SUMMARY
An essential step in pre-mRNA splicing is the release of the mRNA product from the spliceosome. The DEAH-box RNA helicase Prp22 catalyzes mRNA release by remodeling contacts within the spliceosome that involve the U5 snRNP. Spliceosome disassembly requires a segment of >13 ribonucleotides downstream of the 3′ splice site. I show here by site-specific cross-linking and RNase H protection that Prp22 interacts with the mRNA downstream of the exon-exon junction prior to mRNA release. The findings support a model for Prp22-catalyzed mRNA release from the spliceosome wherein a rearrangement that accompanies the second transesterification step deposits Prp22 on the mRNA downstream of the exon-exon junction. Bound to its target RNA, the 3′→5′ helicase acts to disrupt mRNA/U5 snRNP contacts, thereby liberating the mRNA from the spliceosome.
INTRODUCTION
Faithful expression of eukaryal genes relies on an elaborate mRNA splicing “machine” that excises introns from primary transcripts. Splicing entails two successive transesterification steps at the intron-exon junctions and intronic branch site. Selection of reactive phosphodiesters in the pre-mRNA is accomplished by a multitude of transacting factors that include snRNPs (U1, U2, U5 and U4/6 snRNAs and associated proteins) and non-snRNP splicing factors (Will and Lührmann, 2006). A network of RNA-RNA and RNA-protein interactions assists in positioning the 2′ OH at the intron branch point and the 3′-5′ phosphodiester at the 5′ exon-intron boundary for the first step, and the 3′ OH of the 5′ exon and the intron-3′ exon junction for the second transesterification step (Valadkhan, 2005). Conformational rearrangements in the splicing complex and changes in its composition accompany spliceosome assembly and the transesterification steps that yield mRNA and excised lariat-intron. The products of the splicing reaction are then released in a series of energy-requiring disassembly steps.
ATPases of the DExD/H box family are key players in remodeling events throughout the splicing cycle (Staley and Guthrie, 1998), and they have been implicated in splicing fidelity (Burgess and Guthrie, 1993; Mayas et al., 2006; Pandit et al., 2006; Xu and Query, 2007). DExD/H proteins are defined by a set of conserved peptide motifs that are important for NTP phosphohydrolase activity (Bleichert and Baserga, 2007). The repertoire of biochemical activities ascribed to DExD/H NTPases includes unwinding of nucleic acid duplexes, displacement of proteins from RNA, and annealing of RNA strands. It is generally thought that the DExD/H enzymes use the energy of NTP hydrolysis to modulate the structure or the composition of RNA-protein assemblies (Jankowsky and Bowers, 2006). However, the molecular mechanism by which they accomplish this and the consequences of their actions are poorly understood.
The yeast DEAH-box splicing factor Prp22 is essential for dissociating mRNA from the spliceosome (Company et al., 1990). Prp22 is an RNA-stimulated NTPase and an NTP-dependent RNA helicase, which unwinds short nucleic acid duplexes that have a 3′ single-stranded RNA tail (Tanaka and Schwer, 2005). The NTPase and helicase activities are required for Prp22’s biological function, insofar as (i) mutations that abolish Prp22’s NTPase activity preclude spliceosome disassembly in vitro and are lethal in vivo, and (ii) mutations that ablate Prp22’s helicase activity without affecting NTP hydrolysis are lethal or cause severe growth phenotypes that correlate with defects in spliceosome disassembly (Schwer and Meszaros, 2000; Campodonico and Schwer, 2002; Schneider et al., 2002, 2004). Thus, the chemical energy of ATP hydrolysis by Prp22 must be coupled to mechanical work during spliceosome disassembly. Prp22 binds to single-stranded RNA with high affinity, but does not bind DNA. The length of the RNA chain influences binding and RNA-stimulated NTP hydrolysis similarly, and optimal activities require RNA oligomers of ≥20 nt (Tanaka and Schwer, 2005). Whereas unwinding of a nucleic acid duplex by Prp22 depends on a 3′ single-stranded RNA tail to provide a binding site for the 3′→5′ helicase, the enzyme is blind with respect to the displaced strand, which can be either RNA or DNA. Thus, the mechanical work performed by Prp22 could plausibly involve unwinding of RNA helices or disruption of RNA-protein contacts, or both.
