Abstract
Isothermal titration calorimetry (ITC) has been applied to the study of proteins for many years. Its use in the biophysical analysis of RNAs has lagged significantly behind its use in protein biochemistry, however, in part because of the relatively large samples required. As the instrumentation has become more sensitive, the ability to obtain high quality data on RNA folding and RNA ligand interactions has improved dramatically. This review provides an overview of the ITC experiment and describes recent work on RNA systems that have taken advantage of its versatility for the study of small molecule binding, protein binding, and the analysis of RNA folding.
Keywords: ITC, thermodynamics, binding, folding
INTRODUCTION
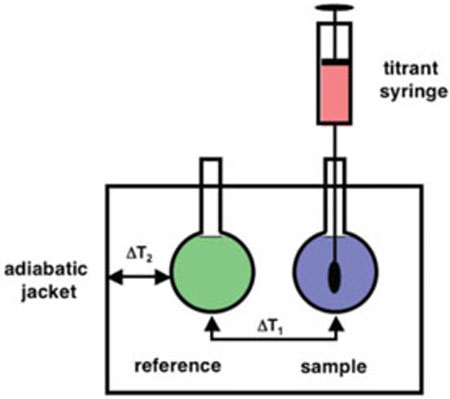
Isothermal titration calorimetry (ITC) is a powerful technique for probing the thermodynamics of molecular interactions. ITC is most commonly used to probe the binding interaction between two molecules. These can be pairs of macromolecules or a macromolecule binding to a small molecule ligand. A schematic diagram of a typical instrument is shown in Figure 1. An electronic feedback circuit is the key component allowing the control of the heating elements adjacent to the two cells. The experiment involves incremental addition of one species (a titrant) into its binding partner (titrate) while monitoring the energy input required to maintain the sample and reference cells at a common temperature. The integration of each peak in the power versus time curve corresponds to the heat associated with each increment of the titration.
FIGURE 1.
Schematic diagram of an isothermal titration calorimeter.
Over the past 15–20 years, the sensitivity of these instruments has improved dramatically.1,2 The result of these advances is that sample requirements are now easily accessible for most types of biological systems. Since reaction heat is probed directly, there is no need to engineer extrinsic labels into the system that might perturb binding in any way, making the methodology extremely versatile and adaptable for a wide range of applications. These instruments are now widely available in shared instrumentation facilities at many universities making it possible to use them without the need to purchase the instruments individually. Thus ITC data are becoming much more commonplace in the analysis of biophysical systems, complementing gel mobility shift assays and thermal melting studies.
This review will focus on the ways in which ITC has been used to study problems associated with RNA biochemistry. With that in mind, I have specifically excluded a large body of excellent work in the area of protein–DNA interactions and protein biophysics. I refer the interested reader to several other current reviews that have summarized the thermodynamics of those interactions.3–9
OVERVIEW OF THE METHODOLOGY—HOW ITCs WORK
Titration calorimeters directly measure the energy of a chemical or biochemical reaction. Most modern instruments work in an energy compensation mode where sample and reference cells are maintained at a constant temperature, relative to the adiabatic jacket. A feedback loop controls the heating of the cells and the instrument monitors the energy required to maintain the temperature as a function of time. During the actual experiment, a small volume of a titrant A (typically 3–20 μL) is added to the titrate B (usually on the order of 1.4 mL). Each injection yields a deflection in the relative power versus time curve as the instrument compensates for the heat (qi) absorbed or released by a given injection and the subsequent binding event (Figure 2a). These peaks are integrated to yield the injection heat qi for a given injection and adjusted for background sources of heat resulting from dilution, stirring, etc. (Figure 2b). The resulting ITC data must then be fit to model dependent equations. It should be noted that the first injection usually under represents the true heat because of issues with the mechanical backlash in the screw that drives the syringe.10.
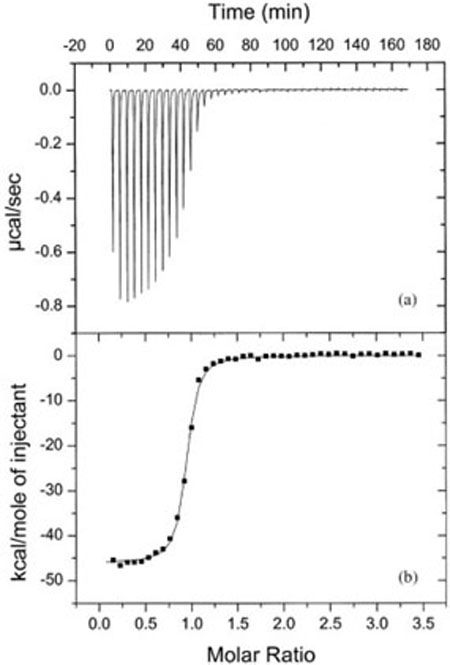
FIGURE 2.
Typical ITC data. (a) Power versus time curve for a titration of a 75 μM solution of a 6 nt RNA titrated into a 5 μM solution of its complement at 15°C in 50 mM HEPES (pH 7.5) and 1M NaCl. (b) The injection enthalpy plotted versus the ratio of reactants and fit to obtain ΔH = −46.1 ± 0.2 kcal/mol, Ka = 4.3 × 107 M−1 and n = 0.91. (Reproduced from Ref. 57, with permission from American Chemical Society).
