Abstract
Pax proteins are a family of transcription factors with a highly conserved paired domain; many members also contain a paired-type homeodomain and/or an octapeptide. Nine mammalian Pax genes are known and classified into four subgroups: Pax-1/9, Pax-2/5/8, Pax-3/7, and Pax-4/6. Most of these genes are involved in nervous system development. In particular, Pax-6 is a key regulator that controls eye development in vertebrates and Drosophila. Although the Pax-4/6 subgroup seems to be more closely related to Pax-2/5/8 than to Pax-3/7 or Pax-1/9, its evolutionary origin is unknown. We therefore searched for a Pax-6 homolog and related genes in Cnidaria, which is the lowest phylum of animals that possess a nervous system and eyes. A sea nettle (a jellyfish) genomic library was constructed and two pax genes (Pax-A and -B) were isolated and partially sequenced. Surprisingly, unlike most known Pax genes, the paired box in these two genes contains no intron. In addition, the complete cDNA sequences of hydra Pax-A and -B were obtained. Hydra Pax-B contains both the homeodomain and the octapeptide, whereas hydra Pax-A contains neither. DNA binding assays showed that sea nettle Pax-A and -B and hydra Pax-A paired domains bound to a Pax-5/6 site and a Pax-5 site, although hydra Pax-B paired domain bound neither. An alignment of all available paired domain sequences revealed two highly conserved regions, which cover the DNA binding contact positions. Phylogenetic analysis showed that Pax-A and especially Pax-B were more closely related to Pax-2/5/8 and Pax-4/6 than to Pax-1/9 or Pax-3/7 and that the Pax genes can be classified into two supergroups: Pax-A/Pax-B/Pax-2/5/8/4/6 and Pax-1/9/3/7. From this analysis and the gene structure, we propose that modern Pax-4/6 and Pax-2/5/8 genes evolved from an ancestral gene similar to cnidarian Pax-B, having both the homeodomain and the octapeptide.
Keywords: paired box, eye evolution, homeobox, DNA binding, Cnidaria
Pax genes, which are defined by the presence of a conserved paired box that codes for a 128-aa paired domain, encode transcription factors involved in developmental control, notably in the formation of the central nervous system. Besides the paired domain, many Pax genes also encode a paired-type homeodomain and/or an octapeptide motif. Nine Pax genes have been identified in human and mouse (1, 2), and six have been identified in Drosophila (3, 4). Based on sequence similarity and gene structure, the nine mammalian Pax genes have been divided into four subgroups: Pax-1/9, Pax-2/5/8, Pax-4/6, and Pax-3/7 (5). However, the evolutionary relationships among the Pax genes remain to be examined in detail.
Pax-6 is of special interest to us because it is involved in eye development in both invertebrates and vertebrates. Mutations in Pax-6 can cause aniridia in humans (6), small eye in rodents (7), and eyeless in Drosophila (4). In Drosophila, ectopic expression of either the Drosophila or the mouse Pax-6 gene induces eye formation (8). This suggests that Pax-6 is the key control gene in the development of the two different types of eye in vertebrates and Drosophila (8), and raises the possibility that a Pax-6 homolog is also used to control eye development in more primitive organisms. Cnidarians are the most primitive organisms that possess eyes, ranging from simple eye spots to complicated lens eyes (9, 10). We selected two cnidarians, a jellyfish (the sea nettle, Chrysaora quinquecirrha) and a hydra (the Hydra littoralis), for the characterization of Pax genes related to Pax-6 or other Pax genes. The sea nettle has simple eye spots. Although the hydra has no eye structure, this may represent an evolutionary loss because fairly sophisticated eye structures are found in the related genera Polyorchis and Spirocodon. Therefore, it is interesting to see whether both the sea nettle and the hydra have a Pax-6 homolog. The hydra has the advantage of being easier to culture and to process for RNA isolation for determining the coding regions of a gene.
Here we describe the isolation and sequencing of two paired box containing genes (Pax-A and Pax-B) from both the sea nettle and the hydra resulting from a search for a Pax-6 homolog and other Pax genes. A detailed analysis of the evolutionary relationships among the Pax genes suggests that the sea nettle and hydra Pax-B genes are more closely related to Pax-2/5/8 and Pax-6 than to Pax-1/9 and Pax-3/7. Consistent with this, a DNA binding assay demonstrated that three of the cnidarian paired domains bind to Pax-5/6 sites. We propose that the modern Pax-2/5/8 and Pax-6 genes have evolved from an ancestral gene similar to the cnidarian Pax-B gene.
MATERIALS AND METHODS
Preparation of Genomic DNA.
