Abstract
The translation initiation factor eIF4E mediates the binding of the small ribosomal subunit to the cap structure at the 5′ end of the mRNA. In Saccharomyces cerevisiae, the cap-binding protein eIF4E is mainly associated with eIF4G, forming the cap-binding complex eIF4F. Other proteins are detected upon purification of the complex on cap-affinity columns. Among them is p20, a protein of unknown function encoded by the CAF20 gene. Here, we show a negative regulatory role for the p20 protein in translation initiation. Deletion of CAF20 partially suppresses mutations in translation initiation factors. Overexpression of the p20 protein results in a synthetic enhancement of translation mutation phenotypes. Similar effects are observed for mutations in the DED1 gene, which we have isolated as a multicopy suppressor of a temperature-sensitive eIF4E mutation. The DED1 gene encodes a putative RNA helicase of the DEAD-box family. The analyses of its suppressor activity, of polysome profiles of ded1 mutant strains, and of synthetic lethal interactions with different translation mutants indicate that the Ded1 protein has a role in translation initiation in S. cerevisiae.
Keywords: RNA helicase, DEAD-box family, cap-dependent initiation, yeast, 4E-BP
In eukaryotes, translation initiation is a multicomponent pathway that positions the 43S preinitiation complex at the initiator AUG codon of a mRNA (1). Translation of most of the eukaryotic mRNAs is dependent on the recognition of the 5′ cap {m7G(5′)ppp(5′)X} by eIF4F, the cap-binding complex, although cap-independent translation is also found in viral and cellular mRNAs (2). In higher eukaryotes, eIF4F is composed of eIF4E, the cap-binding protein, eIF4A, and eIF4G (p220) (1). This complex, together with eIF4B, is thought to promote the binding of the 43S preinitiation complex to mRNAs via the physical interaction between eIF4G and eIF3, which is associated with the small ribosomal subunit (3). The positioning and the subsequent scanning of the small ribosomal subunit are facilitated by the unwinding of secondary structures in the 5′ untranslated region of mRNAs via the helicase activity of eIF4A/eIF4B (4, 5). Although translation in eukaryotes is highly conserved, the yeast Saccharomyces cerevisiae does not possess a cap-binding complex directly equivalent to mammalian eIF4F. Instead, eIF4E (CDC33) is thought to be associated with eIF4G (p150; TIF4631 and TIF4632), p20 (CAF20), and weakly with eIF4B (STM1, also called TIF3). However, an association with eIF4A (TIF1 and TIF2) has not been detected (6, 7).
Much attention has been paid to the yeast cap-binding protein eIF4E. Temperature-sensitive alleles of the CDC33 gene lead to a defect in translation initiation in vivo and an arrest in the G1 phase of the cell cycle (8–11). Recently, the analysis of deletion variants of eIF4E with reduced affinity for the 5′ cap indicated that eIF4E mediates the selectivity of yeast ribosomes for capped mRNAs (12).
In the mammalian and yeast systems, eIF4E is associated with eIF4G. Recently, two other mammalian binding proteins (4E-BP1 and 4E-BP2) were shown to interact with eIF4E and negatively regulate cap-dependent translation initiation (13, 14). It was demonstrated that eIF4G and 4E-BP1 compete for binding to eIF4E (15). The binding of 4E-BP1 to eIF4E is negatively regulated via phosphorylation of 4E-BP1 (13, 14).
So far no equivalent to the 4E-BPs has been described in yeast. A gene (CAF20) encoding a small cap-associated protein, p20, was isolated by screening an expression library with polyclonal antibodies raised against eIF4F (16). This protein is phosphorylated to various degrees under different growth conditions, suggesting a role in translational control analogous to the mammalian 4E-BPs (17). However, there is no clear evidence for such a role of the p20 protein.
Here, we present genetic data demonstrating that the p20 protein acts as a general negative regulator of translation in yeast. We also describe a putative RNA helicase, Ded1p, as a suppressor of temperature-sensitive cdc33 mutant alleles. Our results indicate a genetic interaction between Ded1p and eIF4E, and a role of Ded1p in translation initiation.
MATERIALS AND METHODS
Yeast Strains, Media, and Genetic Methods.
CW04 was used as the genetic background (MATα ura3–1 ade2–1 his3–11,15 leu2–3,112 trp1–1), (18). The strains SS13–3A (tif1–1), RCB1–1A (stm1::ADE2) and RCB1–1C (stm1::ADE2) were previously described (19). CBY1.1 (tif4631::LEU2) and CBY1.2 (tif4631::LEU2) were a gift from C. Berset (University of Bern, Switzerland). CDK44–6A was obtained by introducing the prt1–1 mutation from T92 (M. Altmann, University of Bern) by three subsequent crosses. CDK33 was generated by transforming ASZ3, a diploid derivative of CW04, with a HindIII fragment from pUC9-cdc33::LEU2. CDK33–10B and CDK33–7C are meiotic products of CDK33. CDK35 was generated by transforming ASZ3 with a HindIII fragment from pUC19-cdc33::TRP1 (see below). CDK35–4A and CDK35–4B are meiotic products of CDK35. The plasmid shuffling technique (20) was used to introduce cdc33 alleles on different plasmids in either CDK33–10B, CDK33–7C, CDK35–4A, or CDK35–4B. CDK36 was generated by transforming ASZ3 with an EcoRI fragment from pUC9-caf20::TRP1 (M. Altmann). CDK36–1A and CDK36–1B are meiotic products of CDK36. Correct integrations were verified by Southern blot analysis. The wild-type strain DBY747 (MATa ura3–52 his3Δ1 leu2–3,112 trp1–289) and its mutant derivatives DJY105 (ded1/spp81–3), DJY106 (ded1/spp81–2), and DJY112 (dbp1::TRP1) were a gift from J. Beggs (University of Edinburgh, U.K.). CDK105–1A was obtained by introducing the ded1/spp81–3 mutation from DJY105 into the CW04 background by three subsequent crosses. CDK112–1A is a dbp1::TRP1 strain in the CW04 background.