As the new generation of calorimeters were first being developed, a series of mathematical expressions were derived that allowed facile analysis for the common binding phenomena encountered with biological samples [Eqs. (1)–(3)].2,10–12 The integrated heat of each injection (qi) is a function of the fractional saturation of the binding reaction (F), the stoichiometry of the binding (n), the total concentration of titrant A (AT), the binding enthalpy (ΔH), and the cell volume (V). So long as one knows the initial concentration of the titrate in the cell (BT), one can rewrite Eq. (1) as a quadratic relationship. Upon solving for F, one gets Eq. (3), which is now in a form that can be used directly in nonlinear least-squares fitting to obtain ΔH, KB, and n. ΔG is readily obtained from KB, allowing determination of ΔS. Thus, from a single titration one gets a complete thermodynamic characterization of the binding interaction.
| 1 |
| 2 |
| 3 |
Where
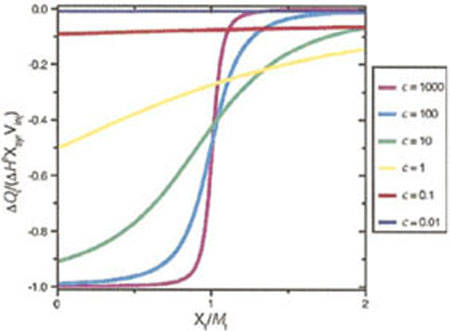
The product nKB · AT, is sometimes referred to as the “c value.” The magnitude of c in a given experiment can be controlled by adjusting the titrant concentration and has a profound impact on the data quality.14,15 Simulated data show what a typical integrated ITC data set looks like over the range of 0.01 < c < 1000 (Figure 3). To determine a binding constant by ITC, the c-value must be between 1 and 1000, and ideally between 10 and 100. If AT is too small, many times the cell volume is required to saturate binding and dilution of the titrate over the course of the experiment makes it impossible to resolve the transition. When AT it is too large, the titration appears like a delta function where stoichiometric binding occurs in the ith injection, but no binding occurs in injection i + 1 because binding is already saturated. Since the concentrations are limited at the low end by the need for measurable amounts of heat to be released during each injection, one finds that there is a practical limit of KB ≥ 10 nM for most ITC measurements unless one resorts to displacement techniques.16 A continuous injection method has also been reported that circumvents some of these limitations and extends the accessible binding constants down to around 10 pM while simultaneously reducing the data collection time.17 Relatively few studies have used this methodology to date, but the software necessary to analyze the power versus time curves for simple binding models is now supplied with new ITC instruments. One concern raised regarding the single injection method has been the time required to achieve complete equilibration in the calorimeter and whether this response time affects the data quality in single-injection mode.18.
FIGURE 3.
The shape of an ITC curve as a function of the 0.01 < c <1000 where c is the product of the binding constant KB and the titrate concentration. (Reproduced from Ref. 14, with permission from American Chemical Society).
Joel Tellinghuisen has published extensively on the analysis of ITC data and the optimization of collection parameters to minimize these errors.15,19,20 His recommendations include (a) minimizing the number of injections; (b) maximizing the concentration of the titrate in the cell; and (c) setting the titrant concentration so that at the end of the titration one reaches the ratio [B]T/[A]T = 6.4/(c0.2) + 13/c. In practice, this means that the titrant must typically be at a concentration 15–30 times that of the titrate, depending on the desired c-value and the total volume of titrant to be added. Thus, if solubility is an issue, one should plan accordingly such that the least soluble material is used as the titrate. When assessing ITC data quality, one should look specifically for a return to a linear baseline between each injection, the presence of a well-defined inflection and very modest background heat from the final injections where little or no binding is occurring.
For the specific cases of RNAs, there are a few additional considerations that should be taken into account during experimental design. Buffer match is critical because of the high ionic strength conditions typically used for nucleic acids. Mismatched buffers leads to large background heats during the titration because of the dilution of the salts, which increases the error in the data unless due care is taken. Another issue that must be considered extensively is the energetics of refolding and structural homogeneity. This detail is particularly important in temperature dependent studies or studies at elevated temperature. In many cases, the end state of the titration (protein- or ligand-bound RNA) is thermally more stable than the unbound state. For better or worse, ITC measures all heat taken up or released during the reaction, not just that portion associated with binding. Thus, at even mildly elevated temperatures, the measured enthalpy can include both a binding component and a folding component. The effects due to folding transitions become significant in the measured enthalpy changes even 20°C below the Tm of the melting transition of the RNA and should be accounted for in the data analysis. Finally, be cognizant of the stoichiometry from the fitting. In many cases, noninteger stoichiometry is a strong indication of the data quality and the accuracy with which the titrate and the titrant concentrations have been measured. Errors in concentration determination or poorly behaved systems may make subsequent interpretation of the data difficult.