Prior genetic analyses, especially allele-specific suppression of cold-sensitive growth phenotypes arising in helicase-deficient Prp22 mutants, have implicated the U5 snRNP component Prp8 and also the conserved loop 1 in the U5 snRNA in contacts that are broken by Prp22 helicase (Schneider et al., 2004; Aronova et al. 2007). Loop 1 of U5 snRNA pairs with exon bases adjacent to the splice sites to juxtapose the exons for catalysis, while Prp8 assists in stabilizing the U5-exon contacts (Newman, 1997; Crotti et al., 2007; Teigelkamp et al., 1995a). It has been proposed that the interactions between the U5 snRNP and exon bases persist after exon-joining and that Prp22 helicase breaks these contacts to release mRNA from the spliceosome (Aronova et al., 2007). An outstanding question concerns the single-stranded RNA within the spliceosome that serves as a loading dock for the 3′→5′ helicase. A candidate is the mRNA downstream of the exon-exon junction, considering that the snRNAs are likely to be more structured and that Prp22 does not exhibit overt sequence specificity. However, it is not known whether the 3′ exon is important for spliceosome disassembly.
A 3′ exon is not required for spliceosome assembly or the first transesterification step. Truncating pre-mRNAs between the branchpoint and 3′ splice site leads to the accumulation of lariat-intermediates and 5′ exons in both yeast and mammalian in vitro splicing systems (Rymond and Rosbash, 1985; Anderson and Moore, 1997). The 3′ splice site PyAG↓ needs to be identified and placed in the catalytic center of the spliceosome for the second transesterification reaction. The DExD/H NTPase Prp16 plays an important role in this transition (Schwer and Guthrie, 1991). NTP hydrolysis by Prp16 elicits a conformational change in the spliceosome that results in protection of the 3′ splice site from oligo-directed RNase H cleavage (Schwer and Guthrie, 1992). This protection, which likely reflects binding of the 3′ splice site PyAG↓ in the active site of the spliceosome, requires the splicing factors Slu7, Prp18 and Prp22 (Schwer and Gross, 1998; B.S., unpublished). The function of Prp22 during the second step is independent of Prp22’s ability to hydrolyze NTP, and it is not essential for splicing of all pre-mRNA in vitro (Schwer and Gross, 1998)
The present study addresses outstanding questions regarding the requirement and the disposition of the 3′ exon during pre-mRNA splicing. Experiments to probe the RNP structure of the 3′ exon before and after exon-joining revealed a change in the accessibility of the mRNA downstream of the splice junction. Specifically, a rearrangement in the spliceosome that accompanies the second transesterification step affords protection of an ~ 23 nt segment downstream of the exon-exon junction when spliceosome disassembly is blocked by an NTPase-inactivating Prp22 mutation. Site-specific crosslinking experiments showed that Prp22 binds to the 3′ exon prior to mRNA release. Together with the demonstration that a segment of >13 ribonucleotides downstream of the 3′ splice site is required for spliceosome disassembly, the findings support a model for Prp22-catalyzed mRNA release from the spliceosome wherein the 3′→5′ helicase binds to the mRNA downstream of the splice junction and then acts to disrupt mRNA/U5 snRNP contacts.
