Abstract
Animals have evolved diverse appendages adapted for locomotion, feeding and other functions. The genetics underlying appendage formation are best understood in insects and vertebrates. The expression of the Distal-less (Dll) homeoprotein during arthropod limb outgrowth and of Dll orthologs (Dlx) in fish fin and tetrapod limb buds led us to examine whether expression of this regulatory gene may be a general feature of appendage formation in protostomes and deuterostomes. We find that Dll is expressed along the proximodistal axis of developing polychaete annelid parapodia, onychophoran lobopodia, ascidian ampullae, and even echinoderm tube feet. Dll/Dlx expression in such diverse appendages in these six coelomate phyla could be convergent, but this would have required the independent co-option of Dll/Dlx several times in evolution. It appears more likely that ectodermal Dll/Dlx expression along proximodistal axes originated once in a common ancestor and has been used subsequently to pattern body wall outgrowths in a variety of organisms. We suggest that this pre-Cambrian ancestor of most protostomes and the deuterostomes possessed elements of the genetic machinery for and may have even borne appendages.
Appendages develop as outgrowths of the body wall orthogonal to the primary body axes, and possess a third, proximodistal patterning axis. Genetic studies have revealed a number of similarities in the signaling molecules and regulatory genes that organize growth and patterning in insect and vertebrate limbs (N. Shubin, C. Tabin, and S. B. Carroll, unpublished data). For example, the Dll gene is expressed in the primordia, and later in the distal regions, of the developing limbs of all arthropods (2–5) (Fig. 1 a and b). Dlx genes are broadly expressed early in developing mouse (Fig. 2 a and b) and chicken limb buds (not shown), and later at high levels in the apical ectodermal ridge (4, 6–11) (Fig. 2c). These similarities are puzzling because arthropod and vertebrate appendages have such vastly different anatomies and evolutionary histories (N. Shubin, C. Tabin, and S. B. Carroll, unpublished data). This implies that the deployment of Dll/Dlx genes in the distal domains of each limb could be coincidental. On the other hand, it is possible that this reflects some relationship between these structures at a more fundamental level as body wall outgrowths (12). With information from only two highly divergent taxa, it is difficult to distinguish between the possibilities. However, characters shared among multiple taxa are less likely to reflect convergence. Therefore, we have examined Dll/Dlx expression in the outgrowths of animals in other metazoan phyla using an antibody that specifically recognizes both Dll and Dlx proteins (4, 5, 13) (see below).
Figure 1.
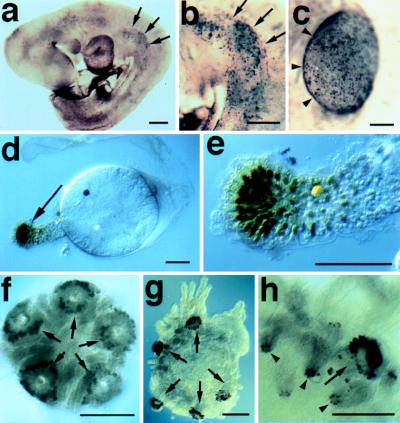
Dll expression in representative protostomes. (a) Lateral view of a late stage Precis coenia butterfly embryo stained with the Dll antibody. Arrows (➞) point to the distal tips of the left abdominal prolegs. Dll expression is detected in central nervous system in the brain (br) and in the ventral nerve cord (➤). (b) Higher magnification image of abdominal prolegs (➞) from an embryo similar to that shown in a. (c) Ventral view of a late stage Peripatopsis capensis onychophoran embryo stained with the Dll antibody. The antennae (ant), oral papilla (➤) and lobopods express Dll. The lobopods shown in higher magnification in e are indicated with ➞. (d) Right halves of two segments from a young P. capensis embryo stained with the Dll antibody. Dll expression is detected in the ectoderm of the presumptive lobopods prior to the formation of visible buds. (e) Higher magnification view of the lobopods indicated in d. (∗) The neurogenic ectoderm, which also expresses Dll. (f) Polychaete annelid Chaetopterus variopedatus, ventral view of larva just prior to metamorphosis. Dll expressing cells are visible in parapodial rudiments (➞), antennae (out of focus on opposite dorsal surface, (➤), prospective feeding organs (⊓), and in the neurogenic ectoderm (∗). (g) C.variopedatus, same specimen at a higher magnification, showing Dll reactive ectodermal nuclei in prospective distal cells of the anterior parapodia (➞) and in the neurogenic region (∗). (h) Later stage C. variopedatus larva showing staining in distal portions of two anterior parapodia (➞). Anterior is to the left in all panels. (Bars = 0.1 mm.)