REVIEW OF LITERATURE
The vast majority of the ITC literature has been in the area of small molecules ligands binding to proteins, driven to a great extent by the drug discovery field.3,7,21,22 Protein–DNA6,23–25 and small molecule–DNA21,26 binding have also been studied extensively but are outside the scope of this review. The literature in the area of RNA biochemistry is much more modest. To illustrate the utility of ITC in analyzing RNA biochemistry, I will showcase a few studies below on ways in which ITC has been utilized to answer problems relevant to the RNA community. These studies fall into three general categories: small molecule binding, RNA–protein interactions and fundamental RNA folding studies.
Small Molecule Binding to RNAs
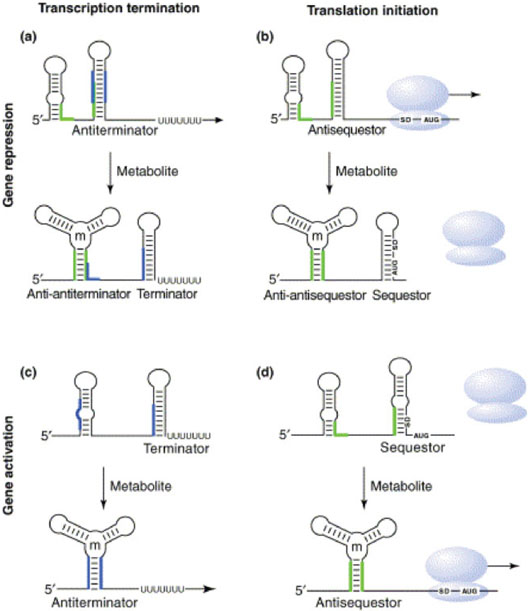
The binding of small molecules to nucleic acids is an exceedingly important phenomenon. Riboswitches, for instance, are metabolite sensing elements present in some mRNAs that regulate their own transcription or translation (Figure 4).27–30 In the guanine sensitive riboswitch, the guanosine binding induces the RNA to fold in a manner that presents a stable terminator hairpin whereas under low guanosine conditions, the riboswitch core is destabilized allowing an antiterminator hairpin to form that permits the RNA polymerase to read through the regulatory domain. ITC has been used to study ligand binding to both the natural purine riboswitch31,32 as well as a synthetic riboswitch that responds to tetracycline.33 Binding has been shown to be enthalpically favorable and entropically opposed for all of the species that can be accommodated into these riboswitches. The ITC data on the purine system shows a highly nonlinear temperature dependence of ΔH and a heat capacity change (ΔCP) in great excess of what one would predict based on buried surface area approximations.31 Such data are probably indicative of a significant structural rearrangement in the unbound state over the measured temperature range.
FIGURE 4.
Diagram illustrating the structural transitions associated with the action of metabolite sensing riboswitches in gene regulation. (a) Repression at the transcriptional level. (b) Repression at the translational level. (c) Upregulation at the transcriptional level. (d) Upregulation at the translational level. (Reproduced from Ref. 27, with permission from Elsevier Science. All rights reserved.)
ITC analysis has also been used to analyze the substrate specificity of the binding pocket.31 As part of a series of guanosine derivatives, Batey and coworkers showed that hypoxanthine and purine bound rather poorly whereas 2,6-diaminopurine bound almost as well as the natural substrate. The extensive characterization of these analogs with the systematic removal of hydrogen bonding substituents showed significant nonadditive behavior. Thus cooperative interactions in the binding pocket facilitate ligand recognition and discrimination between structurally similar nucleobases.
Another group of small molecules–RNA interactions that has been studied extensively by ITC are the aminoglycoside antibiotics. These agents are known to bind the 16S rRNA,34,35 hammerhead ribozymes,36 group I introns,37 HIV Rev response elements38,39 and many other RNAs. The Pilch lab has looked at aminoglycoside binding to ribosomal RNAs in great detail revealing several surprises. As with the guanosine binding to the riboswitches, the heat capacity changes observed for binding were quite significant. The authors attributed these to structural rearrangements in the target RNA upon binding rather than electrostatic or solvation changes.40.
The analysis of aminoglycoside binding also illustrates the importance of performing controls to assess thermodynamic linkage when one is doing ITC. Neomycin binding to the 16S rRNA shows strong pH and buffer sensitivity.40–42 This result epotimizes thermodynamic coupling of a binding equilibrium to a protonation/deprotonation event. By repeating the ITC measurements under several buffer conditions, one can correct the raw ITC data for the buffer contribution by using Eqs. (4) and (5) and the buffer component proton dissociation enthalpy data in Table I. Here, ΔHcorr is the buffer independent binding enthalpy, ΔHobs1 and ΔHobs2 are observed enthalpies under different binding conditions, ΔHion is the proton dissociation enthalpy of the relevant buffer, and Δn is the number of protons associated with the binding transition.
Table I.