Sea nettles were collected from Galveston Bay, Texas. High molecular weight genomic DNA was isolated from several medusae by the method of Dilella et al. (11). Polyps of the hydra were obtained from Carolina Science and Math (Burlington, NC). Genomic DNA was isolated several times by the same method, each time from about 100 whole polyps.
Isolation and Characterization of Genomic Clones.
A genomic library of the sea nettle was constructed using λGEM-12 XhoI half-site vector arms (Promega) according to the manufacturer’s instruction. Plaques (2.4 × 105) were screened at low stringency [4× SSPE (standard saline phosphate/EDTA: 0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA), 64°C] using a PCR-generated probe that contained the human Pax-6 paired box sequence (bases 373–718) (6) from a cDNA clone provided by G. Saunders (University of Texas M. D. Anderson Cancer Center, Houston). A labeled probe was synthesized by amplification of 0.1 μg of paired box DNA in a reaction containing 10 pmol of the primer jffor [GTCACAG(CT)GGAGT(GA)AA(TC)CAGCT]/7.5 μM dATP, dGTP, and dTTP/50 μCi (1 Ci = 37 GBq) [α-32P]dCTP/50 mM Tris⋅HCl, pH 9.0/2 mM Mg2+. After 5 min at 94°C, 1.5 units of Taq polymerase was added, and the reaction was cycled 10 times (94°C, 30 s; 61°C, 30 s; 72°C, 1 min). Selected clones were restriction mapped, and fragments containing paired box sequences were identified by Southern blot analysis with the Pax-6 probe. Fragments were cloned into the pBluescript SK vector (Stratagene) and fully sequenced on both strands by generating nested deletions with exonuclease III (Erase-a-base kit, Promega).
PCR Amplification of Paired Box Fragments.
Degenerate PCR primers corresponding to the most conserved regions of the paired box sequence were designed and used for nested PCR amplification of paired box sequences from sea nettle and hydra. The PCR conditions were 200 ng of genomic DNA/0.2 mM each dNTP/0.8 μM each primer/1.5 units of Taq polymerase/1× PCR buffer (Promega) in a 50 μl reaction volume. After 5 min at 94°C, Taq polymerase was added, and the reaction cycled 30 times (94°C, 30 s; 42°C, 30 s; 72°C, 1 min). The first PCR was carried out using primers prdf3 [(GC)(GA)(AGCT)GT(AGC)AA(TC)CA(AG)(TC)T(AGCT)GG] and prdr4 [(CT)(TG)(AGCT)TC(TCG)C(TG)(AGT)AT(TC)TCCCA]. Two microliters of the reaction was then used as a template in a second PCR with the primers prdf2 [GG(AGCT)GT(AGCT)TT(TC)(AG)T(CG)AA(TC)GG] and prdr3 [AT(TC)TCCCA(AGCT)(GC)(CTA)(AG)AA(AGCT)AT].
cDNA Cloning of Hydra Paired Box Sequences.
Full-length cDNAs of two paired box containing genes from the hydra were obtained by rapid amplification of cDNA ends (RACE)–PCR (12). Internal primers were designed based on the sequences of the paired boxes obtained by PCR. RNA was isolated from 50 to 100 polyps by lysis in 1 ml of TRIzol (Life Technologies, Gaithersburg, MD), followed by addition of 200 μl of chloroform and recovery of the RNA by isopropanol precipitation from the aqueous phase. For 3′ RACE, one-tenth of one RNA preparation was reverse transcribed using Superscript reverse transcriptase and aprt [GGCCACGCGTCGACTAGTAC(T17)] as the primer following the manufacturer’s instructions (Life Technologies). One-tenth of this reaction was then used as the template for nested PCR. To recover the genes containing the two different paired boxes isolated by genomic PCR, two different primer sets were used. The set 1 primers were hy1rt (CTACCTGATTACATGAGACATC) and uaprt (GGCCACGCGTCGACTAGTAC) for the first PCR; hy1rt2 (GTATAATAGAACTGGCGCAAT) and oligo(dT) [GTAC(T17)] for the second PCR. The set 2 primers were hy3rt (CTCGTTCGTAGAAAAATTGTTG) and uaprt for the first PCR; hy3rt2 (AGTTGGCACATCCAGGTGTCC) and oligo(dT) for the second PCR. Two microliters of the first PCR products was used as the template for the second PCR. To recover the 3′ terminus of the gene amplified by the set 2 primers, a third round of amplification was performed using the primers hy3rt3f (CATGATTCTTCAGCTGCTTCA) and oligo(dT).
