Abstract
Insights into the function of a gene can be gained in multiple ways, including loss-of-function phenotype, sequence similarity, expression pattern, and by the consequences of its misexpression. Analysis of the phenotypes produced by expression of a gene at an abnormal time, place, or level may provide clues to a gene’s function when other approaches are not illuminating. Here we report that an eye-specific, enhancer–promoter present in the P element expression vector pGMR is able to drive high level expression in the eye of genes near the site of P element insertion. Cell fate determination, differentiation, proliferation, and death are essential for normal eye development. Thus the ability to carry out eye-specific misexpression of a significant fraction of genes in the genome, given the dispensability of the eye for viability and fertility of the adult, should provide a powerful approach for identifying regulators of these processes. To test this idea we carried out two overexpression screens for genes that function to regulate cell death. We screened for insertion-dependent dominant phenotypes in a wild-type background, and for dominant modifiers of a reaper overexpression-induced small eye phenotype. Multiple chromosomal loci were identified, including an insertion 5′ to hid, a potent inducer of apoptosis, and insertions 5′ to DIAP1, a cell death suppressor. To facilitate the cloning of genes near the P element insertion new misexpression vectors were created. A screen with one of these vectors identified eagle as a suppressor of a rough eye phenotype associated with overexpression of an activated Ras1 gene.
Mutational inactivation is a powerful tool for understanding the role of a gene product in a specific process. However, this approach is limited by the fact that the majority of genes do not have an easily assayable loss-of-function phenotype; that is, mutations in most genes are phenotypically silent under laboratory conditions. Second, any observed phenotype only reflects that part of a gene’s function that is not compensated for by other genes and pathways. Finally, many genes are required for multiple aspects of normal development or adult function. These limitations make it difficult to address the function of a gene late in development, or in the adult if the gene is also required at an earlier stage for cell proliferation, cell survival, or differentiation (reviewed in ref. 1).
An alternative approach to understanding gene function is to characterize phenotypes resulting from tissue-specific expression of individual genes in tissues where they are not normally expressed or are expressed at elevated levels at a normal site of expression. Such misexpression may create phenotypes, whereas mutational inactivation does not. Misexpression also provides a way of asking if a gene product is able to direct a particular process or alter the output of a signaling pathway in a particular tissue. Genes identified in one tissue as signal modifiers by overexpression phenotypes are likely to be important regulators, even if the gene is normally not expressed in that tissue. These genes might be useful in gene therapy, where a major goal is to identify genes that can modify signaling pathways in novel contexts. Misexpression of individual, cloned genes is a valuable approach for identifying developmental regulators or signaling molecules, but it requires that one have the full-length candidate gene in hand and that these genes be introduced into the germ line one at a time. Also, selection of appropriate candidate genes requires prior knowledge about what genes are likely to be important for the process under study. In contrast, misexpression of random genes allows one to search for genes that can affect a process without preconceptions.
Important developmental regulators and oncogenes have been identified as a result of fortuitous tissue-specific gene overexpression due to chromosomal aberrations or insertions of retrotransposons or transposable elements that bring genes under the control of novel transcriptional regulators (2–5). Screens designed to identify genes based on misexpression-dependent phenotypes can be carried out in several ways. In transfectable single-cell organisms such as yeast, or in mammalian cell culture, overexpression of random clones from cDNA libraries can be used to identify genes that can alter cell fate or modify the output of specific signaling pathways (6–8). In intact plants and animals, tissue-specific overexpression of unknown genes can be brought about by using insertions of transposable elements containing tissue-specific enhancers or enhancer–promoters to drive the expression of nearby genes. The molecular markers provided by the insertions facilitate cloning of the expressed genes. This “activation tagging” approach has also led to the identification of developmental regulators and oncogenes (9–11). Drosophila is an ideal system in which to carry out activation tagging screens because transposable elements (P elements) can be mobilized throughout the genome at a high frequency, in a controlled fashion (12), and because mutagenic P elements have a preference for insertion near the 5′ end of a gene (13).
Previously we described a P element expression vector pGMR that drives eye-specific expression of cloned genes (14). Here we show that sequences within pGMR are also able to drive the eye-specific expression of endogenous genes near the site of P element insertion. We generated 500 new insertions of the empty GMR vector. Four percent of these insertions are associated with dominant eye phenotypes, and another 1.6% act as modifiers of an excess eye cell death phenotype resulting from expression of the apoptosis inducers reaper (rpr) or hid. We have created a new P element vector, GMREP, that facilitates cloning of genes identified through eye misexpression phenotypes.