Enthalpy and Heat Capacity Changes for Proton Dissociation from Buffers in 0.1M KCl at 25°Ca
| Buffer | pK* | ΔH (kcal mol−1) | ΔCP (Cal mol−1 K−1) | δΔCP/ΔT (10−3 cal mol−1 K−2) |
|---|---|---|---|---|
| Acetate | 4.62 | 0.117 ± 0.005 | −30.5 ± 0.5 | |
| MES | 6.07 | 3.709 ± 0.007 | 3.8 ± 0.5 | |
| Cacodylate | 6.14 | 5.245 ± 0.005 | −18.6 ± 0.5 | |
| Glycerol | 6.26 | 4.949 ± 0.005 | −42.8 ± 0.5 | 0.19 ± 0.09 |
| PIPES | 6.71 | 2.735 ± 0.010 | 4.5 ± 1.0 | |
| ACES | 6.75 | 7.502 ± 0.012 | −6.4 ± 1.0 | |
| Phosphate | 6.81 | 1.223 ± 0.007 | −44.7 ± 0.7 | 0.48 ± 0.05 |
| BES | 7.06 | 6.012 ± 0.017 | 0.5 ± 1.2 | |
| MOPS | 7.09 | 5.212 ± 0.007 | 9.3 ± 0.7 | |
| Imidazole | 7.09 | 8.739 ± 0.014 | −3.8 ± 1.2 | |
| TES | 7.42 | 7.820 ± 0.007 | −7.9 ± 0.7 | |
| HEPES | 7.45 | 5.018 ± 0.017 | 11.7 ± 1.2 | |
| EPPS | 7.87 | 5.147 ± 0.012 | 13.4 ± 1.0 | |
| Triethanolamine | 7.88 | 8.023 ± 0.010 | 11.5 ± 0.7 | |
| Tricine | 8.00 | 7.636 ± 0.012 | −10.7 ± 1.0 | |
| Bicine | 8.22 | 6.461 ± 0.012 | 0.5 ± 1.0 | |
| TAPS | 8.38 | 9.910 ± 0.014 | 5.5 ± 1.2 | |
| CAPS | 10.39 | 11.594 ± 0.017 | 6.9 ± 1.4 |
Data taken from Ref. 43.
| 4 |
| 5 |
One of the strengths of ITC analysis is the ability to measure the binding under a wide range of solution conditions. The only limitation is that the titrant and titrate must be in the same buffer system. Pilch and coworkers used this flexibility to assess a variety of factors that might impact aminoglycoside binding, including ionic strength (for electrostatic contribution) and osmolytes (for solvation contribution).40 This array of studies allowed them to partition the binding free energy into a series of component free energy terms, including: rotational and translational motions, conformational flexibility, hydration, polyelectrolyte, and direct molecular contacts. This type of deconstruction has been championed by Brad Chaires to parameterize drug interactions in a manner that will facilitate quantitative structure–activity relationships.21 The Pilch analysis of neomycin binding showed that the specific molecular contacts provide the dominant favorable contribution toward binding with the most significant unfavorable contribution deriving from restricted rotational and translation motions. Thus, they conclude that the aminoglycoside antibiotics should be amenable to structure-based drug design to alter their affinity and/or specificity given the correct set of target RNAs. The caveat in this whole process is that the species with the highest affinity to a model target does not always correlate with the best pharmacological results.40
Protein Binding
Gel shift assays and filter binding studies are probably the most common means to analyze RNA–protein interactions because of their relative simplicity and their small sample requirements.44 However, ITC provides some significant benefits over those methodologies. The flexibility and control with respect to condition space is a significant advantage as is the ability to obtain directly ΔH, ΔS, and stoichiometry information in addition to the Kd. The difficulty, however, comes at the level of analysis of these extra parameters. In many cases, both the RNA and the protein undergo significant local rearrangements upon binding. This phenomenon can make interpreting the thermodynamic values in a constructive manner challenging. Nonetheless, significant insights have derived from ITC studies probing RNA–protein complex formation. While not an exhaustive survey of the published studies, two examples will be used to illustrate how ITC has helped to develop a deeper understand of the systems being analyzed.
Recht and Williamson have used ITC to probe individual steps in the assembly pathway of the small subunits of bacterial ribosomes.45,46 In these studies, they dissected the binding of 5 proteins (S6, S8, S11, S15, and S18) to a central domain fragment of the 16S rRNA. The most current E. coli 30S assembly map shows that S8, S11, and S15 bind in independent steps followed by binding of the S6/S18 heterodimer and then the S21 protein.47 The ITC analysis showed that S8 and S11 are thermodynamically independent of the other proteins, but that S15 binding is cooperatively linked to the S6/S18 heterodimer.46 A striking result was how well the sum of the individual protein binding steps agreed with an aggregate reaction where binding of all of the components occurred during a single injection. These data showed that a deconstructive approach was justified in probing ribosome assembly.