Figure 2.
Dll expression in representative deuterostomes. (a) Nine-day mouse embryo stained with the Dll antibody. Arrows (➞) point to medial border of cells expressing one or more Dlx genes in the presumptive forelimb. Dlx expression can be detected in developing mouse limbs as the bud forms from the flank, and somewhat earlier than previously reported for mice or other vertebrates (6–11). (b) Higher magnification view of the forelimb indicated in A. (c) Dorsal view of the forelimb of a 10-day mouse embryo stained with the Dll antibody. (➤) The position of the apical ectodermal ridge. (d) Three-day Molgula occidentalis ascidian larva from which an ampulla is extending. Cells at the distal tip of the ampulla express Dll (➞). (e) Higher magnification view of the ampulla shown in d. (f and g) Metamorphosing Strongylocentrotus droebachiensis sea urchin larvae stained with Dll antibody. Cells at the distal tip of the tube feet (➞) express Dll prior to (f) and during (g) extension from the body wall. (h) Higher magnification view of a tube foot (➞) and spines (➤) from an S. droebachiensis larva similar to that shown in g. Cells at the distal tip of the developing spines, as well as the tube feet express Dll. (Bars = 0.1 mm.)
METHODS
Production and characterization of the Dll homeodomain antibody was described in Panganiban et al. (4). The antibody recognizes the Dll antigen in all arthropods (4, 5) and stains only Dll-expressing cells in amphioxus (13). At least five vertebrate Dlx proteins are recognized by the antibody (Marc Ekker, personal communication) which does not stain cells in which Dlx genes are not expressed. Together these data suggest that the antibody is both broadly cross-reactive among phyla, and specific for Dll/Dlx gene products. While it is likely that all of the staining we report here reflects Dll/Dlx expression patterns, it is possible that some reflects only crossreactivity of the Dll antibody to other proteins.
The antibody staining was carried out as previously described (4), with the following modifications: ascidian, echinoderm, lepidopteran, and onychophoran specimens were permeabilized by addition of 5% dimethyl sulfoxide to the PBT blocking solution (PBS with 0.1% Triton X-100 and 2% BSA), and 1% dimethyl sulfoxide to the primary and secondary antibody solutions. For optimal antibody penetration, it was necessary to sonicate (4) the lepidopteran and onychophoran embryos while they were in blocking solution. Detailed protocols are available upon request. The nematode embryos were frozen and cracked on polylysine treated slides, then fixed for 5 min in −20°C methanol, and for 30 min at room temperature in 2% paraformaldehyde in PBS, prior to staining using standard methods (14).
RESULTS AND DISCUSSION
Dll Is Expressed in the Appendages of Animals in Many Phyla.
Among the protostomes, we find that Dll is expressed in the distal portion of the developing antennae and lobopods of the onychophoran Peripatopsis capensis (Fig. 1 c–e), and in the prospective distal portions of all developing parapodia and the antennae in the polychaete annelid Chaetopterus variopedatus (Fig. 1 f–h). In Peripatopsis, the onset of Dll expression in the ectoderm of the presumptive lobopods precedes their outgrowth from the body wall (Fig. 1d). In Chaetopterus, Dll expression is first detected in segmentally reiterated sets of ectodermal cell nuclei before metamorphosis. At this time, expressing cells are flush with the contiguous epidermis. At metamorphosis, parapodial structures evaginate from the body wall and the Dll-expressing cells comprise the distal portions of all appendages, including parapodia, antennae, and specialized feeding structures (Fig. 1 f–h).
Among the deuterostomes, there are other phyla besides the vertebrates (Fig. 2 a–c) whose members possess prominent body wall outgrowths. These include the urochordates, in which two Dll/Dlx genes have been identified (15), and the echinoderms. Within the urochordates, the tunicates or ascidians possess a free-swimming larval form that metamorphoses into a sessile adult. At the onset of metamorphosis, the larvae attach to the substratum via newly formed ampullae which may also have a respiratory function (16). These ampullae express Dll prior to and during their outgrowth in what will become the distal region of the developing ampullae (Fig. 2 d and e). Subsequently, metamorphosing ascidians develop one other type of outgrowth, the siphons, through which they pump seawater and extract oxygen and food. The siphons also express Dll distally as they form during metamorphosis (L.S.C. and B.J.S., unpublished work).