MATERIALS AND METHODS
Vector Construction.
pGMREP (Fig. 1) was created by ligating an XhoI–PstI fragment of pGMR that contains the binding sites for the transcription factor GLASS and the hsp70 TATA box into XhoI–PstI cut Bluescript (Stratagene). Sites downstream of the GLASS binding sites present in the pGMR fragment were removed by cutting with SalI and BamHI, followed by blunting with T4 DNA polymerase and ligation to create GMR-Bluescript. To create a 5′ splice donor site downstream of the hsp70 TATA box, a fragment of glass genomic DNA surrounding the first exon 5′ splice donor site (nucleotides 4301–4481; ref. 15) was amplified using primers 5′-CGCTGCAGCTACTTAAAACCGAGTCTTCG and 5′-GGAGATCTTTTCTTTCTTCTTTTTTATTGCAGATTT. The product was cut with BglII, treated with T4 DNA polymerase, and then cut with PstI. The product was then ligated into GMR-Bluescript that had been cut with XbaI, treated with T4 DNA polymerase, and then cut with PstI. A KpnI–NotI fragment containing the glass binding sites, TATA box, and 5′ splice donor site was shuttled into the vector pUAST (16) and removed as an XbaI–BglII fragment, which was cloned into the P element transformation vector PEG117 (17) that was cut with XbaI and BamHI. Restriction endonuclease sites 5′ to the enhancer–promoter were removed by cutting with XbaI and KpnI, treating with T4 DNA polymerase and then religation. The NotI site 3′ to the enhancer–promoter complex was removed by cutting with NotI, blunting with T4 DNA polymerase, and religation. To facilitate plasmid rescue of genomic DNA flanking the 3′ P element end, a fragment containing the Bluescript polylinker was amplified by PCR using the Bluescript universal and reverse primers, blunted with T4 DNA polymerase, and cloned into PEG117 containing the enhancer–promoter construct cut with SacII and blunted with T4 DNA polymerase. All sites in the polylinker except PstI and HindIII are usable for plasmid rescue of genomic DNA flanking the 3′ P element end. The polylinker is oriented such that sequencing with the T3 primer will read into genomic sequence following plasmid rescue.
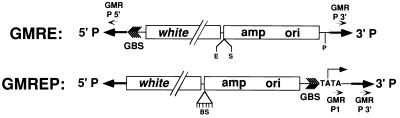
Figure 1.
Maps of the vectors GMRE and GMREP. The positions of the white marker gene (white), sequences for plasmid rescue (amp and ori), and the pentamer of GLASS binding sites (GBS) are indicated. Restriction endonuclease sites for plasmid rescue are represented by short vertical lines. Primer sequences used for PCR are indicated by small arrowheads (GMR P1 and GMR P 3′). In GMREP, the TATA box and 5′ splice donor sequences are indicated. The Bluescript polylinker (BS) is indicated as a triangle with vertical lines.
pGMRE (Fig. 1) was created by cloning an XhoI–SalI fragment of pGMR that contained the pentamer of the GLASS binding sites into XhoI–SalI-cut Bluescript in a way that the XhoI and SalI sites are retained. A NotI–KpnI fragment of Bluescript that contains the GLASS binding sites was isolated and ligated into NotI–KpnI cut PEG117. EcoRI and SacII can be used for plasmid rescue of genomic DNA flanking the 3′ P element end. PstI can be used to rescue genomic DNA flanking the 5′ P element end.
The intracellular domain of the human FAS transmembrane receptor is able to transduce a cell death signal on multimerization (18). To create a potentially constitutively active FAS receptor we fused a sequence coding for the extracellular domain of the influenza hemagglutinin (HA), which forms trimers (19), to the transmembrane and intracellular domain of human FAS, thereby generating a construct known as HAFAS, which we hoped would transduce a ligand independent FAS death signal (details provided on request).
Transformation and Screening.
Flies transgenic for pGMR, pGMRE, and pGMREP were generated using standard procedures (20), and insertions of each of these elements on the X chromosome were identified. Autosomal insertions were generated by crossing females with insertions on the X chromosome with males carrying a stable source of P element transposase activity (21). Progeny were then outcrossed to w1118. Insertions on the autosomes were identified as red-eyed males in the subsequent generation. Males and females with dominant eye phenotypes were balanced for the appropriate chromosome. Most autosomal insertions did not give rise to dominant phenotypes at the level of the dissecting microscope. These insertions were kept outcrossed to w1118. Individual transformant males (heterozygotes for the insertion) were crossed to flies that were either GMR-rprM/TM6B, GMR-rprS/TM6B (22), or sev-Ras1V12 inserted on a CyO chromosome (CR2/Adv; see ref. 23). The progeny were scored for the presence of flies whose eye phenotype differed from that of GMR-rprM, GMR-rprS, or CR2 alone. Modifiers were balanced and characterized as described in the text. th4 and th5 were used to test the 72D insertions GMR228 and GMR355 for allelism (22). eg1 and eg2 were used to test 79A3-4 GMRE28 excision lines for allelism (24).
RNA and DNA Isolation and Characterization.