For 5′ RACE, 25% of an RNA preparation was reverse transcribed using either hy1rt1r (GCAAGGATTTTCCTCTTTTAA) or hy3rt1r (TGTTGGGTTATGCTGTTTGTA) as a primer. The reverse transcription products were purified by phenol–chloroform extraction and ethanol precipitation. The pellets were resuspended in 1× terminal deoxynucleotidyltransferase buffer containing 1 mM dATP and 17 units terminal deoxynucleotidyltransferase (United States Biochemical), and incubated for 30 min at 37°C. Two microliters of these reaction mixes was used as the templates for nested PCR amplification. In the first round, the hy1rt1r primed cDNA was amplified using hy1rt1r and aprt as primers. In the second round, 2 μl of the first round product was amplified with the primer pairs hy1rt2r (GACAATTCGACATACCACTTT) and uaprt. For the hy3rt1r primed cDNA, the first and second round primer pairs were hy3rt1r and aprt and hy3rt2r (CTTGTATTTTAGCAACAACAG) and uaprt, respectively. Since these did not give a satisfactory amplification, a third round of PCR using the primers hy3rt3r (ACTGACACATCCATGAGAAAC) and oligo(dT) was performed. The PCR conditions were 30 cycles of 94°C, 30 sec; 56°C, 30 sec; 72°C, 2 min.
Electrophoretic Gel-Shift Assay.
The complete paired boxes of the sea nettle and hydra, as well as the human PAX-6 paired box, were cloned in frame into the pCITE-4b(+) vector and in vitro transcribed and translated using a single tube system (Novagen) following the manufacturer’s instructions. The binding of these proteins to Pax binding sites was examined by a gel-shift assay (13). The double-stranded oligonucleotides used in the assay were H2A22 (CAGGGTTGTGACGCAGCGGTGGGTGACGACTGTCGG) and CD19–1 (CCCCGCAGACACCCATGGTTGAGTGCCCTCCAGGCC) (13). For each oligonucleotide, the two complementary strands were mixed in 2× SSC/10 mM Tris⋅HCl (pH 8.0), boiled for 5 min, and cooled to room temperature. After radiolabeling with 32P, 3 fmol of DNA probe was incubated for 30 min at room temperature with 5 μl of in vitro translated paired domain peptide in a 20-μl reaction mix containing 2 μg of poly(dI⋅dC)/10 mM Hepes, pH 7.9/100 mM KCl/4% Ficoll/1 mM EDTA/1 mM dithiothreitol. Protein–DNA complexes were analyzed on native 6% polyacrylamide gels containing 0.25% TBE.
RESULTS
Characterization of Cnidarian Pax Gene Sequences.
To search for cnidarian Pax genes related to human PAX-6, a sea nettle genomic DNA library was constructed and a low stringency screen of the library was performed using a human PAX-6 paired box probe. This screen identified six positive clones. Based on their restriction maps, these clones corresponded to two distinct genes (data not shown). A 4.2-kb HindIII fragment from clone I and a 3.7-kb PstI–BamHI fragment from clone II were subcloned and sequenced. Surprisingly, these two clones encoded an identical paired domain peptide sequence with only two synonymous substitutions in 374 bases. However, they carried two quite distinct regions flanking the paired box. In the 5′ flanking region (776 bp), there were 30 differences and 9 insertions/deletions, whereas in the 3′ flanking region (363 bp) there were four differences. Therefore, they could not simply correspond to different alleles of the same gene, and probably correspond to two distinct genes that arose from a recent duplication. We named this paired domain sea nettle paired domain A (Pax-A) and the two genes Pax-A1 and -A2. Like Pax-1 and -9 but unlike other vertebrate Pax genes, both sea nettle genes contain no intron in the paired box.
To isolate additional cnidarian paired box sequences, degenerate primers for PCR amplification of genomic DNA were designed based on the most conserved regions of the paired box sequence. Alignment of 24 paired box sequences in GenBank identified the conserved peptide sequences G(E/R)VNQLG and GVFV(I)NG encoded by the 5′ end of the paired box, and the sequence M(I)FAWEIRD(E/A)R(K/T/Q) encoded by a sequence close to the 3′ end. The primers prdf3 and prdf2, and prdr4 and prdr3 were synthesized according to the 5′ and 3′ end conserved coding sequences, respectively. By using these primers for nested PCR, we were able to recover three types of paired box products from the sea nettle and two from the hydra. Two of the three sea nettle products corresponded to the paired box A1 and A2 identified from the genomic library clones. The third sea nettle product was used as a probe to rescreen the sea nettle genomic library. Two clones were obtained. The restriction maps of these two clones with four different restriction enzymes were identical. A 1.2-kb HindIII fragment from the first positive clone was sequenced. This paired domain was distinct from sea nettle paired domain A, and was designated paired domain B. The paired box of this gene contains no intron. A conserved octapeptide sequence was found downstream of the paired domain in the same reading frame.