Standard yeast genetic techniques and media were as described in (21). Yeast transformation was carried out according to Gietz et al. (22).
Plasmid Constructions.
Escherichia coli DH10B was used for plasmid propagation. Most plasmids used in this study are derivatives of the low (YCplac33 and YCplac111) or high (YEplac195 and YEplac181) copy number plasmids described in ref. 23. A 1.9-kb PstI fragment from pUC13-TIF2 (24) was cloned into the PstI site of the above vectors; a 3.5-kb EcoRI fragment from pUC19-STM1 (19) was cloned into the EcoRI site; a 1.9-kb HindIII–PstI fragment of pUC9-CDC33 (M. Altmann) was cloned into the HindIII–PstI sites; a 6.7-kb EcoRI fragment from YCG206 containing the TIF4631 gene (25) was cloned into the EcoRI site; and a 1.85-kb EcoRI fragment from YEp131–2 containing the CAF20 gene (M. Altmann) was cloned into the EcoRI site.
The 1.4-kb KpnI–SalI fragments containing either cdc33–42, cdc33–43, or cdc33–44 alleles from the pMDA101 plasmids (10) were cloned into YCplac33 or YEplac195. A 1.4-kb BamHI fragment of pMDA101-cdc33–1 was cloned into the BamHI site of YCplac33, YEplac195, and pRS413 (26). A 1.4-kb EcoRV fragment of YEplac181-CDC33 was cloned into the EcoRV site of pRS413. pRS413-cdc33–42 was constructed by digesting pMAD101-cdc33–42 with SalI, treating with T4 DNA polymerase, and digesting with SmaI; the resulting 1.4-kb blunt-ended fragment was cloned into the EcoRV site of pRS413. To construct pUC19-cdc33::TRP1, the 2.1-kb HindIII fragment from pUC9-CDC33 was cloned into the HindIII site of pUC19. A BglII fragment containing the TRP1 marker from pFL39 (27) was cloned in the NcoI–HpaI-digested pUC19-CDC33, with all the sites made blunt-ended by T4 DNA polymerase. The CAF20 gene was amplified by PCR using the oligonucleotides 5′-CGATGGATCCTTTATTTAATTTCACGACATG-3′ and 5′-CGATGGTACCGAATTCAGAAAAGTGAAGC-3′. The BamHI and KpnI sites (underlined) were used to clone the PCR product under control of the CYC1-GAL1 promoter of pGAL (28) or YEPL1 (P.L., unpublished data). The cloned PCR fragments were verified by sequencing.
A 3.5-kb SalI–KpnI fragment from pYDJ5 (J. Beggs) harboring DBP1 was cloned into YCplac111 and YEplac181. The yeast genomic library in YEplac181 was constructed from a Δstm1 strain (D.K., unpublished data). A 2.7-kb HindIII–SalI fragment including DED1 was subcloned into YCplac33, YCplac111, YEplac181, and YEplac195.
Polysome Profile Analysis.
Polysome preparations were done according to Foiani et al. (29). Cultures were harvested at OD600 = 0.8. Cycloheximide was added to a final concentration of 0.1 mg/ml immediately prior to harvesting. Eight A260 units of extract were layered onto 11.2 ml of 7–50% linear sucrose gradients that were centrifuged at 39,000 r.p.m. in a Beckman SW41 rotor at 4°C for 3 h. High-salt conditions were achieved by adding NaCl to a final concentration of 0.7 M in the gradients to dissociate the nontranslating 80S ribosomes (29). Gradient analysis was performed using an ISCO UV-6 gradient collector and continuously monitored at A254.
Ribosome subunit quantification was done in low-Mg2+ gradients. Cells were grown to an OD600 of 0.8 and harvested after a 20-min treatment with 1 mM NaN3. Cycloheximide was omitted to produce a polysome run-off (29). Analysis of 7–50% linear sucrose gradients was done as described above.
In Vivo Protein Labeling.
Cultures of CDK105–1A and DJY106, harboring either YCplac33 or YCplac33-DED1, were grown in SD-Met medium at 30°C. At OD600 of 1, the cultures were shifted to 15°C for 1–3 h. At 0, 1, and 3 h, 1 ml of each culture was collected, and cells were resuspended in the same medium containing 10 μCi L-[35S]methionine (1,000 Ci/mmol; 1 Ci = 37 GBq) and incubated for 10 min. Cell extracts were prepared as described in ref. 30. Labeled proteins were analyzed by SDS/PAGE followed by autoradiography.