To isolate DNA surrounding the site of pGMR insertion, inverse PCR was performed. Genomic DNA from P element lines was cut with Sau3A, diluted, and circularized with T4 DNA ligase. Primers 5′-GCATGTCCGTGGGGTTTGAAT (Pry4) and 5′-CTTGCCGACGGGACCACCTTATGTTATT (GMR P 3′) extend in opposite directions within a Sau3A fragment that contains the 3′ P element end. PCR was carried out with these primers; products were blunted with T4 DNA polymerase and cloned into SmaI cut Bluescript. They were sequenced by the chain termination method (25) using the Automated Laser Fluorescence system (Pharmacia).
To determine the location of the GMRE28 P element with respect to the eagle gene, plasmid rescue of flanking genomic DNA was carried out using SacII. By probing DNA blots of EcoRI digests of cosmids 8 and 27 (see ref. 24) with this plasmid rescue fragment we were able to place the P element 400–500 bases 5′ to the eagle transcription unit, with the 3′ P element end closest to eagle (see Fig. 5G).
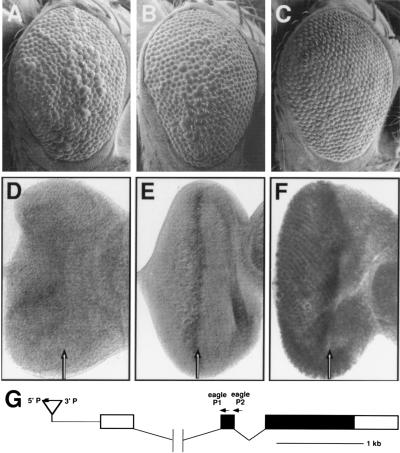
Figure 5.
A GMRE insertion near the eagle gene acts as a suppressor of the sev-Ras1V12-dependent rough eye phenotype by directing eagle expression in the developing eye. The following genotypes are shown: sev-Ras1V12 (CR2)/+ (A); sev-Ras1V12 (CR2)/GMRE28 (B); sev-Ras1V12 (CR2)/GMR-eagle (C); wild type (D); GMRE28/+ (E); GMR-eagle/+ (F). Expression of sev-Ras1V12 (CR2) results in a rough eye phenotype (A). This phenotype is mildly suppressed in the presence of the GMRE28 chromosome (B) and more strongly suppressed by GMR-eagle expression (C). The degree of sev-Ras1V12 rough eye suppression is correlated with the level of eagle expression: wild-type eye imaginal discs express little if any eagle (D) discs from GMRE28 flies express eagle at higher levels in the morphogenetic furrow (arrow), and to some extent posterior to the morphogenetic furrow (E); and discs from GMR-eagle flies express high levels of eagle in and posterior to the morphogenetic furrow (F). (G) Map of the eagle genomic region. GMRE28 is inserted approximately 500 bases 5′ to the eagle transcription unit as indicated. Noncoding cDNA sequences are indicated by open boxes and coding sequences by filled boxes.
PCR assays were performed to determine if chimeric transcripts extending from within the P element into the surrounding genomic region were being generated. RNA was isolated from 50 eye–antennal imaginal discs using the Micro-Scale Total RNA separator kit (Clonetech, Palo Alto, CA). cDNA was generated from this RNA using the Clonetech Marathon cDNA amplification kit. Gene-specific primers were used to prime cDNA synthesis: 5′-AATATATTGTTCTTGTGTCCCGTC (hid P2, see Fig. 3D); 5′-TTGAATTTGAGGACTTGGGTGCGC (DIAP P2, see Fig. 4G); and 5′- GCAGCCTTCACATGTAAATGCC (eagle P2, see Fig. 5G). PCR was then carried out using this cDNA as a template. One primer of each primer pair was a gene-specific primer located 3′ to a large intron in the gene of interest, extending toward the 5′ end of the gene: 5′-AACCGTCACAACAGTTGGCCAAGTGAA (hid P1, see Fig. 3C); 5′-TGGCGCAGGCCACCACATGACCGC (DIAP P1, see Fig. 4G); and 5′-GCACACTTTGCACAGCTGGTTCAT (eagle P1, see Fig. 5G). The second primer was either 5′-CGTCGCTAAGCGAAAGCTAAGCAA (GMR P1), present just 3′ to the hsp70 transcription start site, or 5′-CTTGCCGACGGGACCACCTTATGTTATT (GMR P3′), present just 5′ to the 3′ P element end (see Fig. 3C).
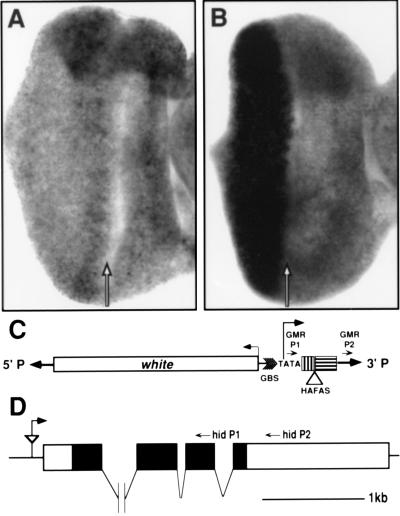
Figure 3.