The two PCR products obtained from the hydra were the homologs of the sea nettle paired domains A and B. The complete coding sequences of the two hydra gene products were obtained by reverse transcriptase–PCR cDNA cloning. The two cDNAs contained 641 and 608 codons, respectively, and were named hydra Pax-A and Pax-B. Hydra Pax-B contained both the octapeptide and a complete homeodomain, whereas Pax-A contained neither.
DNA Binding Site-Specificity of Cnidarian Paired Domains.
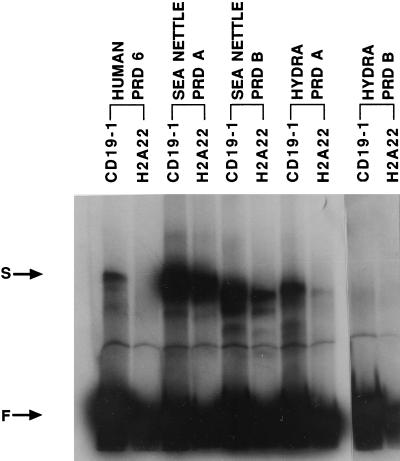
Since the paired domain is a sequence-specific DNA binding domain, we performed gel-shift DNA binding assays to examine the functionality of the four cloned cnidarian paired domains. To provide a preliminary assessment of the binding specificity, we selected two human PAX binding sites, CD19–1 and H2A22 (13). The CD19–1 site is bound by human PAX-5 and -6 paired domains, while H2A22 is only bound by PAX-5. As shown in Fig. 1, the two sea nettle paired domains and hydra paired domain A bound both sites, whereas hydra paired domain B bound neither. The human PAX-6 paired domain was used as a positive control, demonstrating the expected binding specificities.
Figure 1.
DNA binding assays. The binding specificity of in vitro translated human paired domain 6 (human prd 6), sea nettle paired domain A and B (sea nettle prd A and B), and hydra paired domain A and B (hydra prd A and B) with PAX-5 binding sites CD19–1 and H2A22 were analyzed. Shifted band (S) and free probe (F) are shown by arrows.
Conserved Regions.
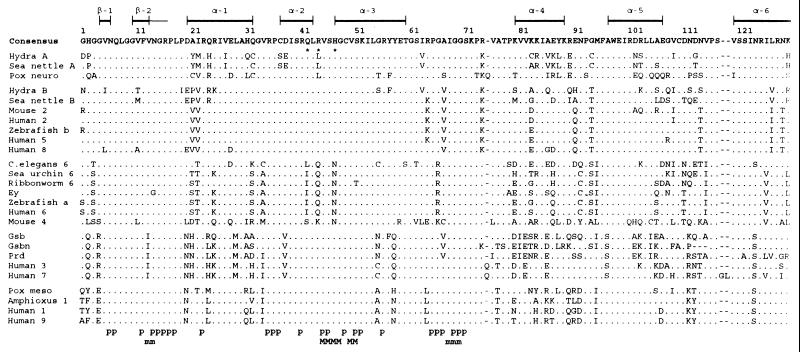
Fig. 2 shows an alignment of 26 paired domain sequences. The length of the alignment is 126 aa; we have excluded the last 2 aa residues because they are not available for some of the sequences.
Figure 2.
Alignment of the paired domain amino acid sequences. The domain includes a β sheet (β-1), a β sheet+turn (β-2), and six α helices. The DNA contact positions with the sugar phosphate backbone (p) and major/minor grooves (M/m) are indicated at the bottom of the figure. Amino acids identical to the consensus sequence are indicated by dots and insertions/deletions are indicated by dashes. The GenBank accession numbers for the sequences are: pox neuro, X58917X58917; mouse Pax-2, X55781X55781; human PAX-2, L25597L25597; zebrafish b, X63961X63961; human PAX-5, M96944M96944; human PAX-8, L19606L19606; Caenorhabditis elegans pax-6 (vab-3), U31537U31537; sea urchin pax-6, U14621U14621; ribbonworm pax-6, X95594X95594; Drosophila ey, X79493X79493; zebrafish a, X61389X61389; human PAX-6, M93650M93650; mouse Pax-4, P32115P32115 (Swiss-Prot database); gsb, M14944M14944/2; gsbn, M14943M14943; prd, M14548M14548; human PAX-3 and -7, U02368U02368 and X15042X15042/15250/1; pox meso, X16992X16992; amphioxus Pax-1, U20167U20167; human PAX-1, X15044X15044; and human PAX-9, L09745L09745. Xenopus laevis Pax-6; chicken Pax-1 and -9; quail Pax-6; rat Pax-8; mouse Pax-1, -3, -5, -6, -7, -8, and -9; and dog Pax-8 are identical to their human homologs and are not presented in the alignment.