Other Analytical Methods.
DNA manipulations and immunoblotting were carried out as described in ref. 31. Total yeast protein extracts were prepared according to ref. 32. Protein concentration was determined by the Bradford method (33). Rat antibodies against p20 were a gift from M. Altmann. Blots were decorated with peroxidase-conjugated anti-rat immunoglobulin G and developed using the ECL detection kit (Amersham). Quantification analysis was done by using the wincam 2.1 (Cybertech, Berlin) computer application.
DNA sequence comparisons were performed at the Saccharomyces Genome Database (Stanford University) and at the National Center for Biotechnology Information.
RESULTS
The p20 Protein Plays a Negative Role in Translation Initiation.
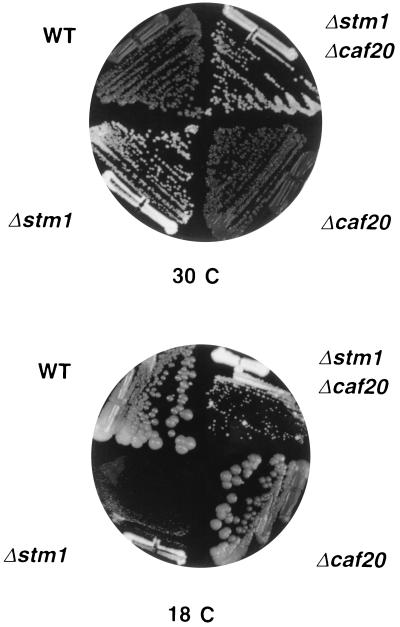
The CAF20 gene encoding the cap-associated factor p20 was isolated by reverse genetics (16). To understand the function of this protein in translation initiation, we analyzed genetic interactions between some translation initiation factors and p20. Temperature-sensitive or null mutants of the corresponding genes were crossed with each other and with the Δcaf20 null mutant, and double-segregant mutants were isolated to study synthetic interactions. The concept of synthetic lethality is based on the observation that certain double mutants are inviable under conditions where the parental single mutants are viable (34), which suggests a biochemical function in the same pathway. Indeed, different combinations of translation factor mutations were found to be lethal or to show a synthetic enhancement (Table 1). In contrast, none of the mutations were found to be synthetically lethal with the Δcaf20 allele (Table 1). Instead, the deletion of CAF20 partially suppressed the growth defect of some mutants (as shown for Δstm1 in Fig. 1). The results obtained on solid media were confirmed by measuring the growth rates in liquid media (Table 2). The most significant improvement was observed for eIF4B (Δstm1) and eIF4G (Δtif4631) mutants. This weak suppressor effect was lost when the double mutant strains were transformed with a CEN-CAF20 plasmid (data not shown).
Table 1.
Synthetic lethal interactions
| Mutation 1 | Mutation 2 | Synthetic enhancement* |
|---|---|---|
| tif1-1 | stm1::ADE2 | Lethal |
| tif1-1 | tif4631::LEU2 | Lethal |
| tif1-1 | cdc33-1 | Lethal† |
| tif1-1 | cdc33-42 | Lethal† |
| tif1-1 | prt1-1 | ND |
| stm1::ADE2 | tif4631::LEU2 | Lethal |
| stm1::ADE2 | cdc33-1 | Lethal |
| stm1::ADE2 | cdc33-42 | Slow growth |
| stm1::ADE2 | prt1-1 | None |
| tif4631::LEU2 | cdc33-1 | Lethal |
| tif4631::LEU2 | cdc33-42 | Slow growth |
| tif4631::LEU2 | prt1-1 | None |
| cdc33-1 | prt1-1 | None |
| cdc33-42 | prt1-1 | None |
| tif1-1 | caf20::TRP1 | None |
| stm1::ADE2 | caf20::TRP1 | None |
| tif4631::LEU2 | caf20::TRP1 | None |
| cdc33-1 | caf20::TRP1 | None |
| cdc33-42 | caf20::TRP1 | None |
| prt1-1 | caf20::TRP1 | None |
tif1 (eIF4A), stm1 (eIF4B), cdc33 (eIF4E), tif4631 (eIF4G), prt1 (eIF3-Prt1p), and caf20 (p20). ND, not determined.
Inability to grow in absence of a plasmid carrying a wild-type copy of one of the mutated genes or inviability of the spores (
).
Figure 1.
Suppression of the Δstm1 phenotype by Δcaf20. CDK36–1A (Δcaf20) and RCB1–1C (Δstm1) were crossed and subsequently sporulated. A representative tetratype tetrad is shown on rich medium (yeast peptone dextrose) at 30°C and 18°C. The plates were incubated for 3 and 6 days, respectively.
Table 2.