Misexpression of the hid gene by the GMR-HAFAS110 insertion. The hid expression pattern in eye-antennal discs from wild type (A) and GMR-HAFAS110/GMR-HAFAS110 (B) larvae are shown. The hid transcript is expressed uniformly in the wild-type eye imaginal disc (A) and at high levels in and posterior (left) to the morphogenetic furrow (arrow) in GMR-HAFAS110 eye imaginal discs (B). (C) Map of the GMR-HAFAS construct. The expression vector pGMR contains the white marker gene (white), a multimer of glass binding sites (GBS), TATA sequences from the hsp70 promoter (TATA), approximately 200 bases of 5′ untranslated region (box marked with vertical bars), a polylinker into which the HAFAS construct was cloned (HAFAS is not drawn to scale), and the hsp 70 3′ untranslated region (box marked with horizontal bars). The direction of transcription from the hsp70 promoter is indicated. The P element ends are indicated as boldface arrows. The location of the P element primers used to detect chimeric transcripts are indicated with small arrowheads (GMR P1 and GMR P2). (D) Map of the genomic region at the site of insertion of the GMR-HAFAS110 P element. GMR-HAFAS110 is inserted 131 base pairs 5′ to the longest hid cDNA (described in ref. 26). The P element is indicated by the triangle and is oriented such that transcription from the hsp70 TATA box reads through the P element 3′ end and into the 5′ end of the hid transcription unit as indicated by the arrow. The locations of primers used for cDNA synthesis (hid P2) and PCR (hid P1) are indicated on the map. The introns are not drawn to scale.
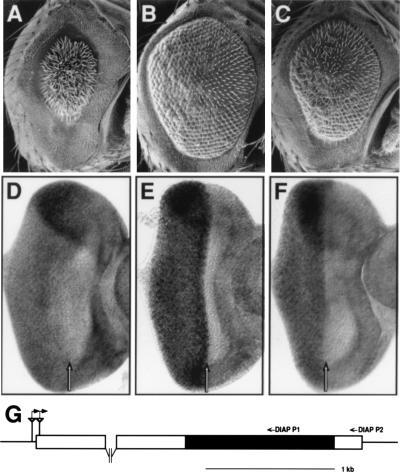
Figure 4.
GMR insertions near the DIAP1 gene act as strong suppressors of a GMR-rpr-dependent small eye phenotype and express high levels of the DIAP1 gene in the developing eye. The following genotypes are shown: GMR-rprS/TM6B (A); GMR-rprS/GMR228 (B); GMR-rprS/GMR355 (C); wild type (D); GMR228/TM6B (E); GMR355/TM6B (F). GMR-rpr expression results in a cell death-dependent small eye phenotype (A), which is suppressed in the presence of the GMR228 (B) and GMR355 (C) chromosomes. DIAP1 transcript levels are uniform throughout the wild-type eye-imaginal disc (D) but are elevated posterior to the morphogenetic furrow in the GMR228 (E) and GMR355 (F) lines. (G) Diagram of the DIAP1 chromosomal region. The P element in the GMR228 line is inserted after base 34 in the DIAP1 5′ untranslated region and 70 bases upstream of the DIAP1 5′ untranslated region in the GMR228 line. Both P elements (triangles) are oriented with the 3′ P end nearest the DIAP1 gene. The direction of hsp70 transcription is indicated by the raised, rightward pointing arrows. Small arrows above the map indicate the primers used to prime cDNA synthesis (DIAP P2) and to carry out PCR (DIAP P1) to detect the presence of chimeric DIAP1 transcripts.
Histology.
Scanning electron microscopy (26), fixation and sectioning of adult eyes (27), and tissue in situ hybridizations (28) were performed as described previously.
RESULTS AND DISCUSSION
We are interested in identifying and characterizing cell death signal transduction pathways in the fly. One approach we have taken is to express, specifically in a nonessential tissue, such as the eye, molecules that might be expected to alter the normal pattern of cell death. If expression of these molecules alters normal cell death signaling we can use the resulting phenotypes present in the adult as backgrounds in which to carry out genetic screens for Drosophila cell death regulators. To carry out this approach we made the pGMR expression vector, which contains a pentamer of binding sites for the GLASS transcription factor derived from the Rh1 promoter and TATA box sequences from the Drosophila hsp70 promoter, cloned into the CaSpeR-hs vector (14). Sequences placed downstream of these sites are transcribed in a similar pattern to glass expression, in and posterior to the morphogenetic furrow during larval and pupal eye development (29). Pattern formation in the eye occurs during this same period as a series of inductive events in and posterior to the morphogenetic furrow during which cells must choose to differentiate, proliferate, or die (30).