Since the most important function of the paired domain is DNA binding, it is interesting to see whether the DNA binding contact positions with the sugar phosphate backbone (Fig. 2, p) are well conserved. Indeed, with a few exceptions, all the 25 DNA contact positions are completely conserved among all the known sequences, even though not all of these contact positions are clustered together. The positions for contacting the major and minor grooves are also well conserved, though not so well as the phosphate backbone DNA contact positions. Generally speaking, the regions that cover these DNA contact positions are better conserved than other regions. In particular, the region from positions 6 to 19, which includes nine DNA contact positions, a β sheet (β-1), and a β sheet+turn (β-2), and the region from positions 35 to 54, which includes 13 DNA contact positions and two α helices (α-2 and α-3), are two of the best conserved regions. The α-5 and α-6 regions are moderately conserved, but the α-1 and α-4 regions are not well conserved.
In Fig. 2, we classify the sequences into five groups according to their sequence similarity. Clearly, sea nettle and hydra Pax-A are more similar to Drosophila pox neuro than to other sequences, whereas sea nettle and hydra Pax-B are more similar to vertebrate Pax-2/5/8. Note that in each group there are well conserved blocks of amino acids. For example, the block from positions 25 to 62 is almost completely conserved in the Pax-B/2/5/8 group. Note also that the sequences in each group share many unique amino acids. For example, amino acids R, I, M, V, Q, E, and S at positions 4, 13, 29, 37, 58, 81, and 96 are uniquely shared by the group gsb/gsbn/prd/Pax-3/Pax-7, but not by any other sequence.
Hydra Pax-B is quite divergent from both Pax-5 and the consensus sequence, which may explain why it did not bind the two Pax-5 sites tested. Mouse Pax-4 is even more divergent. In fact, it differs from the consensus sequence by 44 positions among the 126 positions aligned and it differs from the other member in the ey/Pax-4/Pax-6 group, on average, by 37 positions. It is possible that mouse Pax-4 is on the way to become a pseudogene. For this reason, this gene will not be discussed in detail in the rest of the paper.
Fig. 3 shows an alignment of octapeptide sequences. It is noted that hydra Pax-B has an octapeptide identical to that of human PAX-5, whereas sea nettle Pax-B has an octapeptide identical to those of human PAX-2 and zebrafish zf-b. On the other hand, hydra Pax-A does not have a recognizable octapeptide sequence; sea nettle Pax-A sequence is not yet available for this region.
Figure 3.
Alignment of octapeptide sequences. The octapeptide is absent in hydra Pax-A, vertebrate Pax-6, sea urchin and ribbonworm Pax-6, Drosophila ey, C. elegans vab-3, and Drosophila prd.
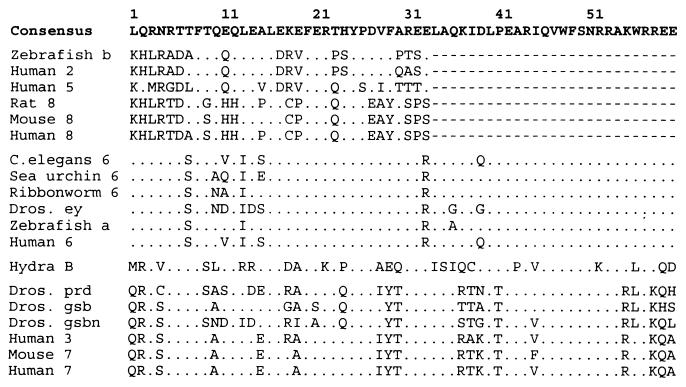
Fig. 4 shows an alignment of the known paired-type homeodomain sequences. As is already known, the Pax-2/5/8 group members have only a partial homeodomain. Fig. 4 reveals that the partial domain can barely be recognized; it varies considerably among the members and differs greatly from the consensus sequence. In contrast, the ey/Pax-6 group has a complete, well-conserved homeodomain, especially in the following regions: positions 1–6, positions 16–32, and positions 40–60. The gsb/gsbn/prd/Pax-3/Pax-7 group also has a complete homeodomain, although it is slightly less well conserved than that of the ey/Pax-6 group. The only cnidarian sequence included in Fig. 4 is from hydra Pax-B because hydra Pax-A does not possess the homeodomain and because sea nettle Pax-A and -B have not yet been sequenced in this region. In this sense, hydra Pax-B is different from the Pax-2/5/8 group but similar to the ey/Pax-6 and prd/gsb/gsbn/Pax-3/Pax-7 groups because it has a complete homeodomain. In terms of sequence similarity, it is more similar to the prd/gsb/gsbn/Pax-3/Pax-7 group than to any other sequence.
Figure 4.
Alignment of the paired-type homeodomain sequences. Pax-2, Pax-5, Pax-8, and zebrafish b have only a partial homeodomain; the missing part is indicated by dashes. Amino acids identical to the consensus sequence are indicated by dots. For sequence sources, see Fig. 2.