RESULTS
A 3′ exon is required for mRNA release from the spliceosome
Spliceosome assembly and the 1st transesterification reaction can occur in the absence of a 3′ exon (Rymond and Rosbash, 1985). Fig. 1A depicts the experimental strategy used here to determine whether the 3′ exon is required for the 2nd transesterification step and for mRNA release. Splicing of 32P-labeled ACT1 precursor RNA (Fig. 1B) was carried out in yeast extract depleted of the essential second step factor Prp16. Under these conditions, splicing was arrested prior to step 2 so that lariat-intermediates and 5′ exons accumulated (Fig. 1C, lane 1). Aliquots of the reaction mixture were then supplemented with DNA oligonucleotides complementary to the 3′ exon RNA segments from +3 to +17 (oligo 2-1), or +21 to +34 (oligo 2–4) (Fig. 1B). This resulted in cleavage of the RNA/DNA hybrid in pre-mRNA and lariat-intermediate by RNase H present in the extract (Fig. 1C, lanes 2 and 3). Then, purified Prp16 protein was added to aliquots of the mixtures and the products were analyzed by denaturing PAGE. The appearance of newly spliced RNAs that migrated more slowly than the 5′ exon in Prp16-supplemented reactions (Fig. 1C, lanes 4 and 5) showed that truncated lariat-intermediates were substrates for the 2nd transesterification step. The length heterogeneity of spliced products is likely due to RNase H cleavage at various positions within the hybrid regions.
Fig. 1.
The influence of 3′ exon length on mRNA release. (A) Experimental outline. (B) Schematic depiction of the ACT1 pre-mRNA. The 5′ exon and intron are depicted as a grey can and a black line, respectively and the sequences of the 3′ exon and the DNA oligos 2-1 and 2–4 are shown. (C) Analysis of the reaction products formed after 20 min of splicing in Δprp16 extract (lane 1), after incubation with oligos (lanes 2 and 3), and after incubation with Prp16 protein (lanes 4 and 5) by denaturing PAGE. Lane M: 5′-32P-labeled Msp1 fragments of pBR322 DNA, the lengths for several fragments are indicated at right. The symbols at the left indicate the positions of the following nucleic acid species (from top to bottom): lariat-intermediate, lariat-intermediate after RNase H digestion, excised lariat-intron, pre-mRNA, cleaved pre-mRNA, mRNAs and 5′ exon. (D) and (E) Glycerol gradient sedimentation of the reaction mixtures after addition of Prp16. The odd numbered fractions (# 3–29) were analyzed by denaturing PAGE. An aliquot of each reaction mixture before sedimentation was analyzed in the left-most lane (In). The arrows at left mark the positions of spliced products. (D) Analysis of the reaction mixture, in which cleavage of the 3′ exon was directed by DNA oligo 2-1 prior to the addition of Prp16; (E) Analysis of the reaction mixture, in which cleavage of the 3′ exon was targeted by DNA oligo 2–4. Autoradiograms of the dried gels are shown.
To test whether spliced mRNAs with truncated 3′ exons were released from the spliceosome, the reaction products were analyzed by glycerol gradient sedimentation (Fig. 1D, E). Spliced RNA cleaved close to the 3′ splice site migrated near the bottom of the gradient in fractions 17–25, indicating that it was retained in the spliceosome (Fig. 1D). In contrast, spliced RNA cleaved further away from the 3′ splice site was released and sedimented at the top of the gradient (Fig. 1E). Thus, shortening the ACT1 3′ exon does not prevent the 2nd transesterification reaction; however, an RNA segment downstream of the splice junction is necessary for the release of spliced product from the spliceosome.
More than 13 ribonucleotides downstream of the exon/exon junction are required for mRNA release
A set of chimeric pre-mRNAs was generated to investigate the 3′ exon requirements with respect to composition and length. Unlabeled ACT1 RNAs that encompassed the 5′ exon, the intron and truncated 3′ exons (6, 13, 18 and 31 nt in length) were synthesized by T7 RNA polymerase and then ligated to a 32P-labeled 30-nt DNA oligonucleotide (Fig. 2A). Splicing was carried out in wild-type extract and the reaction products were analyzed by glycerol gradient sedimentation. Spliced products in which the 3′ exons were composed of 6 or 13 nt RNA plus 30 nt DNA remained associated with spliceosomes and sedimented close to the bottoms of the gradients (Fig. 2B). Increasing the length of the RNA segment in the chimeric 3′ exon to 18 and 31 nt allowed 72% and 95% of the spliced product to be released (Fig. 2C). Thus, an RNA segment of >13 nt downstream of the splice junction is needed for release of the mature mRNA from the spliceosome.