Some echinoderms, such as indirectly developing sea urchins, also possess a free-swimming larval form, the pluteus, a portion of which, the rudiment, gives rise to the radially symmetric adult. Among the earliest structures to develop from the rudiment are five tube feet, protrusions from the body that allow the animal to move over the substratum. Remarkably, the distal cells within these developing tube feet express Dll (Fig. 2 g and h). Expression in the tube feet also is detectable before these structures grow out from the body wall (Fig. 2f). A second major type of outgrowth in metamorphosing sea urchins, the spines, also express Dll at their distal tips (Fig. 2h).
Conservation vs. Convergence as Explanations for Dll Expression Patterns.
In the six coelomate taxa we examined, the prospective apical ectoderm of body wall outgrowths expressed Dll/Dlx. Given the phylogenetic relationships between these animals (Fig. 3) there are two scenarios that can account for Dll/Dlx expression in such diverse structures. The Dll/Dlx genes could have been co-opted independently for appendage formation from some other function in these lineages. Or, the more parsimonious interpretation, Dll expression along the proximodistal axes arose once in a common ancestor of these animals and has been utilized repeatedly in body wall outgrowths ever since. We favor the latter explanation, and if true, this has implications for the evolutionary relationships among appendages and other types of outgrowths. Specifically, these appendages/outgrowths must be either derived from preexisting appendages/outgrowths, or are novel structures formed by the ectopic activation of existing appendage/outgrowth-forming pathways. To distinguish between these possibilities for any particular structure, we must refer to the fossil record and current concepts of metazoan phylogeny.
Figure 3.
The evolution of animal appendages. The cladogram depicts the relationships between a selected subset of animal phyla and is based upon a combination of sources (see ref. 17 for review; other phylogenies are possible but do not alter the basic inferences drawn here). Branch lengths are not scaled. Taxa for which Dll expression data is presented here are shown in bold along with the appearance of the various appendages in these groups. The appearance of various appendages within the phyla studied here are indicated. Only the jointed limbs of arthropods and tetrapod limbs arose by modification of a pre-existing appendage. The immediate ancestors of polychaetes, Onychophora, echinoderms, urochordates, and vertebrates are not thought to have borne appendages that would be homologous to the structures analyzed here. The deuterostome ancestor (b) may have had one Dll/Dlx gene since cephalochordates have only one (13). However, the presence of two Dlx genes in ascidians (15) leaves open the possibility that at least one Dlx duplication event occurred earlier in the deuterostome lineage. The ancestor of the appendage-bearing protostome clade and the deuterostomes possessed the Dll gene and all of the features of the proposed Urbilaterian (18), and may have borne appendages (a). The Dll gene predates this ancestor and is found in nematodes and expressed in the CNS, which may be an older site of function than body wall outgrowths. There is a report of a possible cnidarian Dll ortholog (19), but the similarity of the cnox3 homeodomain to Drosophila Dll (56%) is not significantly higher than that of nonorthologous homeodomains, and the Dll antibody does not recognize this gene product. We therefore believe the origin of Dll is unresolved.
Evolutionary Relationships Among Parapodia, Lobopodia, and Arthropodia.
Morphological analyses of Cambrian lobopodans and arthropods have led to the widely held view that arthropod limbs probably evolved from an ancestral lobopod (20, 21). The patterns of Dll expression in modern terrestrial onychophora, a probable sister group to Cambrian lobopods, support the idea that arthropod limb development is derived from mechanisms present in lobopodans. However, annelids are not a sister taxon to either onychophora or arthropods (22–29), therefore, polychaete parapodia could not be the forerunners of lobopodia, as has sometimes been proposed (30), and must have evolved independently. One plausible scenario for the origin of parapodia and lobopodia is that they were independently derived from cephalic feeding or sensory outgrowths in the annelid and lobopodan/onychophoran lineages. This idea appears to be supportable on paleontological and phylogenetic, as well as developmental genetic evidence. For instance, Cambrian polychaetes (31), Onychophora (32), and arthropods (33) possessed antenniform outgrowths suggesting that this outgrowth is primitive. In addition, phylogenetic schemes based on both morphological characters (34) and molecular evidence (17) are consistent with the existence of a common antennae-bearing ancestor of these protostomes (Fig. 3, see c). Furthermore, studies of Hox genes in Drosophila and other insects suggest that the “ground state” for appendage identity is the antennae because loss of Hox gene function in legs transforms tissue to antennal identity (35) and loss of all Hox genes in Tribolium transforms all body segments to antennae-bearing metameres (36). We propose, then, that the ancestor of these higher protostome taxa bore antenniform outgrowths, and that these structures were duplicated and transformed on the trunk to parapodia and lobopodia after the divergence of the annelid and onychophoran lineages (Fig. 3).