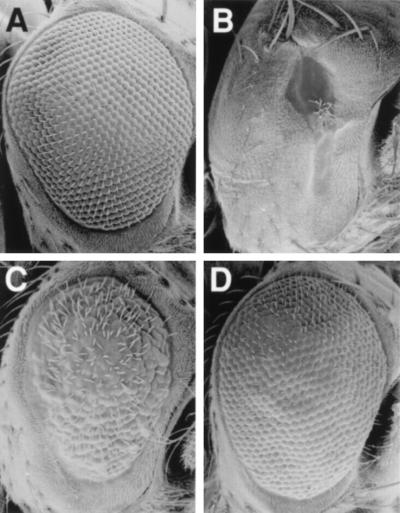
As a part of our efforts to activate cell death signaling in the fly, we generated a large number of GMR transformants (about 100) expressing a transcript coding for a chimeric protein called HAFAS (GMR-HAFAS; described in Materials and Methods) that we hoped would transduce a cell death signal. Most lines transgenic for this construct have no visible adult eye phenotype. However, in one line (GMR-HAFAS110) the flies have very small eyes (Fig. 2B). This phenotype is suppressed by decreasing the dose of glass (Fig. 2C), suggesting that it is the result of glass-dependent transcription. The phenotype is completely suppressed, and eye size restored to normal, by coexpression of the baculovirus cell death inhibitor, p35 (Fig. 2D). P35 blocks cell death in multiple contexts in Drosophila, including death due to overexpression of the cell death activators rpr, hid, and grim (22, 31–33). These results indicate that glass-dependent activation of a cell death signaling pathway is occurring in the GMR-HAFAS110 line. Because most insertions of this construct have no visible phenotype in the adult, it is unlikely that cell death activation is due to expression of the HAFAS chimera. The GMR-HAFAS110 P element is located in the 75C1-2 cytological region. This region contains the rpr, hid, and grim genes, each of which is capable of inducing a cell death-dependent small eye phenotype when overexpressed (22, 31–34). The GMR-HAFAS110 P line is semilethal in trans to a chromosomal deletion for the 75C region, suggesting that it has inserted near an essential gene. Sequencing of genomic DNA surrounding the insertion site shows that it is inserted 131 bases 5′ to the longest hid cDNA described (ref. 31; Fig. 3C). Tissue in situ hybridizations to third instar eye imaginal discs with a hid cDNA probe show that in wild-type eye imaginal discs hid is expressed at uniform low levels (Fig. 3A) but at much higher levels in and posterior to the morphogenetic furrow in eye imaginal discs from GMR-HAFAS110 flies (Fig. 3B). These observations suggest that the multimerized GLASS binding sites present in the GMR-HAFAS110 line are acting to drive eye-specific expression of the endogenous hid gene. By carrying out PCR on cDNA generated from GMR-HAFAS110 eye–antennal disc total RNA using P-element- and gene-specific primers (see Materials and Methods and Fig. 3C), we were able to detect chimeric transcripts containing sequences from the P element 3′ end and the hid coding region. We were unable to detect chimeric transcripts initiating at the transcription start site downstream from the hsp70 TATA box, but we were able to detect chimeric transcripts that contained P element sequences closer to the 3′ P end. These chimeric transcripts may initiate at the hsp70 promoter, but be unstable, perhaps due to the presence of sequences from the hsp70 3′ untranslated region; alternatively, chimeric hid transcripts may be generated using an cryptic promoter closer to the 3′ P element end.
Figure 2.
Scanning electron micrographs of adult eyes of the following genotypes are shown: wild-type (A); GMR-HAFAS110/+ (B); GMR-HAFAS110/gl60j (C); GMR-HAFAS110/GMR-p35 (D). Flies with the GMR-HAFAS110 insertion (B) have small eyes due to excess cell death in the developing eye. This phenotype is partially suppressed by removing one copy of gl (C) and completely suppressed by coexpressing baculovirus p35 (D).
Because mutagenic P element insertions tend to occur in or near the 5′ end of genes (13), our observations with the HAFAS110 line suggest that it should be possible to sample a significant fraction of the genome for genes that can affect some aspect of eye development when overexpressed by mobilizing pGMR throughout the genome.
To test this idea we carried out several screens in which new insertions of empty pGMR were created. In one screen we looked for dominant phenotypes as a result of mobilization of pGMR to new sites on the autosomes. In a second screen these autosomal insertions were then scored for their ability to act as dominant modifiers of rpr overexpression-induced small eye phenotypes (GMR-rprM and GMR-rprS) used previously in a screen for genes in which reduction in function modified the extent of cell death (22). A dominant eye phenotype was observed in 4% of the lines tested (19 of 500), ranging from a very small eye to various degrees of roughness (data not shown). These phenotypes are suppressed by removing one copy of glass, therefore indicating that they are due to glass-dependent expression, presumably of nearby genes.