Effect of caf20::TRP1 on growth rates
| Strain (relevant genotype) | Doubling time, h
|
||
|---|---|---|---|
| 30°C | 33°C or 35°C | 18°C | |
| Wild type | 2.0 | 1.9 | 5.7 |
| caf20::TRP1 | 2.0 | 1.9 | 5.6 |
| stm1::ADE2 | 3.7 | 4.5 | 14.1 |
| stm1::ADE2 caf20::TRP1 | 3.0 | 3.6 | 8.3 |
| tif4631::LEU2 | 2.9 | 2.9 | 10.2 |
| tif4631::LEU2 caf20::TRP1 | 2.5 | 2.4 | 8.0 |
| cdc33-1 | 4.5 | >20* | ND |
| cdc33-1 caf20::TRP1 | 4.0 | >20* | ND |
| cdc33-42 | 3.0 | 8.1* | ND |
| cdc33-42 caf20::TRP1 | 2.7 | 7.2* | ND |
| tif1-1 | 3.2 | 5.0* | ND |
| tif1-1 caf20::TRP1 | 3.0 | 4.8* | ND |
| prt1-1 | 4.0 | >20* | ND |
| prt1-1 caf20::TRP1 | 3.5 | >20* | ND |
| ded1/spp81-3 | 4.5 | ND | ND |
| ded1/spp81-3 caf20::TRP1 | 3.7 | ND | ND |
Cultures were grown in yeast peptone dextrose medium at the indicated temperatures and growth measured at OD600. Data are the average of at least three different experiments. Standard deviation was less than 0.3 h. ND, not determined.
Growth at 33°C.
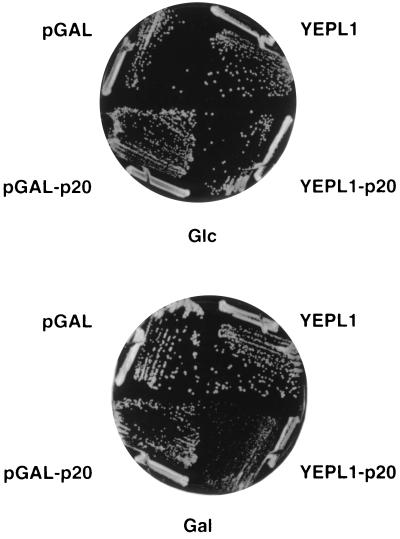
The results suggest that p20 plays a negative role in translation initiation. Therefore, overexpression of p20 is expected to inhibit the growth of the above mutants. To test this, CAF20 was cloned under the control of a CYC1-GAL1 promoter in either a mono- (pGAL-p20) or a multicopy (YEPL1-p20) plasmid. Quantitative Western blot analyses showed that, in galactose medium, p20 was overexpressed about 2-fold in the former case and around 3- to 5-fold in the latter one (data not shown). A negative effect was observed for cdc33–1 and Δtif4631 with the pGAL-p20 plasmid. Using the YEPL1-p20 plasmid, a remarkable decrease in growth rate was found particularly for the Δstm1, Δtif4631, and cdc33 mutants (Fig. 2; Table 3). As estimated by quantitative Western blot analyses, p20 was overexpressed at the same level in the different strains. Therefore, the observed variations in growth rate in these experiments are not due to strain-to-strain differences in the steady-state levels of p20. Taken together, these data strongly support a negative regulatory role of the p20 protein. By analogy with the mammalian 4E-BPs, p20 may inhibit translation via an interaction with eIF4E.
Figure 2.
Inhibition of Δstm1 growth by p20 overexpression. The RCB1–1C (Δstm1) strain was transformed with either the pGAL-p20 (CEN-URA3) or YEPL1-p20 (2μ-URA3) plasmids that contain the CAF20 gene under a CYC1-GAL1 promoter or as a control with pGAL or YEPL1. Transformants were grown for 4 days on SD-Ura or on SGal-Ura at 30°C.
Table 3.
Effect of p20 overexpression on growth rates
| Strain (relevant genotype) | Doubling time, h
|
|
|---|---|---|
| YEPL1 vector | YEPL1-p20 | |
| Wild type | 3.0 | 3.4 |
| caf20::TRP1 | 3.0 | 3.6 |
| stm1::ADE2 | 4.6 | 8.5 |
| tif4631::LEU2 | 4.2 | 7.6 |
| cdc33-1 | 7.8 | 14.0 |
| cdc33-42 | 5.0 | 8.9 |
| tif1-1 | 5.0 | 5.6 |
| prt1-1 | 4.8 | 5.3 |
| ded1/spp81-3 | 7.0 | 12.1 |
Precultures were grown in SD-Ura and diluted into SGal-Ura medium. Cultures were incubated at 30°C and growth measured at OD600. Data are the average of at least three different experiments. Standard deviation was less than 0.3 h.
Isolation of Multicopy Suppressors of a Temperature-Sensitive eIF4E Mutant.
To learn more about the cap-binding protein eIF4E, we carried out a screen for multicopy suppressors of the temperature-sensitive cdc33–42 allele. We transformed the CDK35–4A (cdc33::TRP1) strain, carrying the cdc33–42 allele on a CEN-URA3 plasmid, with a YEplac181-based multicopy library. After an 8-h incubation at a permissive temperature (30°C), the transformants were incubated for 6 days at a nonpermissive temperature (35°C). To exclude clones that received a wild-type CDC33 gene, growing colonies were streaked on selective medium and on minimal medium containing fluoro-orotic acid (5-FOA) at 30°C. Only those candidates unable to segregate the resident plasmid were selected. Plasmid DNA was isolated from the thermo-tolerant candidates, rescued in E. coli, and back-transformed into CDK35–4A. Two clones out of the initial 60,000 transformants were found to reproducibly suppress the temperature sensitivity of the CDK35–4A strain at 35°C. Restriction analysis of the two suppressor plasmids revealed two different clones, both having 2.9-kb PstI and 1.7-kb BamHI fragments.