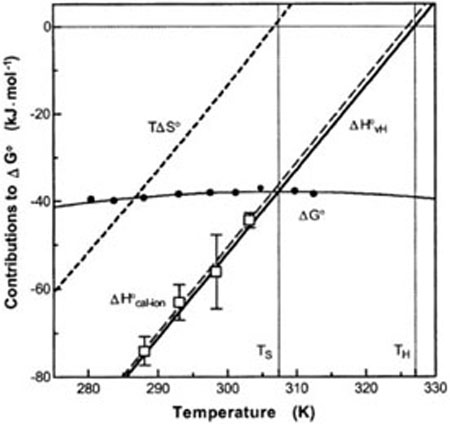
Another study used ITC to probe eIF4E binding to a 7-methyl-GpppG cap analog.48–50 As is typical of RNA–protein complexes, the temperature dependent analysis showed significant enthalpy–entropy compensation (Figure 5). Such compensation is a manifestation of heat capacity changes associated with the interaction.51,52 Previous work with DNA–protein complexes has shown that a sizeable negative ΔCP is typically associated with base unstacking transitions that are coupled to protein binding.53 In addition, linkage to protonation equilibria also typically contributes significantly to the apparent ΔCP of nucleic acid–protein complexes.54,55 In the case of eIF4E, Niedzwiecka and colleagues deconvoluted both the linked protonation event and the singlestranded stacking transition, but still observed a very large positive ΔCP of +464 cal mol−1 K (as opposed to the negative ΔCPs of most other systems).49 While the experimental values are pretty clear, the interpretation of the heat capacity changes is less obvious. The authors attributed the positive value to additional hydration of polar surfaces in the bound complex. The authors have not completely ruled out the possibility that the protein undergoes significant structural changes in response to cap binding leading to the unusual heat capacity changes in this system.
FIGURE 5.
Enthalpy–entropy compensation that accompanies the binding of eIF4E to 7-methyl-GpppG. Solid circles represent measured binding free energies (ΔG°) and open squares show calorimetry enthalpies (ΔH°cal-ion) corrected for ligand dilution, protein activity, and buffer ionization. (Reproduced from Ref. 48, with permission from American Chemical Society).
Fundamental Folding Studies
Protein folding is typically a unimolecular event making it impossible to study by ITC with the exception of a few systems where folding is dependent upon metal ion binding. Nucleic acid folding is fundamentally different in that secondary structures can be either unimolecular, such as in the case of a hairpin, or bimolecular. In vitro, clever re-engineering can typically convert a unimolecular folding event into a biomolecular reaction. Such modification opens the door for extensive use of ITC to study RNA folding.
The basic thermodynamics of RNA folding is still an active area of study despite the ever increasing acumen of the nearest neighbor algorithms inside programs like mFold, and RNAStructure.56,57 Part of the issue derives from the need to understand RNA structures in experimental solutions or cellular milieus, and these programs are only as good as the extrapolation of the measured thermodynamic values to the specific conditions of interest. ITC offers the ability to measure structure formation under almost any specific solution condition of interest. Furthermore, as the data is a direct measurement of the binding enthalpy, it is unaffected by issues related to non-two-state behavior the way many other techniques are. Thus, it has an important place in the study of RNA folding energetics. Several labs have begun to investigate simple duplexes52,58,59 and more complicated structures like helical junctions60–62 using ITC. The common thread that runs through these analyses is the need to carefully consider the ground state conformations prior to the binding reaction.
In the case of thermal melting experiments, the ground state is an ensemble of nearly completely unfolded species. Residual structure is often minimal because of the thermal energy of the system. In ITC, the single stranded state is typically at lower temperature and consists of an ensemble of many conformations. The extent of residual structure is much more significant and is highly dependent upon the ionic conditions and the sequences involved. This is both a strength and a weakness for ITC analysis. One can measure the Kd and ΔH of virtually any duplex at a given temperature using ITC, but whether or not the value matches parameters derived from UV melting is highly condition dependent. The reason is that the energetics of unfolding any unimolecular structure is coupled to the formation of the duplex. If one studies the temperature dependence of the transitions by ITC, apparent heat capacity changes derive from the melting of the single stranded structures of the titrate and titrant.
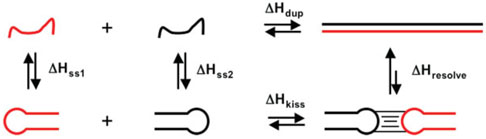
This phenomenon can best be illustrated for the extreme case of hairpins that can resolve into an extended bimolecular duplex (Figure 6). Such structural transitions are biologically relevant since reactions like HIV genome dimerization promoted through the interaction of the dimerization initiation sequence63,64 and the pairing reactions of small noncoding RNAs with structured regions of their mRNA targets to mediate post-transcriptional gene regulation in bacteria rely on these structural rearrangements.65,66 If one considers the ramifications of such a system of equilibria, one can express the ITC derived enthalpy change for the reaction at any given temperature based on Eqs. (6) and (7). At temperatures sufficiently low that the hairpins are fully formed (1 — α ≈ 1), the measured enthalpy for the formation of the bimolecular duplex at temperature is a function of the intrinsic enthalpy of duplex formation and the unfavorable unfolding enthalpies of the two hairpins . As the temperature rises toward the Tm of the any of the species, however, the unfolded state of the hairpin becomes populated. At sufficiently high temperature, even the extended duplex fails to form completely and the measured value for should drop again at high temperature. The result is that one should observe highly nonlinear plots of ΔH versus T passing through a maximum at intermediate temperature. This temperature dependence of ΔH reflects not just the intrinsic heat capacity change for the system, but also the structural transition of the single stranded species. Given sufficient temperature dependent data, plots can be fit to models that account for thermal melting in the single stranded and duplex states [Eqs. (6) and (7)] where one explicitly considers the thermal unfolding of each species over the whole range of data collection temperatures.59 While the data are most dramatic for species with highly structured single stranded states, such as hairpins, the methodology holds for all levels of structure. Thus, linear approximations for ΔCP are only valid in regions outside the temperature ranges where structure transitions of the component species occur.