Fig. 2.
Splicing and release of RNA/DNA chimeras. (A) The splicing substrates. The 5′ exons (in grey) and the introns (indicated by a line) are identical for each of the precursor RNAs, and the sequences of the chimeric 3′ exons are shown. The star (*) marks the position of the single 32P within the 3′ exons. (B) and (C) Glycerol gradient sedimentation and analysis of odd-numbered fractions (# 3–29) by denaturing PAGE. An aliquot of the reaction mixtures prior to sedimentation (In) was analyzed at the left of each gel. The symbols at the left indicate the positions of the following nucleic acid species (from top to bottom): lariat-intermediate, precursor and spliced product. Autoradiograms of the dried gels are shown.
A change in the 3′ exon RNP structure accompanies the second transesterification reaction
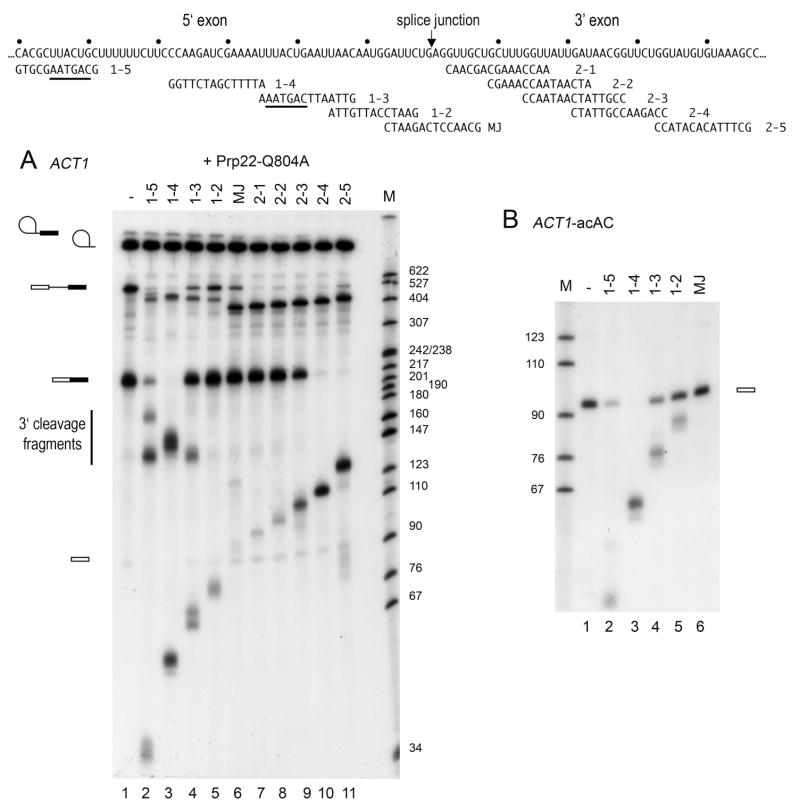
A set of overlapping DNA oligonucleotides complementary to segments in the ACT1 3′ exon was employed to test whether the 3′ exon is accessible to targeted RNase H digestion after the 2nd transesterification step and prior to mRNA release, i.e. at the stage when the RNA segment downstream of the 3′ splice site is functionally important. To prevent mRNA release, splicing mixtures were supplemented with dominant-negative Prp22 mutant proteins. Prior work had shown that ATPase/helicase-defective mutants compete with wild-type Prp22 for spliceosome binding but then fail to catalyze mRNA release (Schwer and Meszaros, 2000; Schneider et al. 2002, 2004). Splicing of 32P-body-labeled ACT1 precursor was carried out in wild-type extract in the absence or presence of the ATPase-defective Prp22-Q804A protein (Fig. 3B). Increased levels of excised lariat-intron RNA signified that spliceosome disassembly was indeed inhibited by Prp22-Q804A (compare lanes 1 and 10). DNA oligos complementary to regions in the 3′ exon or the intron (Fig. 3A) were then added to aliquots of the mixtures to allow cleavage by endogenous RNase H. The products were analyzed by denaturing PAGE (Fig. 3B).