Phylogenetic Relationships.
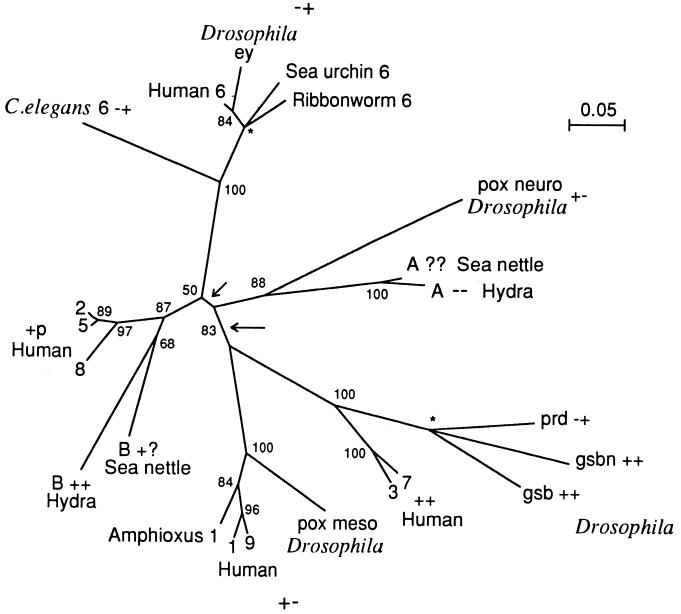
Fig. 5 shows a tree constructed from the paired domain sequences. The distances used were computed using Dayhoff’s (14) PAM matrix. Mouse Pax-4 was not included in the tree because, as mentioned, it is highly divergent from all other sequences yet it is clearly more closely related to the ey/Pax-6 group than to any other sequence (Fig. 2). All other sequences in Fig. 2 were used in the tree construction. However, the vertebrate sequences are, for simplicity, represented only by the human sequences, because the vertebrate homologs are highly similar to one another. For example, zebrafish b differs from human PAX-5 by only two amino acids and zebrafish a differs from human PAX-6 by only one amino acid (Fig. 2).
Figure 5.
Phylogenetic tree of Pax genes. The tree was inferred from the paired domain sequences (Fig. 2) using Dayhoff’s (14) distances between proteins and the neighbor-joining method (15). The numbers near each branching node denote the bootstrap values. ∗, A trifurcation point. The long arrow points to the most plausible root of the tree, whereas the short arrow points to an alternative root of the tree. The two plus-minus symbols next to a gene name signify the presence (+) or absence (−) of the octapeptide and the paired-type homeodomains, respectively; p, partial; ?, unknown.
Several clusters in Fig. 5 are well supported by bootstrap values. First, human PAX-3 and -7 are closely related and they are clustered with Drosophila prd, gbs, and gsbn; however, the branching order for Drosophila prd, gbs, and gbsn is very uncertain, being close to a trichotomy. Second, the divergence between human Pax-1 and -9 occurred long after the separation of vertebrates from amphioxus, the commonly believed predecessor of the vertebrates. These two genes and amphioxus Pax-1 are clearly clustered with Drosophila pox meso. Third, the preceding two groups (i.e., prd/gsb/gsbn/Pax-3/Pax-7 and poxm/Pax-1/Pax-9) form a supergroup that is separated from the other Pax genes. Since the bootstrap technique tends to give severe underestimates of the true statistical confidence (16–18), the 83% bootstrap value for this major division may represent a fairly high statistical confidence. Fourth, Drosophila ey and all Pax-6 genes belong to one cluster, though the branching order within this cluster apparently involves some errors, e.g., the clustering of human Pax-6 with Drosophila ey rather than with sea urchin Pax-6 is incorrect. Fifth, sea nettle and hydra Pax-A are closely related and are clustered with Drosophila pox neuro (88% bootstrap value). Finally, sea nettle and hydra Pax-B belong to one cluster and their clustering with Pax-2/5/8 is supported by a bootstrap value of 87%. However, the phylogenetic position of the Pax-A/poxn subgroup is very uncertain. In Fig. 5 this subgroup is an outgroup to the Pax- 2/5/8/Pax-B and ey/Pax-6 groups, but it was clustered with the Pax-2/5/8/Pax-B group if the Poisson distances instead of the Dayhoff’s distances were used.