Evolutionary Relationships Among Deuterostome Appendages.
The origins of the deuterostomes and of their outgrowths are uncertain. It is not believed that any of the protostomes surveyed here would include the deuterostome ancestor (37). The substrate-gripping ampullae of the ascidians and the multifunctional tube feet of the echinoderms might be derived from a body wall outgrowth present in a common ancestor (Fig. 3). However, these appendages, as well as the ascidian siphons and the echinoderm spines, may be new structures that exploited an existing genetic circuit to make body wall outgrowths (Fig. 3). Similarly, the origin of fish fins is unclear. However, other vertebrate outgrowths such as the branchial arches express Dlx genes and key limb bud signaling molecules, and limb and craniofacial tissues can substitute for one another in recombination experiments (38). These observations suggest that fins also evolved by exploiting extant outgrowth-forming mechanisms.
The Question of Homology and the Origin of Metazoan Appendages.
Recent discoveries that homologous genes in insects and vertebrates control eye (39), heart (40, 41) and dorsoventral axis formation (42–44) have prompted the reconsideration of ideas concerning the origin and evolution of these structures and the relationships among the animals that possess them (12). For example, DeRobertis and Sasai (18) have suggested that all of these common features date to a common ancestor of the bilateria. As demonstrated here, a common genetic component underlying the formation of secondary axes (i.e., appendages and other types of body wall outgrowths) of both protostomes and deuterostomes also appears to have a deep historic basis. Whether one considers various eyes, hearts, or limbs to be homologous clearly depends on how one defines “homology.” As pointed out by Bolker and Raff (45), “meaningful assignments of homology must specify a biological level.”
Thus, while some of the appendages/outgrowths described here certainly are not homologous in the classical sense (i.e., directly derived from a common structure), our findings suggest that they are homologous at a more fundamental level. Their development utilizes similar genetic pathways. The most straightforward explanation for these observations is that the last common ancestor of the protostomes and deuterostomes had some primitive type of body wall outgrowth, e.g., a sensory or perhaps a simple locomotory appendage, and that the genetic circuitry governing the outgrowth of this structure was deployed at new sites many times during evolution.
We do not know what this pre-Cambrian creature looked like, since no definitive body fossils are known from this period. However, trace fossils in sediments indicate some form of peristaltic or pedal movement made by animals that are believed to have been more advanced than flatworms (17, 46) (Platyhelminthes, Fig. 3). The possession of sensory and/or of locomotory appendages by a coelomic, triploblastic animal is therefore plausible, and may have been key to the success of its’ many descendants (34).
The Ancestral Function of the Dll Gene.
It is likely that the Dll gene is older than the putative outgrowth-bearing ancestor. The expression of arthropod Dll genes in the central nervous system (CNS) and peripheral nervous system including the brain optic lobes (47), and of vertebrate Dlx genes in the CNS, including parts of the brain involved in optic function (6, 48), suggest that Dll/Dlx functions could have originated in tissues other than appendages/outgrowths. To consider the origin and ancestral functions of the Dll gene, it is necessary to identify Dll orthologs in more primitive taxa.
Recent molecular phylogenies based on limited data sets place the nematodes basal to the protostomes and the deuterostomes (29, 49) (Fig. 3). We have found a strong candidate for a Dll ortholog in the C. elegans genomic sequence database (C28A5.4, refs. 50 and 51; “Ce-Dll”) that encodes a homeodomain with 74% identity to Drosophila Dll and with an intron at the same position as the Dlx genes of Drosophila and ascidians (15, 50) (Fig. 4a). This is much more similar than ceh-23 (50% homeodomain identity to Drosophila Dll), which was previously considered a possible Dll ortholog (1), and compares favorably with the amino acid identities of the four C. elegans Hox gene homeodomains to their Drosophila orthologs (69–79%) (1). Ce-Dll is expressed in the CNS of nematode embryos (Fig. 4 b and c). Since Dll/Dlx genes are expressed in the annelid and onychophoran CNS as well (Fig. 1 c, e–g), Dll/Dlx function may well have arisen in the CNS before becoming involved in body wall outgrowths. Nematodes also possess orthologs of genes in the various signaling pathways involved in appendage formation and patterning. This suggests that genetic components required for the genetic machinery for appendage formation evolved long before these structures arose.