FIGURE 6.
Schematic diagram illustrating the importance of accounting for single-stranded folding in the ITC analysis of RNA structural rearrangements.
| 6 |
| 7 |
The real value of ITC experiments becomes evident in the regime of intermediate structure. Stable hairpins exhibit significant hyperchromicity and can be readily studied by UV melting.67,68 Thus, one could account for hairpin unfolding computationally. The impact of single stranded structure on duplex formation is evident even when weakly structured states are present at reduced temperature. While this is not a problem for thermal melting analysis because the thermodynamic properties are measured at the Tm, it does play a more important role for RNA folding at ambient temperatures. ITC analysis allows further interrogation of the structural transitions of these single stranded states that have been difficult to observe in any other way.
Several groups have also begun to apply ITC to the analysis of higher order RNA structures such as helical junctions.60,62,69 One can classify these studies into two types, those that initiate folding by the mixing of two RNA species in a common buffer and those that titrate Mg(II) into preannealed RNAs. Both types of reactions have benefits and limitations and they focus on somewhat different aspects of the folding process. In the case of the ion induced folding, the thermodynamics are strongly linked to the movement of ions into and out of the condensation layer with Mg2+ typically displacing Na+ in the diffuse binding region. The energetics of the RNA rearrangements then superimpose on that effect. The Lilley Lab used ITC to study metal ion binding to the minimal hammerhead ribozyme.69 They observed two sequential binding events where the first one was typically more favorable enthalpically and the second one more favorable entropically. ΔH values ranged from −4 to +6 kcal/mol range for ion binding.69 Equivalent metal binding was observed even in mutant species A14G and dG5, which are catalytically inactive.
In the alternative experiment, one mixes two components of the junction together under conditions that allow it to fold into a native conformation. The resulting values are composites, including the contribution of the duplex regions as well as that of the tertiary element of interest. One then must deconvolute contributions of the individual helical elements from the total enthalpy to obtain the contribution from a junction or other structure. Helical junctions have been probed in this way by the Feig62 and Turner60 labs. In the study from the Turner lab, ITC was used to avoid problems with non-two-state behavior evident in the optical melting data. They used the data to focus on total free energy contributions over a series of junction sequences and found ΔG°37 values ranging from 1 to 5 kcal/mol for the sequences tested. The free energies for these multibranch loops were highly sequence dependent and appeared idiosyncratic, but that may be the result of an insufficient numbers of examples to make the underlying patterns evident. The Feig lab study focused more directly on a single junction from the minimal hammerhead ribozyme and dissected the enthalpic and entropic contributions to folding. As with the Turner study, the absolute ΔG was small and slightly unfavorable (+3.3 kcal/mol), but resulted from the compensation between a highly favorable entropic term and a highly unfavorable enthalpic term. In both cases, very large ΔCPs were reported for the junctions. These heat capacity changes most likely originate in the same types of structural transitions in the single-stranded state that were described earlier in relation to studies of simple duplexes. Additional studies will be required to decipher the precise energetics of these tertiary structures and further unravel their energetic mysteries.
CONCLUSIONS
ITC analysis comes with both advantages and limitations relative to the spectroscopic and thermal scanning methods typically used to measure the thermodynamics of RNA folding and ligand binding. On the positive side, it allows one to measure binding without the need for the chemical modifications. Furthermore, since heat is a universal detector, it can provide information on linked equilibria (such as protonation/deprotonation) that are very hard to deconvolute using other methodologies. On the negative side, the experiment typically requires significantly more sample than thermal melting analysis. The ability to probe effects in heat capacity changes as well as issues relating to single-stranded structure transitions however, makes it a very useful addition to the repertoire of thermodynamic measurements for RNA and RNP analysis.