DISCUSSION
The clustering of sea nettle and hydra Pax-B with Pax-2/5/8, though supported by a bootstrap value of 87% (Fig. 5), is not very certain because Pax-2/5/8 possess only a poorly conserved partial homeodomain, whereas hydra Pax-B, like the ey/Pax-6 group, possesses a complete homeodomain. In this sense, Pax-B is more similar to Pax-6. [Although in terms of sequence similarity in the homeodomain, Pax-B is more similar to Pax-3/7 (Fig. 4), Fig. 5 suggests that Pax-B is only distantly related to Pax-3/7.] The clustering of Pax-B with Pax-2/5/8 is also supported by the fact that all these genes contain the octapeptide (Fig. 3). However, the octapeptide, whose function is unclear, apparently can be lost in a relatively short time. For example, Drosophila prd has lost this motif since its separation from Drosophila gsb and gsbn (Fig. 5). Therefore, the presence or absence of the octapeptide might not readily correlate with the overall phylogeny of the Pax genes. Because of these uncertainties, we consider three possible relationships among Pax-B, Pax-2/5/8, and ey/Pax-6. First, Pax-B could in fact be more closely related to Pax-2/5/8 than to ey/Pax-6. In this case, a Pax-6 homolog that controls eye development in the sea nettle has yet to be found. Of course, it is possible that this Pax-6 homolog has been lost in the sea nettle, but this could have happened only if Pax-B or another gene can control eye development in the sea nettle. Second, sea nettle and hydra Pax-B might not be clustered with Pax-2/5/8 and their ancestor branched off before the divergence of the Pax-2/5/8 and ey/Pax-6 groups. In this case, Pax-B is the homolog of both ey/Pax-6 and Pax-2/5/8 and it should function as the key regulator of eye development in the sea nettle. Third, sea nettle and hydra Pax-B might actually be more closely related to ey/Pax-6 than to Pax-2/5/8, although their paired domains are less similar to those of ey/Pax-6 because this domain has evolved faster in ey/Pax-6 than in Pax-2/5/6 (Figs. 2 and 5). In this case, Pax-B is in fact the Pax-6 homolog in Cnidaria. In all cases, the common ancestor of ey/Pax-6 and Pax-2/5/8, like Pax-6 and Pax-B, should have a complete homeodomain. It should also have the octapeptide, because assuming the loss of this motif in the ey/Pax-6 group is easier than assuming the gain of this motif in Pax- B and Pax-2/5/8. In other words, we propose that both ey/Pax- 6 and Pax-2/5/8 were derived from a common ancestor with a gene structure similar to that of hydra Pax-B, that is, it had both the homeodomain and octapeptide as well as the paired domain.
Noll (3) has suggested that the Pax genes could be classified into four groups: poxm/Pax-1/Pax-9, prd/gsb/gsbn/Pax-3/Pax-7, poxn/Pax-2/Pax-5/Pax-8, and ey/Pax-6. Our phylogenetic analysis supports the first two groupings. The other two were also supported if the Poisson distances were used. However, if Dayhoff’s distances were used, Pax-2/Pax-5/Pax-8 was closer to ey/Pax-6 than to pox neuro (Fig. 5). In support of this, we note that pox neuro is clustered with sea nettle and hydra Pax-A (Fig. 5) and that pox neuro and hydra Pax-A do not have the homeodomain, whereas ey/Pax-6 has a complete homeodomain and Pax-2/Pax-5/Pax-8 has a partial homeodomain. However, the grouping of Pax-2/Pax-5/Pax-8 with ey/Pax-6 remains to be substantiated because its bootstrap value is only 50%. With these caveats in mind, we propose the following four groupings: poxm/Pax-1/Pax-9, prd/gsb/gsbn/Pax-3/Pax-7, Pax-A/poxn, and Pax-B/Pax-2/Pax-5/Pax-8/Pax-6/ey (where Pax-A and -B refer to the cnidarian Pax-A and -B). If Pax-B is in fact closer to Pax-2/5/8, as suggested by Fig. 5, then the last group should be divided into Pax-B/Pax-2/Pax-5/Pax-8 and ey/Pax-6.
Our phylogenetic analysis suggests that the four groups proposed above can be merged into two supergroups (Fig. 5, long arrow): Supergroup I consists of Pax-A/poxn and Pax-B/Pax-2/Pax-5/Pax-8/Pax-6/ey and supergroup II consists of poxm/Pax-1/Pax-9 and prd/gsb/gsbn/Pax-3/Pax-7. This major division is supported by a bootstrap value of 83%. The DNA binding experiments with cnidarian paired domains reported here show binding affinity for a Pax-5/6 site and a Pax-5 site. This functional result is consistent with the proposed major groupings. The lack of a detectable mobility shift with hydra paired domain B and either of the two Pax sites tested suggests that this domain has a binding specificity distinctly different from the other cnidarian paired domains. However, at present we cannot exclude the possibility that the in vitro translated peptide might not be in the correct conformation to bind DNA. A third possibility is that the function of hydra Pax-B has changed. Further studies will be required to resolve this question. Note that in each supergroup, the homeodomain is present in some members (e.g., Pax-3 and -7) but absent in the others (e.g., Pax-1 and -9). This observation implies either multiple gains or multiple losses of the homeodomain during the evolution of these genes. Since gaining a homeodomain is probably more difficult than losing it, we propose that the common ancestral gene contained the homeodomain. By the same reasoning, we propose that the ancestral gene also contained the octapeptide.