Figure 4.
Identification and expression of a nematode Dll gene. (a) Alignment of the homeodomains of Drosophila (52), mouse (48, 53), and nematode Dll/Dlx (50) gene products. The amino acid sequence of the Ce-Dll homeodomain is 74% identical to that of Drosophila Dll. (➤), The position of an intron conserved between the fly (54), ascidian (15), and nematode (50) genes. (b and c) Lateral views of 100-cell (b) and comma (c) stage Caenorhabditis elegans embryos stained with the Dll antibody (green) and an antibody (O1C1D4) that recognizes P-granules (blue). (d) Dorsolateral view of a three-fold stage C. elegans embryo stained with the Dll antibody (green) and an antibody (3NB12) that recognizes pharyngeal cells (red). The Ce-Dll expressing cells in b and c are the precursors of the nerve ring cells that express Ce-Dll in d. Anterior is to the left in b–d, and the magnification in b–d is the same. (Bar = 10 μm.)
Genetic and Cellular Features of Secondary Axis Formation.
One testable prediction of the hypothesis that the ancestor of the protostomes and the deuterostomes possesses body wall outgrowths is that the similarities in the developmental regulatory mechanisms governing metazoan appendage formation will be more extensive than would be expected from their diverse morphologies and functions. The expression of Dll/Dlx genes in such a diverse array of outgrowths suggests that there are cellular processes common to the formation of all appendages that may be under Dll/Dlx control. The identification of genes regulated by Dll in model organisms such as Drosophila may, therefore, reveal universal aspects of secondary axis formation.
Acknowledgments
We thank Jim Valentine and Doug Eernisse for crucial discussions on phylogeny and early metazoans; David Fitch and Kelley Thomas for discussions of nematode phylogeny; Susan Strome for the O1C1D4 antibody; H. Okamoto and J. N. Thompson for the 3NB12 antibody; B. Kay Simandl for the mouse embryos; Jim Valentine, B. Kay Simandl, and Dave Lewis for comments on the manuscript; Leanne Olds for artwork; and Jamie Wilson for help with manuscript preparation. For logistic assistance to M.H.W. with permits and collection of Onychophora from the Cape of Good Hope, Chief Directorate of Nature Conservation and Dr. Mike Picker, University of Cape Town, are gratefully acknowledged. This work was supported by a National Science Foundation (NSF)/Sloan Foundation Fellowship in Molecular Evolution (G.P.), NSF Grant IBN-9623453 (S.M.I), National Institutes of Health Grant HD32551 (J.F.F.), NSF Grant IBN-9304958 (B.J.S.), NSF Grant IBN-9506855, the Sloan Foundation (G.A.W.), and the Howard Hughes Medical Institute (J.K. and S.B.C.).