The Suppressor Gene Encodes the Ded1 Protein.
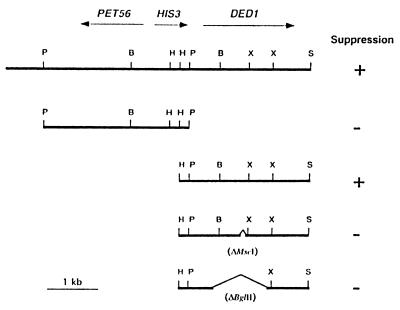
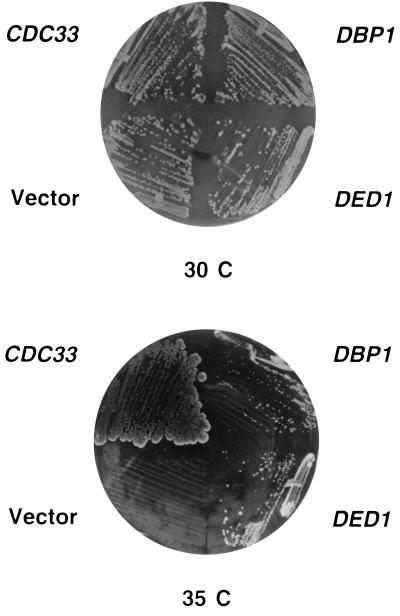
The flanking sequences of the DNA fragments that conferred suppressor activity were determined and compared with the yeast genome database. Both suppressor clones contained DNA from chromosome XV corresponding to the PET56-DED1 region (Fig. 3). Further subclonings into YEplac181 defined the minimal region required for suppression as a 2.7-kb HindIII–SalI DNA fragment containing basically the ORF of the DED1/SPP81 gene (Figs. 3 and 4). This essential gene codes for a putative RNA helicase of the DEAD-box family (35). This minimal fragment could suppress the cdc33–42 mutation both in high and low copy number, indicating that even a few copies of DED1 were able to confer suppression. To confirm that DED1 is indeed the suppressor gene, two internal deletions (ΔMscI and ΔBglII; Fig. 3) of the DED1-ORF were constructed. Both constructs no longer suppressed the cdc33–42 mutation.
Figure 3.
The PET56-DED1 region of chromosome XV. The original fragment carrying the suppressor activity and four relevant subclones are shown. B, BamHI; H, HindIII; P, PstI; S, SalI; X, XbaI. The SalI site belongs to the polylinker of YEplac181.
Figure 4.
Suppression of the cdc33–42 mutation by DED1 and DBP1. CDK35–4A was transformed with either YEplac181, YEplac181-CDC33, YEplac181-DED1, or YEplac181-DBP1. Transformants were grown on SD-Leu at 30°C or 35°C for 3 and 6 days, respectively.
The highly homologous DBP1 gene, which has been isolated as a suppressor of a ded1/spp81 mutation (36), was also found to suppress the cdc33–42 mutation, but only when present on a multicopy plasmid (Fig. 4).
Analysis of the Suppression by DED1 and DBP1.
To test the allele specificity of the suppressor effect, we transformed the CDK35–4A strain carrying either the cdc33–1, cdc33–43, or cdc33–44 allele with multicopy plasmids bearing either DED1 or DBP1. Variable levels of suppression were observed. The cdc33–1 allele was suppressed at 35°C only by DED1, whereas cdc33–43 and cdc33–44 were suppressed by either DED1 or DBP1 at 37°C. Thus, the suppression is not allele specific. This suppression is not due to an increased expression of the cdc33 alleles (data not shown).
Among the other mutations tested (tif1–1, Δstm1, Δtif4631, prt1–1), only tif1–1 was suppressed at 35°C by multicopy DED1. Moreover, no reciprocal suppression of two slow-growth cold-sensitive ded1 mutants could be observed by multicopy plasmids carrying either TIF2, CDC33, TIF4631, or STM1. These results suggest that neither Ded1p nor Dbp1p are interacting directly with eIF4E, but they interact functionally with the translation initiation machinery.
The Ded1p Protein Is Required for Translation Initiation.
To investigate whether Ded1p is involved in translation, we analyzed the two ded1 mutants for protein synthesis rate by in vivo [35S]methionine incorporation studies. As controls, we also used the same mutants carrying a wild-type DED1 gene on a CEN-plasmid. The amount of [35S]methionine incorporated into proteins in the ded1 mutants was clearly reduced compared with the isogenic wild-type strains at permissive temperature (30°C). After a 1-h shift to a nonpermissive temperature (15°C), the reduction was more pronounced, at about 50% of the incorporation of the wild-type strains (data not shown). These results suggested a protein synthesis defect in the ded1 mutants.