Fig. 3.
Accessibility of the 3′ exon to oligo-directed RNase H cleavage. (A) A segment of the ACT1 pre-mRNA is shown; the branchpoint and the 3′ splice site (3′ ss) are marked. DNA oligos complementary to the intron and 3′ exon are shown below the pre-mRNA. (B) ACT1 pre-mRNA was spliced in wild-type extracts in the absence (-) and presence (+) of Prp22-Q804A protein during 20 min at 28°C. Aliquots of the mixtures were then supplemented with the indicated DNA oligo and incubated for 10 min at 22°C. The reaction products were analyzed by denaturing PAGE and the autoradiograms of the dried gels are shown. Lane M: 5′-32P-labeled Msp1 fragments of pBR322 DNA. (C) In the ACT1-acAC pre-mRNA, the 3′ splice site ag↓AG is changed to ac↓AC. Otherwise, ACT1-acAC is identical to ACT1 pre-mRNA used in (B). ACT1-acAC was spliced in wild-type extracts supplemented with buffer (-), or with dominant-negative Prp22-Q804A protein (+). To aliquots of the reaction mixtures, DNA oligos were added as indicated, and the RNAs were analyzed by denaturing PAGE. The 3′ splice site mutations are marked by * in the symbols for lariat-intermediate and pre-mRNA at the left. A triangle marks a product that migrates at the position of mRNA and is formed in small amounts that vary between experiments. Based on prior studies (Vijayraghavan et al., 1986) showing that the ac↓AC pre-mRNA is spliced accurately in vivo and in vitro, albeit with low efficiency, I surmise that the RNA species marked by the triangle is mRNA.
In the absence or presence of added Prp22-Q804A protein, DNA oligos complementary to intron sequences (i-1 and i-3), to the intron-3′ exon junction (LI-ag), and to 3′ exon sequences (2-1 to 2–5) directed cleavage of the pre-mRNA to yield incrementally longer 5′ fragments. (The 3′ fragments are degraded by 5′→3′ exonucleases in the extract.) As expected, the spliced mRNA product was immune to RNase H in the presence of the intron-targeting oligos i-1 and i-3 in the absence or presence of Prp22-Q804A (lanes 2, 3, 11 and 12). The bulk of spliced mRNA (≥80%) that was formed in wild-type extract and subsequently released from the spliceosome (-Prp22-Q804A) was cleaved when RNase H was targeted to regions in the 3′ exon (lanes 4–9). In contrast, when mRNA release was blocked (+ Prp22-Q804A), the mRNA remained intact upon addition of oligos LI-ag, 2-1 and 2–2 (lanes 13–15). DNA oligos 2–3 and 2–4, 2–5 directed partial and complete cleavage of the mRNA, respectively (lanes 16–18). Thus, a segment of ~23 nt downstream of the splice junction was protected from oligo-directed RNase H cleavage when spliceosome disassembly was blocked by a dominant-negative Prp22 mutant. [Note that the margin of protection (~23 nt) is only an approximation that is based on the finding that mRNA remained intact in the presence of oligo 2-2, complementary to positions +9 to +23 downstream of the splice junction.] Protection of the mRNA segment downstream of the splice junction was also observed when Δprp22 extract was supplemented with ATPase-deficient Prp22-K512A or helicase-deficient (but ATPase-proficient) Prp22-H606A mutant proteins (not shown).