ABBREVIATIONS
- Dll
distal-less
- Dlx
vertebrate Dll genes
- CNS
central nervous system
References
- 1.Wang B B, Muller-Immergluck M M, Austin J, Robinson N T, Chisholm A, Kenyon C. Cell. 1993;74:29–42. doi: 10.1016/0092-8674(93)90292-x. [DOI] [PubMed] [Google Scholar]
- 2.Cohen S M. Nature (London) 1990;343:173–177. doi: 10.1038/343173a0. [DOI] [PubMed] [Google Scholar]
- 3.Panganiban G, Nagy L, Carroll S B. Curr Biol. 1994;4:671–675. doi: 10.1016/s0960-9822(00)00151-2. [DOI] [PubMed] [Google Scholar]
- 4.Panganiban G, Sebring A, Nagy L, Carroll S B. Science. 1995;270:1363–1366. doi: 10.1126/science.270.5240.1363. [DOI] [PubMed] [Google Scholar]
- 5.Popadic A, Rusch D, Peterson M, Rogers B T, Kaufman T C. Nature (London) 1996;380:395. [Google Scholar]
- 6.Dolle P, Price M, Duboule D. Differentiation (Berlin) 1992;49:93–99. doi: 10.1111/j.1432-0436.1992.tb00773.x. [DOI] [PubMed] [Google Scholar]
- 7.Beauchemin M, Savard P. Dev Biol. 1992;154:55–65. doi: 10.1016/0012-1606(92)90047-k. [DOI] [PubMed] [Google Scholar]
- 8.Bulfone A, Kim H-J, Puelles L, Porteus M H, Grippo J E, Rubenstein J L R. Mech Dev. 1993;40:129–140. doi: 10.1016/0925-4773(93)90071-5. [DOI] [PubMed] [Google Scholar]
- 9.Akimenko M-A, Ekker M, Wegner J, Lin W, Westerfield M. J Neurosci. 1994;14:3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simeone A, Acampora D, Pannese M, D’Esposito M, Stornaiuolo A, Gulisano M, Mallamaci A, Kastury K, Druck T, Huebner K, Boncinelli E. Proc Natl Acad Sci USA. 1994;91:2250–2254. doi: 10.1073/pnas.91.6.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrari D, Sumoy L, Gannon J, Sun H, Brown A M, Upholt W B, Kosher R A. Mech Dev. 1995;52:257–264. doi: 10.1016/0925-4773(95)98113-o. [DOI] [PubMed] [Google Scholar]
- 12.Raff R. The Shape of Life. Chicago: Univ. of Chicago Press; 1996. [Google Scholar]
- 13.Holland N D, Panganiban G, Henyey E L, Holland L Z. Development (Cambridge, UK) 1996;122:2911–2920. doi: 10.1242/dev.122.9.2911. [DOI] [PubMed] [Google Scholar]
- 14.Miller D M, Shakes D C. Methods Cell Biol. 1995;48:365–389. [PubMed] [Google Scholar]
- 15.DiGregorio A, Spagnuolo A, Ristoratore F, Pischetola M, Aniello F, Branno M, Cariello L, DiLauro R. Gene. 1995;156:253–257. doi: 10.1016/0378-1119(95)00035-5. [DOI] [PubMed] [Google Scholar]
- 16.Bates W. Dev Growth Differ. 1991;33:401–411. doi: 10.1111/j.1440-169X.1991.00401.x. [DOI] [PubMed] [Google Scholar]
- 17.Valentine J W, Erwin D H, Jablonski D. Dev Biol. 1996;173:373–381. doi: 10.1006/dbio.1996.0033. [DOI] [PubMed] [Google Scholar]
- 18.DeRobertis E M, Sasai Y. Nature (London) 1996;380:37–40. [Google Scholar]
- 19.Schummer M, Scheurlen I, Schaller C, Galliot B. EMBO J. 1992;11:1815–1823. doi: 10.1002/j.1460-2075.1992.tb05233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snodgrass R E. Evolution of the Annelida, Onychophora and Arthropoda. Washington, DC: Smithsonian Inst.; 1938. [Google Scholar]
- 21.Manton S M. The Arthropoda: Habits, Functional Morphology and Evolution. Oxford: Clarendon; 1977. [Google Scholar]
- 22.Field K, Olsen G, Lane D, Giovannoni S, Ghiselin M, Raff E, Pace N, Raff R. Science. 1988;239:748–753. doi: 10.1126/science.3277277. [DOI] [PubMed] [Google Scholar]
- 23.Halanych K M. Mol Phylogenet Evol. 1995;4:72–76. doi: 10.1006/mpev.1995.1007. [DOI] [PubMed] [Google Scholar]
- 24.Lake J. Proc Natl Acad Sci USA. 1990;87:763–766. doi: 10.1073/pnas.87.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turbeville J M, Pfeifer D M, Field K G, Raff R A. Mol Biol Evol. 1991;8:669–686. doi: 10.1093/oxfordjournals.molbev.a040677. [DOI] [PubMed] [Google Scholar]