In crosses to GMR-rprM or GMR-rprS flies five enhancers and two suppressors were identified (Fig. 4 A–C). The two suppressor lines (GMR228 and GMR355) each contain a single P element that maps to the 72D1-2 cytological region. Complementation crosses identify GMR228 as a lethal allele of thread (th), which codes for the DIAP1 protein, a dose-dependent suppressor of rpr- and hid-dependent cell death (22). In contrast to the GMR228 th allele, other lethal alleles of th act as enhancers of rpr and hid-dependent cell death, due to a decrease in DIAP1 activity. The GMR355 line is semilethal when homozygous but complements lethal th alleles for viability. The GMR228 P element is inserted at base 34 of the 5′ untranslated region of the largest DIAP1 cDNA isolated (22), whereas the GMR355 P element is inserted 70 bases 5′ to this cDNA. (Fig. 4G). Tissue in situ hybridizations with a DIAP1 probe show that DIAP1 mRNA is expressed at uniform low levels in wild-type third instar eye-antennal discs (Fig. 4D) but at much higher levels in and posterior to the morphogenetic furrow in discs from GMR228 (Fig. 4E) and GMR355 flies (Fig. 4F). In both lines the P element is inserted such that transcription from the GMR hsp70 TATA box would extend into the 5′ end of the DIAP1 gene (Fig. 4G). PCR using P element and DIAP1 specific primers (see Materials and Methods) show that chimeric transcripts are being created (data not shown). Therefore, the cell death suppression seen in these lines is due to the GMR insertion-dependent expression of chimeric DIAP1 transcripts.
The above results show that insertion of pGMR results in a high frequency of eye phenotypes. Characterization of the above three lines suggests that pGMR is acting as an enhancer–promoter vector; eye-specific transcription is driven through the 3′ P element end and into the surrounding genomic region. pGMR is not ideal for overexpression screens because it contains extraneous sequences (the hsp70 3′ untranslated region) between the hsp70 TATA box and the 3′ P element end and because it lacks plasmid rescue capability. To facilitate further screens of this kind, and to determine if promoter sequences are required to generate a high frequency of GLASS multimer-dependent eye phenotypes, we constructed and tested two new vectors. In one vector, GMREP, the GLASS binding site pentamer, hsp70 TATA box sequences, and a 5′ splice donor sequence are located near the 3′ P element end (Fig. 1). In a second vector, pGMRE, the GLASS binding site multimer alone is present, located near the P element 5′ end (Fig. 1). Both vectors have plasmid rescue sequences and multiple unique restriction endonuclease sites to facilitate the cloning of nearby overexpressed genes.
Mobilization of pGMREP to the autosomes resulted in glass-dependent, dominant phenotypes with a frequency of 8–10%, more than twice that seen with pGMR. This higher frequency may be due to some combination of increased translatability of GMREP transcripts, which have a much shorter 5′ untranslated region, increased transcript stability due to the lack of sequences from the hsp70 3′ untranslated region, or splicing of transcripts originating from GMREP intron insertions to downstream coding exons. Mobilization of pGMRE resulted in a very low frequency (1/1000) of dominant eye phenotypes. GMRE lines were also scored for their ability to modify a GMR-rpr induced small eye phenotype or a rough eye phenotype associated with expression of an activated Drosophila Ras1 gene under control of the sevenless enhancer-promoter (sev-Ras1V12; see ref. 35). Several enhancers of GMR-rprS and one suppressor of sev-Ras1V12 (GMRE28) were identified. The sev-Ras1V12 suppressor was characterized further.
sev-Ras1V12 flies have a rough eye phenotype (Fig. 5A) associated with extra R7 photoreceptors and ommatidial fusions (35). GMRE28 dominantly suppresses this rough eye phenotype (Fig. 5B), and there appear to be fewer ommatidial fusions (data not shown). The GMRE28 suppressor phenotype is P element- associated, because excision of the P element results in a loss of the suppressor phenotype (data not shown); this phenotype is also glass-dependent and dosage-sensitive. Cytologically, the GMRE28 P element maps to 79A3-4. Excision lines were generated with the intention of isolating loss-of-function mutations in the ectopically expressed gene at 79A3–4. A high percentage of these lines exhibited a held-out wing phenotype similar to that associated with mutations- affecting the eagle gene (24). The GMRE28 P element is inserted approximately 400–500 bases 5′ to the eagle transcription unit (Fig. 5G) and complements eagle alleles. The GMRE28-dependent sev-Ras1V12 suppression is not due to a decrease in eagle function because chromosomal deletions for the region and loss of function eagle alleles do not suppress the sev-Ras1V12 phenotype (data not shown).