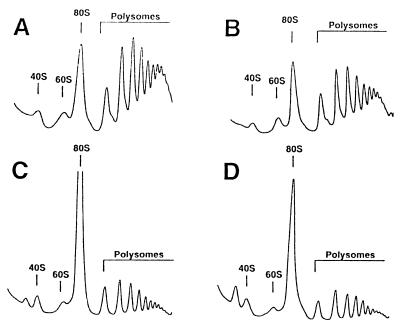
To specify the role of Ded1p in translation, we analyzed polysome profiles of both ded1 mutants and their parental wild-type counterparts. When cells were grown at 30°C, the polysome profiles from the ded1 mutants showed a marked increase in the 80S peak and a reduction in the polysome content (Fig. 5). When the cells were shifted to 15°C for 1 h, a more dramatic increase of the 80S peak and reduction of polysomes was observed. Quantification analyses showed that, although 70% of ribosomes remained as polysomes in the wild-type strain under both conditions, only 30% were in polysomes in the ded1 mutants at 30°C, and this decreased to 15% after a 1-h shift to 15°C (data not shown). For comparison, some translation initiation mutants were also analyzed at 30°C and the polysomes quantified as above (tif1–1, 45%; cdc33–42, 45%; Δstm1, 25%). Complementation of the ded1 mutants with a CEN-plasmid bearing a wild-type DED1 gene resulted in polysome profiles indistinguishable from the ones obtained for wild-type strains (data not shown).
Figure 5.
Polysome analysis of ded1 and dbp1 mutants. Cells were grown in yeast peptone dextrose at 30°C and harvested at an OD600 of 0.8. The peaks of free 40S and 60S ribosomal subunits, 80S, and polysomes are indicated. (A) DBY747, wild-type strain. (B) CDK112–1A, Δdpb1 strain. (C) DJY105, ded1/spp81–3 mutant. (D) DJY106, ded1/spp81–2 mutant.
Salt treatment dissociated most of the 80S ribosomes into 40S and 60S subunits in the ded1 mutants but not in the wild-type strain, indicating that the large 80S ribosome population was mostly nontranslating couples rather than monosomes engaged in translation (data not shown). Because DEAD-box proteins are implicated in a variety of processes involving RNA (37), we examined a role of Ded1p in rRNA processing and ribosomal assembly. To do so, we measured the relative amounts of 40S and 60S subunits on low-Mg2+ sucrose gradients. Similar values in the 60S/40S A254 ratio of around 1.7 were obtained for both ded1 mutants and the parental wild-type strain (data not shown). Thus, since no defect in mRNA synthesis or stability was previously described (35), and since no defect in ribosome biogenesis was detected in the ded1 mutants, we attribute the aberrant polysome profiles to a reduced rate of translation initiation. Altogether, these data indicated that Ded1p is involved in the initiation of protein synthesis in S. cerevisiae.
In contrast, the polysome profile of the null Δdbp1 strain resembled that of the wild-type (Fig. 5). This finding indicated that either Dbp1p is not directly implicated in translation initiation or that Ded1p alone is sufficient to support its translational activity.
Genetic Interactions of DED1 with Known Translation Mutants and with CAF20.
To further demonstrate the role of Ded1p in translation initiation, we analyzed synthetic interactions of ded1/spp81–3 with mutations in known translation factors. The combination of the ded1/spp81–3 allele with either tif1–1, cdc33–1, cdc33–42, Δstm1, or Δtif4631 resulted in inviable double mutants requiring a wild-type copy of one of the mutated genes on a URA3 plasmid for survival. These strains were unable to grow on 5-FOA medium, but regained 5-FOA resistance upon reintroduction of a wild-type copy of either of the mutant genes on another plasmid. The combination of the ded1/spp81–3 allele with the prt1–1 mutation did not result in a synthetic enhancement. Moreover, the combination of this ded1 allele with Δdbp1, which has by itself no apparent phenotype (36), enhanced the slow-growth phenotype of the ded1/spp81–3 mutant. None of the translation initiation factor mutations showed a synthetic interaction with the Δdbp1 mutation.
To substantiate a role of Ded1p in translation initiation, we also analyzed genetic interactions between DED1 and CAF20. Deletion of CAF20 partially suppressed the growth defect of the ded1/spp81–3 mutant (Table 2). This positive effect was similar to that found when CAF20 was deleted in the Δstm1 or Δtif4631 strains (Table 2). Furthermore, overexpression of p20 from YEPL1-p20 in the ded1/spp81–3 mutant resulted in a decreased growth rate similar to that found for Δstm1, Δtif4631, and cdc33 mutants (Table 3).
In conclusion, the genetic data confirmed the positive role of Ded1p and emphasized the negative role of p20 in translation initiation.
DISCUSSION
In this study, we describe the involvement of p20 and Ded1p in translation initiation and, in particular, their functional interaction with eIF4E. This factor is a key element in regulating cap-dependent translation initiation. The mammalian eIF4E is a limiting factor, and its activity is regulated by phosphorylation (1). An additional regulatory mechanism involves the eIF4E binding proteins 4E-BP1 and 4E-BP2 (14). The 4E-BP1 protein competes with eIF4G for binding to eIF4E, thus inhibiting cap-dependent translation (15). The function of 4E-BP1 is inactivated by phosphorylation (14). However, even if mammalian eIF4E substitutes for its yeast counterpart (38), the regulation of yeast eIF4E activity seems to be different. Yeast eIF4E is not a limiting factor (39), and there is no correlation between phosphorylation and translational activity (17). Furthermore, no 4E-BP analogue has been described in yeast. To learn more about yeast eIF4E and its regulating factors, we analyzed p20 as a putative counterpart of 4E-BP1 in yeast and carried out a screen for multicopy suppressors of an eIF4E mutant.