Protection of the 3′ exon coincides with the 2nd transesterification reaction
The 3′ exon is not required to form lariat-intermediates, and consequently, the 3′ exon was accessible to targeted cleavage in splicing complexes arrested at the Prp16-dependent step (Fig. 1). To halt splicing after the Prp16-catalyzed remodeling event but prior to the 2nd transesterification reaction, I employed a mutant ACT1 precursor in which the 3′ splice site uag↓AG is mutated to uac↓AC (↓ indicates the intron-exon junction) (Vijayraghavan et al. 1986). Splicing of 32P-labeled ACT1-acAC pre-mRNA was carried out in wild-type extract and the reaction products were then subjected to RNase H protection analyses (Fig. 3C). Precursor RNA was susceptible to RNase H cleavage in the presence of DNA oligos complementary to intron or 3′ exon sequences. The lariat-3′ exon intermediate RNA was also cleaved when regions within the 3′ exon were targeted by complementary DNA oligos (lanes 4–9). However, RNase H was much less effective in cleaving the RNA within the intron (lanes 2 and 3). The protection patterns were similar whether splicing was carried out in the absence or presence of added Prp22-Q804A (compare lanes 2–9 with lanes 11–18). These findings suggest that a conformational change that leads to protection of the ACT1 mRNA downstream of the 3′ splice accompanies the 2nd transesterification reaction and likely reflects binding of splicing factor(s) required for mRNA release.
The 5′ exon RNase H protection pattern is unchanged during step 2
To gauge the extent to which the splicesome covered the 5′ exon of ACT1 mRNA prior to its release, RNase H protection studies were carried out with DNA oligos complementary to the 5′ exon. 32P-labeled ACT1 pre-mRNA was spliced for 25 min at 28°C in wild-type extract in the presence of the dominant-negative Prp22-Q804A protein to prevent mRNA release. Aliquots of the reaction mixtures were then supplemented with DNA oligos complementary to segments in the mRNA upstream and downstream of the splice junction, incubated for 10 min at 22°C and analyzed by denaturing PAGE (Fig. 4A). Approximately 23 nt downstream (Fig. 4A, lanes 7–11 and Fig. 3A) and a segment of ~15 nt upstream of the splice junction were immune to RNase H (Fig. 4A, lanes 5 and 6). The estimate for the 5′ margin is based on the findings that as much of the mRNA remained intact in the presence of oligo 1–2 complementary to positions −3 to −15 upstream of the splice junction as did in the presence of oligos MJ, 2-1 and 2-2. Addition of oligos 1–3, 1–4 and 1–5 resulted in partial or complete cleavage within the 5′ exon (lanes 2–4). Besides short capped 5′ fragments, the longer 3′ fragments were readily detected, indicating that they were protected by the spliceosome from 5′ to 3′ exonucleolytic activities present in the extract. I surmise that oligo 1–5, which has a 6-nt stretch (CAGTAA) that is identical to oligo 1–3, directed RNase H not only to the region −49 to −60 upstream of the splice junction, but also to −19 to −24 as suggested by the appearance of a shortened 3′ fragment similar in length to that generated in the presence of oligo 1–3 (compare lanes 2 and 4). The bulk of residual unspliced pre-mRNA was accessible to RNase H directed to regions within the 5′ or 3′ exons (lanes 2–11). Similarly, in reactions where mRNA release was not inhibited by a dominant-negative Prp22 protein, the majority of spliced mRNA (≥ 80%) was susceptible to RNase H in the presence of each DNA oligo tested (data not shown and Fig. 3A).
Fig. 4.
Footprint of the spliceosome on mRNA prior to release. The sequence of ACT1 mRNA spanning 60 nt upstream and 47 nt downstream of the splice junction is shown at the top. DNA oligos complementary to positions in the 5′ exon, the 3′ exon and at the exon-exon junction (MJ) are shown beneath the mRNA sequence. (A) ACT1 pre-mRNA was spliced in wild-type extract supplemented with Prp22-Q804A protein to block mRNA release. The indicated DNA oligos were added to aliquots of the reaction mixture. After 10 min of incubation to allow for RNase H cleavage, the products were analyzed by denaturing PAGE and autoradiography. The symbols at the left mark the positions of the splicing intermediates and products and the bar indicates 3′ cleavage fragments. (B) Accessibility of the 5′ exon when the second transesterification reaction was prevented by the 3′ splice site ac↓AC mutation. The symbol at the right marks the position of the 5′ exon.
