooseph et al. 2007). With the spring tide of data from large-scale sequencing projects like the Global Ocean Sampling project, the trustworthiness of signature genes will increase, even if, or better, because some genes thus far thought to be a signature have to be dropped, being discovered in other clades as well.
As our analyses show, signature genes can complement traditional sequence-based methods and classic gene content based on complete genomes in addressing taxonomic questions. Conceptually, this gene-content approach is reminiscent of the slow–fast method (Brinkmann and Philippe 1999), where slowly evolving sites in an amino acid alignment are selected as those positions that have not mutated within predefined clades. These positions are the most reliable for inferring ancient relationships, as fast-evolving sites are likely to be mutationally saturated, obscuring the phylogenetic signal. Signature genes evolve slowly at the gene content level. Especially, the signature genes with high-coverage scores have undergone little loss or HGT and are thus strong indicators of phylogenetic relatedness.
Research aimed at elucidating lineage-specific properties for the clades included in this work will benefit from the list of uncharacterized genes (supplementary table 1, Supplementary Material online), which forms a wealth of suggestions for further experimental investigations into taxon-specific processes. Concluding, signature genes are a promising tool that can be used in a number of research areas, from taxonomic analysis of incomplete genomes and metagenomics data to the identification of clade-specific genes.
Supplementary Material
Supplementary tables 1 and 2, and figure 1 are available at Molecular Biology Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We thank Christian von Mering for the data that maps the metagenomics sequences to STRING 6.3 OGs. This work was supported by the European Union‘s 6th Framework Program, contract number LSHM-CT-2004-503116 (EUMITOCOMBAT) and contract number LSHB-CT-2005-019067 (EPISTEM).
References
- Andersson JO. Lateral gene transfer in eukaryotes. Cell Mol Life Sci. 2005;62:1182–1197. doi: 10.1007/s00018-005-4539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann H, Philippe H. Archaea sister group of Bacteria? Indications from tree reconstruction artifacts in ancient phylogenies. Mol Biol Evol. 1999;16:817–825. doi: 10.1093/oxfordjournals.molbev.a026166. [DOI] [PubMed] [Google Scholar]
- Brochier C, Gribaldo S, Zivanovic Y, Confalonieri F, Forterre P. Nanoarchaea: representatives of a novel archaeal phylum or a fast-evolving euryarchaeal lineage related to Thermococcales? Genome Biol. 2005;6:R42. doi: 10.1186/gb-2005-6-5-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlebois RL, Doolittle WF. Computing prokaryotic gene ubiquity: rescuing the core from extinction. Genome Res. 2004;14:2469–2477. doi: 10.1101/gr.3024704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Di Giulio M. Nanoarchaeum equitans is a living fossil. J Theor Biol. 2006;242:257–260. doi: 10.1016/j.jtbi.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- Dutilh BE, He Y, Hekkelman ML, Huynen MA Forthcoming. Signature: a web server for taxonomic characterization of sequence samples using signature genes. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn277. Web Server Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilh BE, Huynen MA, Bruno WJ, Snel B. The consistent phylogenetic signal in genome trees revealed by reducing the impact of noise. J Mol Evol. 2004;58:527–539. doi: 10.1007/s00239-003-2575-6. [DOI] [PubMed] [Google Scholar]
- Dutilh BE, van Noort V, van der Heijden RT, Boekhout T, Snel B, Huynen MA. Assessment of phylogenomic and orthology approaches for phylogenetic inference. Bioinformatics. 2007;23:815–824. doi: 10.1093/bioinformatics/btm015. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Gocayne JD, White O, et al. (29 co-authors) The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- Gao B, Gupta RS. Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis. BMC Genomics. 2007;8:86. doi: 10.1186/1471-2164-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Paramanathan R, Gupta RS. Signature proteins that are distinctive characteristics of Actinobacteria and their subgroups. Antonie Van Leeuwenhoek. 2006;90:69–91. doi: 10.1007/s10482-006-9061-2. [DOI] [PubMed] [Google Scholar]
- Griffiths E, Ventresca MS, Gupta RS. BLAST screening of chlamydial genomes to identify signature proteins that are unique for the Chlamydiales, Chlamydiaceae, Chlamydophila and Chlamydia groups of species. BMC Genomics. 2006;7:14. doi: 10.1186/1471-2164-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417:63–67. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- Kainth P, Gupta RS. Signature proteins that are distinctive of alpha proteobacteria. BMC Genomics. 2005;6:94. doi: 10.1186/1471-2164-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy SP, Ng WV, Salzberg SL, Hood L, DasSarma S. Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 2001;11:1641–1650. doi: 10.1101/gr.190201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KA, Siefert JL, Yerrapragada S, Lu Y, McNeill TZ, Moreno PA, Weinstock GM, Widger WR, Fox GE. Cyanobacterial signature genes. Photosynth Res. 2003;75:211–221. doi: 10.1023/A:1023990402346. [DOI] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Palla G, Derenyi I, Farkas I, Vicsek T. Uncovering the overlapping community structure of complex networks in nature and society. Nature. 2005;435:814–818. doi: 10.1038/nature03607. [DOI] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, et al. (40 co-authors) The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through Eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. Genome phylogeny based on gene content. Nat Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- Snel B, Huynen MA, Dutilh BE. Genome trees and the nature of genome evolution. Annu Rev Microbiol. 2005;59:191–209. doi: 10.1146/annurev.micro.59.030804.121233. [DOI] [PubMed] [Google Scholar]
- Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Strous M, Pelletier E, Mangenot S, et al. (27 co-authors) Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature. 2006;440:790–794. doi: 10.1038/nature04647. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekaia F, Lazcano A, Dujon B. The genomic tree as revealed from whole proteome comparisons. Genome Res. 1999;9:550–557. [PMC free article] [PubMed] [Google Scholar]
- Tringe SG, von Mering C, Kobayashi A, et al. (13 co-authors) Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, et al. (23 co-authors) Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- von Mering C, Hugenholtz P, Raes J, Tringe SG, Doerks T, Jensen LJ, Ward N, Bork P. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science. 2007;315:1126–1130. doi: 10.1126/science.1133420. [DOI] [PubMed] [Google Scholar]
- von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, Snel B, Bork P. STRING 7–recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters E, Hohn MJ, Ahel I, et al. (22 co-authors) The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc Natl Acad Sci USA. 2003;100:12984–12988. doi: 10.1073/pnas.1735403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yooseph S, Sutton G, Rusch DB, et al. (33 co-authors) The Sorcerer II Global Ocean Sampling expedition: expanding the Universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.