By studying the consequences of the absence or overexpression of p20 in different translation mutants or in a wild-type strain, we show a negative role of p20 on translation initiation (Tables 2 and 3). Our data corroborate the previous hypothesis that p20 is a functional analogue of 4E-BP1, as suggested by different levels of p20 phosphorylation under various growth conditions (17). Furthermore, recent biochemical data have demonstrated that p20 acts similarly to 4E-BP1 in competing with eIF4G for binding to eIF4E (40). In agreement with these data, the absence or the overexpression of p20 significantly affect the growth of mutants of the cap-binding complex, including eIF4E, eIF4G and eIF4B. The latter factor partially suppresses cdc33–1 (J.d.l.C., unpublished data), and it was isolated as a cap-associated factor (41). Interestingly, deletion of CAF20 in cdc33–1 and cdc33–42 mutants did not suppress their growth defect as strongly as expected for a negative regulator that interacts directly with eIF4E. This is in agreement with the reduced interaction observed between these alleles of eIF4E and p20 (40). Nevertheless, overexpression of p20 might favor eIF4E-p20 complex formation, explaining the drastic reduction in the growth rate of the cdc33 mutants. The fact that the effect of p20 is only significant when translation initiation is limited by mutations in certain key factors indicates that either the regulatory role of p20 on translation is minor or that other conditions are necessary to reveal its full inhibitory activity.
In our screening of eIF4E-interacting components, we isolated the DED1 gene as a multicopy suppressor of cdc33–42. This gene was previously identified as an extragenic suppressor of the prp8–1 mutation, which is affected in nuclear pre-mRNA splicing (35). So far, no other data concerning the function of Ded1p have been reported. Our results demonstrate that Ded1p is involved in translation initiation. DED1 is a multicopy suppressor of different cdc33 alleles and tif1–1. Furthermore, the ded1/spp81–3 allele is synthetically lethal with some translation initiation factor mutants. Also, the effects of deletion or overexpression of CAF20 on the growth rate of the ded1/spp81–3 mutant are similar to those found for mutations in eIF4B, eIF4G, or eIF4E. Moreover, ded1 mutants show a translation initiation defect, as detected by polysome analysis (Fig. 5). The profiles obtained are qualitatively and quantitatively similar to those described for other mutations in key translation initiation factors, like eIF4A (D.K., unpublished data), eIF4B (19), or eIF4E (42).
We also show that a DED1 homologue, DBP1 (72% identity), which suppresses in high dosage the growth defects of ded1 mutants (36), also suppresses some cdc33 mutations. However, no polysome defect was found for the null allele of DBP1 (Fig. 5). Similar results are described for other translation initiation factors (eIF4G, eIF5A) whose genes are duplicated, and their products do not contribute equally to growth (25, 43).
Both Ded1p and Dbp1p are putative ATP-dependent RNA helicases of the DEAD-box family (37). In the mammalian system, unwinding of secondary structures in the 5′ untranslated region of mRNAs is attributed to the helicase activity of eIF4A (4, 5). Moreover, overexpression of eIF4E facilitates translation of highly structured mRNAs, presumably by recruiting RNA helicase activity to the mRNA (44). These data indicate the necessity of RNA helicases in translation initiation. Although in yeast, secondary structures do not influence eIF4E binding to the cap structure (39), and partial inactivation of eIF4E does not restrict the translation of mRNAs having different secondary structures (12), our results favor the association of the putative helicase activity of Ded1p with the function of eIF4E. Further experiments are necessary to localize the Ded1 protein in the translation machinery and to test its proposed role in removing cap-proximal secondary structures in mRNAs.
Acknowledgments
We thank M. Rekik for excellent technical assistance and M. Altmann, S. Helliwell, M. Hall, and K. Tanner for critical reading of the manuscript. We are grateful to M. Altmann, H. Trachsel, and T. H. Chang for discussions and communication of unpublished data, and M. Altmann, J. Beggs, and C. Berset for strains and plasmids. J.d.l.C. and I.I. are postdoctoral fellows from the Ministerio de Educación y Ciencia and the European Molecular Biology Organization, respectively. This work was supported by a grant from the Swiss National Science Foundation to P.L. (31-43321.95).