To probe the RNP structure of the 5′ exon before exon-joining, spliceosomes containing lariat-intermediates were formed either in wild-type extracts using the mutant ACT1-acAC precursor, or in Δprp16 extract using ACT1 pre-mRNA during a 25 min incubation at 28°C. DNA oligos were then added to aliquots of the reaction mixtures, and the products were analyzed by denaturing PAGE (Fig. 4B and data not shown). The RNase H protection pattern of the 5′ exon prior to the 2nd transesterification reaction was comparable to that seen after step 2 (Fig. 4A), suggesting that the 5′ margin of the spliceosome remains in place.
Probing the 3′ exon RNP structure of RPS17A mRNA and spliced U3 RNA
To query the generality of the observed change in the RNP structure of the 3′ exon that accompanies the 2nd transesterification reaction in ACT1 splicing, RNase H protection assays were carried out using RPS17A and U3 RNAs (Fig. 5B). 32P-labeled transcripts were spliced in wild-type extract in the absence or presence of dominant-negative Prp22-Q804A. To aliquots of the reaction mixtures, DNA oligos complementary to regions in the 3′ exon, either close to the 3′ splice site (oligo a), or further downstream of the 3′ splice site (oligo b), were added to direct cleavage by RNase H (Fig. 5A). The majority of residual unspliced precursor RNA was cleaved in the presence of each DNA oligo, attesting to their effectiveness in targeting RNase H to the 3′ exons (Fig. 5B). Endogenous RNase H also cleaved the spliced products in the presence of either DNA oligo, when spliceosome disassembly was not inhibited (lanes 2 and 3). However, when product release was blocked (signified by increased levels of lariat-intron RNAs), the RPS17A mRNA and the spliced U3 RNA were immune to RNase H in the presence of oligo a, but not oligo b (compare lanes 5 and 6). The finding that the observed change in the RNP structure of the 3′ exon can be observed for several spliced RNAs is consistent with the idea that it is a requisite step preceding mRNA release.
Fig. 5.
A change in accessibility of the 3′ exon is not unique to ACT1 mRNA and it is a natural intermediate in the pathway. (A) Schematic drawings of pre-mRNA and mRNA, indicating the approximate positions of DNA oligos that were used. (B) 32P-lableled transcripts, RPS17A pre-mRNA and U3 precursor were spliced in wild-type extract in the presence and absence of added Prp22-Q804A protein. Then, aliquots of the mixtures were supplemented with the respective DNA oligos a or b to allow for cleavage by endogenous RNase H. Oligos: RPS17A (a) 5′-TGGTTCTAACTCTACC, (b) 5′-CTTAGAAGCACGCTTGA; U3 snoRNA (a) 5′-CCTATAGAAATGATCC, (b) 5′-GTGACGATTCCTAT. The reaction products were analyzed by denaturing PAGE. (C) Outline of the experimental strategy. The ACT1 pre-mRNA used in this experiment consisted of a 5′ exon that was 98 nt in length, the normal 302-nt intron and a 3′ exon of 195 nt. ACT1 pre-mRNA was spliced in Δprp22 extract to form lariat-intermediates and 5′ exon. The mixture was then depleted of ATP by endogenous hexokinase upon addition of glucose. Aliquots of the reaction mixture were supplemented with wild-type Prp22 protein and ATP as indicated by (+) or buffer (-). DNA oligos were then added to aliquots of each mixture and the RNA products were analyzed upon further incubation. An autoradiogram of the dried gel is shown. 32P-labeled Msp1 fragments of pBR322 DNA were separated in the left-most lane (M). The symbols at the left indicate the positions of the RNA species generated by splicing.