Acknowledgments
Contract grant sponsor: NIH
Contract grant numbers: GM065430, GM075068
REFERENCES
- 1.Spokane R, Gill SJ. Rev Sci Instrum. 1981;52:1728–1733. [Google Scholar]
- 2.Wiseman T, Williston S, Brandts J, Lin L. Anal Biochem. 1989;179:131–135. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 3.Weber PC, Salemme FR. Curr Opin Struct Biol. 2003;13:115–121. doi: 10.1016/s0959-440x(03)00003-4. [DOI] [PubMed] [Google Scholar]
- 4.Ababou A, Ladbury JE. J Mol Recognit. 2006;19:79–89. doi: 10.1002/jmr.750. [DOI] [PubMed] [Google Scholar]
- 5.Ababou A, Ladbury JE. J Mol Recognit. 2007;20:4–14. doi: 10.1002/jmr.803. [DOI] [PubMed] [Google Scholar]
- 6.Plum GE, Breslauer KJ. Curr Opin Struct Biol. 1995;5:682–690. doi: 10.1016/0959-440x(95)80062-x. [DOI] [PubMed] [Google Scholar]
- 7.Haq I. Arch Biochem Biophys. 2002;403:1–15. doi: 10.1016/S0003-9861(02)00202-3. [DOI] [PubMed] [Google Scholar]
- 8.Privalov PL, Dragan AI. Biophys Chem. 2007;126:16–24. doi: 10.1016/j.bpc.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Haq I, Chowdhry BZ, Jenkins TC. Methods Enzymol. 2001;340:109–149. doi: 10.1016/s0076-6879(01)40420-4. [DOI] [PubMed] [Google Scholar]
- 10.Mizoue LS, Tellinghuisen J. Anal Biochem. 2004;326:125–127. doi: 10.1016/j.ab.2003.10.048. [DOI] [PubMed] [Google Scholar]
- 11.Freire E, Mayorga O, Straume M. Anal Chem. 1990;62:950–959. [Google Scholar]
- 12.Ladbury JE, Chowdhry BZ. Chem Biol. 1996;3:791–801. doi: 10.1016/s1074-5521(96)90063-0. [DOI] [PubMed] [Google Scholar]
- 13.Leavitt S, Freire E. Curr Opin Struct Biol. 2001;11:560–566. doi: 10.1016/s0959-440x(00)00248-7. [DOI] [PubMed] [Google Scholar]
- 14.Turnbull WB, Daranas AH. J Am Chem Soc. 2003;125:14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- 15.Tellinghuisen J. J Phys Chem B Condens Matter Mater Surf Interfaces Biophys. 2005;109:20027–20035. doi: 10.1021/jp053550y. [DOI] [PubMed] [Google Scholar]
- 16.Sigurskjold BW. Anal Biochem. 2000;277:260–266. doi: 10.1006/abio.1999.4402. [DOI] [PubMed] [Google Scholar]
- 17.Markova N, Hallen D. Anal Biochem. 2004;331:77–88. doi: 10.1016/j.ab.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 18.Tellinghuisen J. Anal Biochem. 2007;360:47–55. doi: 10.1016/j.ab.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 19.Tellinghuisen J. Anal Biochem. 2005;343:106–115. doi: 10.1016/j.ab.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 20.Tellinghuisen J. Biophys Chem. 2006;120:114–120. doi: 10.1016/j.bpc.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Chaires JB. Biopolymers. 1997;44:201–215. doi: 10.1002/(SICI)1097-0282(1997)44:3<201::AID-BIP2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 22.Ward WH, Holdgate GA. Prog Med Chem. 2001;38:309–376. doi: 10.1016/s0079-6468(08)70097-3. [DOI] [PubMed] [Google Scholar]
- 23.Kozlov AG, Lohman TM. J Mol Biol. 1998;278:999–1014. doi: 10.1006/jmbi.1998.1738. [DOI] [PubMed] [Google Scholar]
- 24.Jelesarov I, Crane-Robinson C, Privalov PL. J Mol Biol. 1999;294:981–995. doi: 10.1006/jmbi.1999.3284. [DOI] [PubMed] [Google Scholar]
- 25.Minetti CA, Remeta DP, Zharkov DO, Plum GE, Johnson F, Grollman AP, Breslauer KJ. J Mol Biol. 2003;328:1047–1060. doi: 10.1016/s0022-2836(03)00365-6. [DOI] [PubMed] [Google Scholar]
- 26.Buurma NJ, Haq I. Methods. 2007;42:162–172. doi: 10.1016/j.ymeth.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Nudler E, Mironov AS. Trends Biochem Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Winkler WC. Curr Opin Chem Biol. 2005;9:594–602. doi: 10.1016/j.cbpa.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 29.Tucker BJ, Breaker RR. Curr Opin Struct Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Batey RT. Curr Opin Struct Biol. 2006;16:299–306. doi: 10.1016/j.sbi.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Gilbert SD, Stoddard CD, Wise SJ, Batey RT. J Mol Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Gilbert SD, Mediatore SJ, Batey RT. J Am Chem Soc. 2006;128:14214–14215. doi: 10.1021/ja063645t. [DOI] [PubMed] [Google Scholar]
- 33.Muller M, Weigand JE, Weichenrieder O, Suess B. Nucleic Acids Res. 2006;34:2607–2617. doi: 10.1093/nar/gkl347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alper PB, Hendrix M, Sears P, Wong C-H. J Am Chem Soc. 1998;120:1965–1978. [Google Scholar]
- 35.Lynch SR, Puglisi JD. J Mol Biol. 2001;306:1037–1058. doi: 10.1006/jmbi.2000.4420. [DOI] [PubMed] [Google Scholar]
- 36.Hermann T, Westhof E. J Mol Biol. 1998;276:903–912. doi: 10.1006/jmbi.1997.1590. [DOI] [PubMed] [Google Scholar]
- 37.von Ahsen U, Davies J, Schroeder R. Nature. 1991;353:368–370. doi: 10.1038/353368a0. [DOI] [PubMed] [Google Scholar]