References
- 1.Merrick W C, Hershey J W B. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 31–69. [Google Scholar]
- 2.Iizuka N, Chen C, Yang Q, Johannes G, Sarnow P. Curr Top Microbiol Immunol. 1995;203:155–177. doi: 10.1007/978-3-642-79663-0_8. [DOI] [PubMed] [Google Scholar]
- 3.Mader S, Lee H, Pause A, Sonenberg N. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rozen F, Edery I, Meerovitch K, Dever T E, Merrick W C, Sonenberg N. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaramillo M, Browning K, Dever T E, Blum S, Trachsel H, Merrick W C, Ravel J M, Sonenberg N. Biochim Biophys Acta. 1990;1050:134–139. doi: 10.1016/0167-4781(90)90154-t. [DOI] [PubMed] [Google Scholar]
- 6.Goyer C, Altmann M, Trachsel H, Sonenberg N. J Biol Chem. 1989;264:7603–7610. [PubMed] [Google Scholar]
- 7.Lanker S, Müller P P, Altmann M, Goyer C, Sonenberg N, Trachsel H. J Biol Chem. 1992;267:21167–21171. [PubMed] [Google Scholar]
- 8.Altmann M, Handschin C, Trachsel H. Mol Cell Biol. 1987;7:998–1003. doi: 10.1128/mcb.7.3.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenner C, Nakayama N, Goebl M, Tanaka K, Tohe A, Matsumoto K. Mol Cell Biol. 1988;8:3556–3559. doi: 10.1128/mcb.8.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altmann M, Sonenberg N, Trachsel H. Mol Cell Biol. 1989;9:4467–4472. doi: 10.1128/mcb.9.10.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altmann M, Trachsel H. Nucleic Acids Res. 1989;17:5923–5931. doi: 10.1093/nar/17.15.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasilescu S, Ptushkina M, Linz B, Müller P P, McCarthy J E. J Biol Chem. 1996;271:7030–7037. doi: 10.1074/jbc.271.12.7030. [DOI] [PubMed] [Google Scholar]
- 13.Lin T A, Kong X, Haystead T A, Pause A, Belsham G, Sonenberg N, Lawrence J., Jr Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 14.Pause A, Belsham G J, Gingras A C, Donzé O, Lin T A, Lawrence J, Jr, Sonenberg N. Nature (London) 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 15.Haghighat A, Mader S, Pause A, Sonenberg N. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altmann M, Krieger M, Trachsel H. Nucleic Acids Res. 1989;17:7520. doi: 10.1093/nar/17.18.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zanchin N I, McCarthy J E. J Biol Chem. 1995;270:26505–26510. doi: 10.1074/jbc.270.44.26505. [DOI] [PubMed] [Google Scholar]
- 18.Banroques J, Delahodde A, Jacq C. Cell. 1986;46:837–844. doi: 10.1016/0092-8674(86)90065-6. [DOI] [PubMed] [Google Scholar]
- 19.Coppolecchia R, Buser P, Stotz A, Linder P. EMBO J. 1993;12:4005–4011. doi: 10.1002/j.1460-2075.1993.tb06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boeke J D, Trueheart J, Natsoulis G, Fink G R. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 22.Gietz R D, Schiestl R H, Willems A, Woods R A. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 23.Gietz R D, Sugino A. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 24.Schmid S R, Linder P. Mol Cell Biol. 1991;11:3463–3471. doi: 10.1128/mcb.11.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goyer C, Altmann M, Lee H S, Blanc A, Deshmukh M, Woolford J L J, Trachsel H, Sonenberg N. Mol Cell Biol. 1993;13:4860–4874. doi: 10.1128/mcb.13.8.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 27.Bonneaud N, Ozier-Kalogeropoulos O, Li G, Labouesse M, Minvielle-Sebastia L, Lacroute F. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 28.Blum S, Mueller M, Schmid S R, Linder P, Trachsel H. Proc Natl Acad Sci USA. 1989;86:6043–6046. doi: 10.1073/pnas.86.16.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foiani M, Cigan A M, Paddon C J, Harashima S, Hinnebusch A G. Mol Cell Biol. 1991;11:3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yaffe M P, Schatz G. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 32.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl A. Current Protocols in Molecular Biology. Vol. 2. New York: Wiley; 1994. pp. 13.0.1–13.14.17. [Google Scholar]
- 33.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 34.Guarente L. Trends Genet. 1993;9:362–366. doi: 10.1016/0168-9525(93)90042-g. [DOI] [PubMed] [Google Scholar]
- 35.Jamieson D J, Rahe B, Pringle J, Beggs J D. Nature (London) 1991;349:715–717. doi: 10.1038/349715a0. [DOI] [PubMed] [Google Scholar]
- 36.Jamieson D J, Beggs J D. Mol Microbiol. 1991;5:805–812. doi: 10.1111/j.1365-2958.1991.tb00753.x. [DOI] [PubMed] [Google Scholar]
- 37.Schmid S R, Linder P. Mol Microbiol. 1992;6:283–292. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 38.Altmann M, Müller P P, Pelletier J, Sonenberg N, Trachsel H. J Biol Chem. 1989;264:12145–12147. [PubMed] [Google Scholar]
- 39.Lang V, Zanchin N I, Lunsdorf H, Tuite M, McCarthy J E. J Biol Chem. 1994;269:6117–6123. [PubMed] [Google Scholar]
- 40.Altmann M, Schmitz N, Berset C, Trachsel H. EMBO J. 1997;16:1114–1121. doi: 10.1093/emboj/16.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altmann M, Müller P P, Wittmer B, Ruchti F, Lanker S, Trachsel H. EMBO J. 1993;12:3997–4003. doi: 10.1002/j.1460-2075.1993.tb06077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnes C A, MacKenzie M M, Johnston G C, Singer R A. Mol Gen Genet. 1995;246:619–627. doi: 10.1007/BF00298969. [DOI] [PubMed] [Google Scholar]
- 43.Schnier J, Schwelberger H G, Smit-McBride Z, Ah Kang H, Hershey J W B. Mol Cell Biol. 1991;11:3105–3114. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koromilas A E, Lazaris-Karatzas A, Sonenberg N. EMBO J. 1992;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]