To determine if the suppression is due to eagle overexpression in the eye we first carried out tissue in situ hybridizations using an eagle probe. In wild-type eye imaginal discs, eagle RNA is present at undetectable levels (Fig. 5D). In GMRE28 eye discs eagle is easily detected in the morphogenetic furrow and is just detectable posterior to the furrow (Fig. 5E). We then fused a full length eagle cDNA directly to the GMR enhancer–promoter and introduced this construct (GMR-eagle) into the germline. GMR-eagle transformants have higher levels of eagle expression in the eye imaginal disc posterior to the morphogenetic furrow (Fig. 5F), and there is a correspondingly higher degree of suppression of the sev-Ras1V12 rough eye phenotype (Fig. 5C). Thus eagle overexpression acts as a sev-Ras1V12 suppressor. The GMRE vector was designed to lack an outwardly pointing promoter and, as expected, RT-PCR experiments (see Materials and Methods) failed to detect hybrid P element-eagle transcripts.
Mutations that alter the sev-Ras1V12 rough eye phenotype include alterations in genes involved in Ras1 pathway signaling, Ras1 posttranslational modification, and sevenless-dependent transcription (23). Our data do not allow us to determine which of these processes is affected by eagle misexpression. Loss-of-function eagle phenotypes in the embryo suggest a requirement for the proper differentiation of a small number of cells in the CNS (24). By tissue in situ hybridization, eagle does not appear to be differentially or transcriptionally activated in photoreceptors or other cells of the eye disc (Fig. 5D). However, loss of function eagle phenotypes have not been characterized in the developing eye, therefore it is not known if eagle plays a role in normal eye development. eagle encodes a zinc finger protein sharing homology with steroid receptor family members, suggesting that it may function to regulate transcription, but its targets are unknown (24).
Concluding Remarks.
We have shown that an eye-specific enhancer–promoter complex, when mobilized throughout the genome in a P element, results in a high frequency of misexpression-dependent phenotypes. In the case of the GMR and GMREP vectors, these phenotypes result from the production of chimeric transcripts, initiating within the P element and extending into the surrounding genomic region. The utility of this approach was demonstrated in screens for genes important in cell death signaling. Because the phenotypes generated by P element insertion-dependent gene activation are due to the production of chimeric transcripts, in most cases the P element will be near the gene being expressed. Loss-of-function mutations in these genes can be made by imprecise excision of the P element. In cases such as the GMR228 insertion into DIAP1, where the normal expression of the gene has been inactivated, the loss-of-function phenotype can be scored directly in tissues other than the eye, where the GMR enhancer–promoter is inactive.
A related approach for large scale random gene misexpression in Drosophila has been described by Rorth (36). In this method, Gal4 expressed in specific patterns is used to drive the expression of genes near the insertion site of a P element containing Gal4 binding sites and a promoter sequence near one P element end. This system is very versatile because it allows one to test gene activating insertions for misexpression-dependent phenotypes in multiple tissues by crossing the Gal4 binding site insertion lines to flies in which Gal4 is expressed in different spatial and temporal patterns. High level Gal4 expression in the eye can disrupt normal development (37), but these effects can be mitigated by using lines that express lower levels of Gal4. Loss of function phenotypes can be characterized in the absence of Gal4 expression. In contrast, when using GMREP, one is primarily limited to screening for phenotypes in the developing eye. However, because insertions can be scored for eye phenotypes directly on generation, screens can be carried out somewhat more quickly. Overexpression-dependent phenotypes in other tissues or at other times can be examined by crossing GMREP lines to flies expressing GLASS under heat shock control (14, 29). The higher frequency of phenotypes seen with GMREP insertions (8–10%) compared with insertions of a Gal4 enhancer–promoter vector in which transcription is driven by sevenless-Gal4 (4%; ref. 36) may reflect the fact that GLASS is expressed earlier than Sevenless during eye development and in more cell types.
Misexpression screens with either system will be useful for identifying developmental regulators or other signaling molecules in which activity is transcriptionally regulated by subtly altering the levels or timing of their expression in tissues in which they are normally expressed. Misexpression screens can also identify genes that can function to regulate development or other cellular functions in a specific tissue, even if the gene is normally not expressed in that tissue. A limitation of the misexpression approach is that many genes can disrupt normal development when misexpressed at high levels. Therefore, a critical factor governing the successful implementation of this approach will be the ability to carry out efficient secondary analyses to identify genes that are directly affecting a process of interest. One way to target a particular pathway is to screen for suppressors of an existing phenotype that has been generated by activation of that pathway, as we have demonstrated here for the GMR-rpr and sev-RasV12 phenotypes. Moreover, by screening for restoration of a more normal eye phenotype, the background of nonspecific effects should be greatly reduced.
Acknowledgments
We thank Todd Laverty for chromosome in situ hybridizations, Paula Sicurello for scanning electron microscopy; Chris Suh for sequencing; Kristen White (Massachusetts General Hospital) for providing some initial mapping data that placed the GMR-HAFAS110 insertion near hid; Henry Chang for preparing the figures; and Alan C. Spradling, Pernille Rorth, and Ilaria Rebay for helpful comments on the manuscript. This work was supported by the Howard Hughes Medical Institute (G.M.R.), and by an American Cancer Society, California Division, Senior Postdoctoral Fellowship (B.A.H.).