- 38.Ennifar E, Paillart JC, Bodlenner A, Walter P, Weibel JM, Aubertin AM, Pale P, Dumas P, Marquet R. Nucleic Acids Res. 2006;34:2328–2339. doi: 10.1093/nar/gkl317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ennifar E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C, Dumas P, Walter P. J Biol Chem. 2003;278:2723–2730. doi: 10.1074/jbc.M205726200. [DOI] [PubMed] [Google Scholar]
- 40.Pilch DS, Kaul M, Barbieri CM, Kerrigan JE. Biopolymers. 2003;70:58–79. doi: 10.1002/bip.10411. [DOI] [PubMed] [Google Scholar]
- 41.Barbieri CM, Pilch DS. Biophys J. 2006;90:1338–1349. doi: 10.1529/biophysj.105.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaul M, Barbieri CM, Srinivasan AR, Pilch DS. J Mol Biol. 2007;369:142–156. doi: 10.1016/j.jmb.2007.02.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukada H, Takahashi K. Proteins. 1998;33:159–166. [PubMed] [Google Scholar]
- 44.Black DL, Chan R, Min H, Wang J, Bell L. In: In RNA: Protein Interactions. Smith CWJ, editor. Oxford University Press; Oxford: 1998. pp. 109–136. [Google Scholar]
- 45.Recht MI, Williamson JR. J Mol Biol. 2001;313:35–48. doi: 10.1006/jmbi.2001.5018. [DOI] [PubMed] [Google Scholar]
- 46.Recht MI, Williamson JR. J Mol Biol. 2004;344:395–407. doi: 10.1016/j.jmb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 47.Talkington MW, Siuzdak G, Williamson JR. Nature. 2005;438:628–632. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niedzwiecka A, Marcotrigiano J, Stepinski J, Jankowska-Anyszka M, Wyslouch-Cieszynska A, Dadlez M, Gingras AC, Mak P, Darzynkiewicz E, Sonenberg N, Burley SK, Stolarski R. J Mol Biol. 2002;319:615–635. doi: 10.1016/S0022-2836(02)00328-5. [DOI] [PubMed] [Google Scholar]
- 49.Niedzwiecka A, Stepinski J, Darzynkiewicz E, Sonenberg N, Stolarski R. Biochemistry. 2002;41:12140–12148. doi: 10.1021/bi0258142. [DOI] [PubMed] [Google Scholar]
- 50.Niedzwiecka A, Darzynkiewicz E, Stolarski R. Biochemistry. 2004;43:13305–13317. doi: 10.1021/bi0491651. [DOI] [PubMed] [Google Scholar]
- 51.Prabhu NV, Sharp KA. Annu Rev Phys Chem. 2005;56:521–548. doi: 10.1146/annurev.physchem.56.092503.141202. [DOI] [PubMed] [Google Scholar]
- 52.Mikulecky PJ, Feig AL. Biopolymers. 2006;82:38–58. doi: 10.1002/bip.20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kozlov AG, Lohman TM. Biochemistry. 1999;38:7388–7397. doi: 10.1021/bi990309z. [DOI] [PubMed] [Google Scholar]
- 54.Kozlov AG, Lohman TM. Proteins. 2000;(Supp 4):8–22. doi: 10.1002/1097-0134(2000)41:4+<8::aid-prot20>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 55.Kozlov AG, Lohman TM. Biochemistry. 2006;45:5190–5205. doi: 10.1021/bi052543x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathews DH, Turner DH. Curr Opin Struct Biol. 2006;16:270–278. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 57.Mathews DH. J Mol Biol. 2006;359:526–532. doi: 10.1016/j.jmb.2006.01.067. [DOI] [PubMed] [Google Scholar]
- 58.Takach JC, Mikulecky PJ, Feig AL. J Am Chem Soc. 2004;126:6530–6531. doi: 10.1021/ja0316263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mikulecky PJ, Feig AL. Biochemistry. 2006;45:604–616. doi: 10.1021/bi0517178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diamond JM, Turner DH, Matthews DH. Biochemistry. 2001;40:6971–6981. doi: 10.1021/bi0029548. [DOI] [PubMed] [Google Scholar]
- 61.Mathews DH, Turner DH. Biochemistry. 2002;41:869–880. doi: 10.1021/bi011441d. [DOI] [PubMed] [Google Scholar]
- 62.Mikulecky PJ, Takach JC, Feig AL. Biochemistry. 2004;43:5870–5881. doi: 10.1021/bi0360657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Windbichler N, Werner M, Schroeder R. Nucleic Acids Res. 2003;31:6419–6427. doi: 10.1093/nar/gkg873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bernacchi S, Ennifar E, Toth K, Walter P, Langowski J, Dumas P. J Biol Chem. 2005;280:40112–40121. doi: 10.1074/jbc.M503230200. [DOI] [PubMed] [Google Scholar]
- 65.Storz G, Altuvia S, Wassarman KM. Annu Rev Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- 66.Majdalani N, Vanderpool CK, Gottesman S. Crit Rev Biochem Mol Biol. 2005;40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- 67.SantaLucia J, Jr., Turner DH. Biopolymers. 1998;44:309–319. doi: 10.1002/(SICI)1097-0282(1997)44:3<309::AID-BIP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 68.Proctor DJ, Ma H, Kierzek E, Kierzek R, Gruebele M, Bevilacqua PC. Biochemistry. 2004;43:14004–14014. doi: 10.1021/bi048213e. [DOI] [PubMed] [Google Scholar]
- 69.Hammann C, Cooper A, Lilley DM. Biochemistry. 2001;40:1423–1429. doi: 10.1021/bi002231o. [DOI] [PubMed] [Google Scholar]