- 26.Backeljau T, Winnepenninckx B, Bruyn L D. Cladistics. 1993;9:167–181. doi: 10.1111/j.1096-0031.1993.tb00216.x. [DOI] [PubMed] [Google Scholar]
- 27.Eernisse D J, Albert J S, Anderson F E. Syst Biol. 1992;41:305–330. [Google Scholar]
- 28.Ballard J, Olsen G, Faith D, Odgers W, Rowell W, Atkinson P. Science. 1992;258:1345–1348. doi: 10.1126/science.1455227. [DOI] [PubMed] [Google Scholar]
- 29.Winnepenninckx B, Backeljau T, Mackey L Y, Brooks J M, Wachter R D, Kumar S, Garey J R. Mol Biol Evol. 1995;12:1132–1137. doi: 10.1093/oxfordjournals.molbev.a040287. [DOI] [PubMed] [Google Scholar]
- 30.Lauterbach K-E. Zool Anat Jahrbuch. 1978;99:64–92. [Google Scholar]
- 31.Morris C S. Philos Trans R Soc London B. 1979;285:227–274. [Google Scholar]
- 32.Hou X, Bergstrom J. Zool J Linnean Soc. 1995;114:3–19. [Google Scholar]
- 33.Briggs D E G, Erwin D H, Collier F J. The Fossils of the Burgess Shale. Washington, DC: Smithsonian Inst.; 1994. [Google Scholar]
- 34.Valentine J. Proc Natl Acad Sci USA. 1994;91:6751–6757. doi: 10.1073/pnas.91.15.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Struhl G. Nature (London) 1981;292:635–638. doi: 10.1038/292635a0. [DOI] [PubMed] [Google Scholar]
- 36.Stuart J, Brown S, Beeman R, Denell R. Nature (London) 1991;350:72–74. doi: 10.1038/350072a0. [DOI] [PubMed] [Google Scholar]
- 37.Gee H. Before the Backbone. London: Chapman & Hall; 1996. [Google Scholar]
- 38.Wall N A, Hogan B L M. Mech Dev. 1995;53:383–392. doi: 10.1016/0925-4773(95)00453-x. [DOI] [PubMed] [Google Scholar]
- 39.Halder G, Callaerts P, Gehring W J. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 40.Bodmer R. Development (Cambridge, UK) 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- 41.Chan-Thomas P S, Thompson R P, Robert B, Yacoub M H, Barton P J. Dev Dyn. 1993;197:203–216. doi: 10.1002/aja.1001970305. [DOI] [PubMed] [Google Scholar]
- 42.Holley S A, Jackson P D, Sasai Y, Lu B, DeRobertis E M, Hoffmann F M, Ferguson E L. Nature (London) 1995;376:249–253. doi: 10.1038/376249a0. [DOI] [PubMed] [Google Scholar]
- 43.Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont L K, DeRobertis E M. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt J, Francois V, Bier E, Kimelman D. Development (Cambridge, UK) 1995;121:4319–4328. doi: 10.1242/dev.121.12.4319. [DOI] [PubMed] [Google Scholar]
- 45.Bolker J A, Raff R A. BioEssays. 1996;18:489–493. doi: 10.1002/bies.950180611. [DOI] [PubMed] [Google Scholar]
- 46.Valentine J W. Proc Natl Acad Sci USA. 1989;86:2272–2275. doi: 10.1073/pnas.86.7.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaphingst K, Kunes S. Cell. 1994;78:437–448. doi: 10.1016/0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 48.Price M, Lemaistre M, Pischetola M, Di Lauro R, Duboule D. Nature (London) 1991;351:748–751. doi: 10.1038/351748a0. [DOI] [PubMed] [Google Scholar]
- 49.Sidow A, Thomas W K. Curr Biol. 1994;4:596–603. doi: 10.1016/s0960-9822(00)00131-7. [DOI] [PubMed] [Google Scholar]
- 50.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, et al. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 51.Stock D W, Ellies D L, Zhao Z, Ekker M, Ruddle F R, Weiss K M. Proc Natl Acad Sci USA. 1996;93:10858–10863. doi: 10.1073/pnas.93.20.10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohen S, Bronner M, Kuttner F, Jurgens G, Jackle H. Nature (London) 1989;338:432–434. doi: 10.1038/338432a0. [DOI] [PubMed] [Google Scholar]
- 53.Porteus M H, Bulfone A, Ciaranello R D, Rubenstein J L R. Neuron. 1991;77:221–229. doi: 10.1016/0896-6273(91)90260-7. [DOI] [PubMed] [Google Scholar]
- 54.Vachon G, Cohen B, Pfeifle C, McGuffin M E, Botas J, Cohen S M. Cell. 1992;71:437–450. doi: 10.1016/0092-8674(92)90513-c. [DOI] [PubMed] [Google Scholar]