ABBREVIATION
- HA
hemagglutinin
References
- 1.Miklos G L G, Rubin G M. Cell. 1996;86:521–529. doi: 10.1016/s0092-8674(00)80126-9. [DOI] [PubMed] [Google Scholar]
- 2.Lewis E B. Nature (London) 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 3.Nusse R. Trends Genet. 1986;2:244–248. [Google Scholar]
- 4.Smith L G, Greene B, Veit B, Hake S. Development (Cambridge, UK) 1992;116:21–30. doi: 10.1242/dev.116.1.21. [DOI] [PubMed] [Google Scholar]
- 5.Tanda S, Leshko L A, Corces V G, Hori S H. In: Drosophila Ananassae: Genetical and Biological Aspects. Tobari Y N, editor. Tokyo: Japan Scientific Societies Press; 1993. pp. 89–138. [Google Scholar]
- 6.Bender A, Pringle J R. Proc Natl Acad Sci USA. 1989;86:9976–9980. doi: 10.1073/pnas.86.24.9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramer S W, Elledge S J, Davis R W. Proc Natl Acad Sci USA. 1992;89:11589–11593. doi: 10.1073/pnas.89.23.11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis R L, Weintraub H, Lassar A B. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi H, Czaja I, Lubenow H, Schell J, Walden R. Science. 1992;258:1350–1353. doi: 10.1126/science.1455228. [DOI] [PubMed] [Google Scholar]
- 10.Haupt Y, Alexander W S, Barri G, Klinken P S, Adams J M. Cell. 1991;65:753–763. doi: 10.1016/0092-8674(91)90383-a. [DOI] [PubMed] [Google Scholar]
- 11.van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 12.Cooley L, Kelley R, Spradling A C. Science. 1988;239:1121–1128. doi: 10.1126/science.2830671. [DOI] [PubMed] [Google Scholar]
- 13.Spradling A C, Stern D M, Kiss I, Roote J, Laverty T, Rubin G M. Proc Natl Acad Sci USA. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hay B A, Wolff T, Rubin G M. Development (Cambridge, UK) 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 15.Moses K, Ellis M C, Rubin G M. Nature (London) 1989;340:531–536. doi: 10.1038/340531a0. [DOI] [PubMed] [Google Scholar]
- 16.Brand A, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 17.Giniger E, Wells W, Jan L Y, Jan Y N. Roux’s Arch Dev Biol. 1993;202:112–122. doi: 10.1007/BF00636536. [DOI] [PubMed] [Google Scholar]
- 18.Itoh N, Nagata S. J Biol Chem. 1993;268:10932–10937. [PubMed] [Google Scholar]
- 19.Kemble G W, Henis Y I, White J M. J Cell Biol. 1993;122:1253–1265. doi: 10.1083/jcb.122.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 21.Robertson H M, Preston C R, Johnson-Schlitz D M, Benz W K, Engels W R. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hay B A, Wassarman D A, Rubin G M. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 23.Karim F D, Chang H C, Therrien M, Wassarman D A, Laverty T, Rubin G M. Genetics. 1996;143:315–329. doi: 10.1093/genetics/143.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higashijima S, Shishido E, Matsuzaki M, Saigo K. Development (Cambridge, UK) 1996;122:527–536. doi: 10.1242/dev.122.2.527. [DOI] [PubMed] [Google Scholar]
- 25.Sanger F, Nicklen S, Coulson A. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimmel B E, Heberlein U, Rubin G M. Genes Dev. 1990;4:712–727. doi: 10.1101/gad.4.5.712. [DOI] [PubMed] [Google Scholar]
- 27.Wolff T, Ready D F. Development (Cambridge, UK) 1991;113:825–839. doi: 10.1242/dev.113.3.825. [DOI] [PubMed] [Google Scholar]
- 28.Dougan S, DiNardo S. Nature (London) 1992;360:347–350. doi: 10.1038/360347a0. [DOI] [PubMed] [Google Scholar]
- 29.Ellis M C, O’Neill E M, Rubin G M. Development (Cambridge, UK) 1993;119:855–865. doi: 10.1242/dev.119.3.855. [DOI] [PubMed] [Google Scholar]
- 30.Wolff T, Ready D F. In: The Development of Drosophila melanogaster. Bate M, Martinez Arias A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1277–1326. [Google Scholar]
- 31.Grether M E, Abrams J M, Agapite J, White K, Steller H. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- 32.White K, Tahaoglu E, Steller H. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- 33.Chen P, Nordstrom W, Gish B, Abrams J M. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- 34.White K, Grether M E, Abrams J M, Young L, Farrell K, Steller H. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 35.Fortini M E, Simon M A, Rubin G M. Nature (London) 1990;355:559–561. doi: 10.1038/355559a0. [DOI] [PubMed] [Google Scholar]
- 36.Rorth P. Proc Natl Acad Sci USA. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freeman M. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]