In summary, there are two plausible scenarios for the evolution of the Pax genes. The first one places the root in between the two supergroups as indicated by the longer arrow in Fig. 5. This scenario implies that the ancestral gene was duplicated before the divergence of supergroups I and II. Therefore, a cnidarian supergroup II gene is yet to be found, unless it has been lost. The supergroup I ancestral gene gave rise to the Pax-A/poxn lineage and the Pax-B/Pax-2/Pax-5/Pax-8/Pax-6/ey lineage. The second scenario places the root on the branch separating the Pax-A/poxn lineage from the Pax-B/Pax-2/Pax-5/Pax-8/Pax-6 lineage (see the shorter arrow in Fig. 5). In this case, the ancestral gene was also duplicated, one copy giving rise to the Pax-A/poxn and the supergroup II lineages, and the other giving rise to the Pax-B/Pax-2/Pax-5/Pax-8/Pax-6 lineage. In Fig. 5, Pax-B is closer to Pax-2/5/8 than to ey/Pax-6, but as noted above, Pax-B might have branched off earlier than the divergence between ey/Pax-6 and Pax-2/5/8, or it might be closer to ey/Pax-6 than to Pax-2/5/8. These possibilities need to be resolved in the future.
Acknowledgments
We thank Dr. Grady F. Saunders for providing us with the human PAX-6 cDNA clones. This study was supported by National Institutes of Health Grant EY10317. H.S. was a recipient of Glenda and Thomas Matney Scholarship Fund.
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. U96195U96195 (sea nettle Pax-A1 paired box), U96196U96196 (sea nettle Pax-A2 paired box), U96197U96197 (sea nettle Pax-B paired box), U96193U96193 (hydra Pax-A), and U96194U96194 (hydra Pax-B)].
References
- 1.Starachan T, Read A P. Curr Opin Genet Dev. 1994;4:427–438. doi: 10.1016/0959-437x(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 2.Tremblay P, Gruss P. Pharmacol Ther. 1994;61:205–226. doi: 10.1016/0163-7258(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 3.Noll M. Curr Opin Genet Dev. 1993;3:595–605. doi: 10.1016/0959-437x(93)90095-7. [DOI] [PubMed] [Google Scholar]
- 4.Quiring R, Walldorf U, Kloter U, Gehring W J. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 5.Walther C, Guenet J L, Simon D, Deutsch U, Jostes B, Goulding M D, Plachov D, Balling R, Gruss P. Genomics. 1991;11:424–434. doi: 10.1016/0888-7543(91)90151-4. [DOI] [PubMed] [Google Scholar]
- 6.Ton C C T, Hirvonen H, Miwa H, Weil M M, Monaghan P, Jordan T, Van Heyningen V, Hastie N D, Meijers- Heijboer H, Drechsler M, Royer-Pokora B, Collins F, Swaroop A, Strong L C, Saunders G F. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 7.Hill R E, Favor J, Hogan B L M, Ton C C T, Saunders G F, Hanson I M, Prosser J, Jordan T, Hastie N D, Van Heyningen V. Nature (London) 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 8.Halder G, Gallaerts P, Gerhring W J. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 9.Land M F. Handbook of Sensory Physiology. 2/6B. New York: Springer; 1981. pp. 471–592. [Google Scholar]
- 10.Land M F, Fernald R D. Annu Rev Neurosci. 1992;15:1–29. doi: 10.1146/annurev.ne.15.030192.000245. [DOI] [PubMed] [Google Scholar]
- 11.Dilella A G, Woo S L C. Methods Enzymol. 1987;152:199–212. doi: 10.1016/0076-6879(87)52021-3. [DOI] [PubMed] [Google Scholar]
- 12.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czerny T, Busslinger M. Mol Cell Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dayhoff M O. Atlas of Protein Sequence and Structure. 5/3. Washington, DC: Natl. Biomed. Res. Found.; 1978. pp. 351–352. [Google Scholar]
- 15.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 16.Zharkikh A, Li W H. J Mol Evol. 1992;9:1119–1147. doi: 10.1093/oxfordjournals.molbev.a040782. [DOI] [PubMed] [Google Scholar]
- 17.Zharkikh A, Li W H. Mol Phyl Evol. 1995;4:44–63. doi: 10.1006/mpev.1995.1005. [DOI] [PubMed] [Google Scholar]
- 18.Hillis D M, Bull J J. Syst Zool. 1993;38:297–309. [Google Scholar]