The conformational change leading to 3′ exon protection reflects an intermediate step in the pathway
The scheme outlined in Fig. 5C was used to inhibit spliceosome disassembly by means other than a dominant-negative Prp22 protein. 32P-labeled ACT1 pre-mRNA was spliced in extracts depleted of Prp22 and the products of the 1st transesterification reaction accumulated (lane 1). Then, the reaction mixture was depleted of ATP by hexokinase present in the extract upon addition of glucose (Horowitz and Abelson, 1993). Aliquots were supplemented with wild-type Prp22 protein (and Mg2+) in the absence or presence of ATP (lanes 4–9). ATP is not required for Prp22′s role during the second step and, consequently, mRNA was formed in the absence and presence of ATP (lanes 4 and 7). However, ATP is required for spliceosome disassembly, and the level of excised lariat-intron RNA was increased in reactions lacking ATP compared to those supplemented with ATP (compare lanes 4 and 7). DNA oligos 2-1 and 2–4 were added to aliquots of each mixture. When RNase H was directed by oligo 2-1 to positions +3 to +17 downstream of the splice junction in mRNA that was formed in the absence of ATP, half of the mRNA remained intact (lane 5). This finding shows that the conformational change coincident with the 2nd transesterification reaction represents a natural intermediate step, insofar as spliceosomes stalled in the presence of wild-type Prp22 can proceed to dissociate when ATP is added. Whereas this conformational step does not require ATP, the extent of protection is reproducibly lower in the absence of ATP, suggesting that ATP might enhance the rearrangement.
Prp22 is situated downstream of the splice junction on spliceosome-associated mRNA
Site-specific cross-linking was used to identify splicing factors that interact with the 3′ exon prior to mRNA release and thereby afford the observed protection of the 3′ exon from targeted RNase H cleavage. A set of pre-mRNAs containing a single 32P four nucleotides upstream of a 4-thiouridine (s4U) was generated via ligation of a 5′-labeled oligonucleotide to T7 transcripts encompassing the ACT1 5′ exon, intron and truncated 3′ exons (Fig. 6A). The 30-mer oligo was composed of 25 ribonucleotides, followed by 5 deoxynucleotides. DNA at the 3′ end does not interfere with splicing or product release (Fig. 2C), but prevents polyadenylation in the extract, resulting in products of discrete lengths (see Supplemental Fig. 1). Pre-mRNAs containing s4U at positions +10, +17, +22 and +35 downstream of the 3′ splice site were spliced in wild-type extract supplemented with the ATPase-defective Prp22-Q804A protein to block mRNA release. Aliquots of the reaction mixtures were withdrawn to gauge mRNA formation (Fig. 6B). The remaining mixtures were placed on ice and exposed to UV light (312 nm). Aliquots were removed after irradiation for 0, 5 or 10 min and each of the samples was then treated with RNase T1 and analyzed by SDS-PAGE (Fig. 6C). Four 32P-labeled polypeptides were detected, migrating at ~40 kDa, ~65 kDa, ~155 kDa and ≥250 kDa, respectively. T1 digestion of the RNA probe should leave an 8-nt 32P-labeled fragment attached to these proteins. Based on size, the ~155 species might be Prp22 (130 kDa), and the largest one might correspond to the U5-snRNP protein Prp8 (280 kDa), which has been shown to interact with the RNA downstream of the 3′ splice site (Teigelkamp et al. 1995a). Cross-linking appears to be position-dependent insofar as the signal for the ~155 kDa polypeptide, normalized to the signal at ~65 kDa present in all reactions, was higher at +17 than at +10 or at +22 and +35 (Fig. 6C, Supplemental Fig. 2). When the reactive s4U was located 35 nt downstream of the splice junction, only two of the labeled species were seen, suggesting that the ~40 kDa and ≥250 kDa polypeptides did not make intimate contact with the RNA in this region. Although the absence of cross-linking does not necessarily reflect the absence of an RNA-protein interaction, the finding that the footprint of the spliceosome did not extend thus far downstream of the exon-exon junction is consistent with the interpretation that splicing components are not stably engaged with this region in the mRNA.
Fig. 6.

Site-specific cross-linking experiments. (A) Precursor RNAs containing a single 32P (marked by *) and an s4U residue (indicated by a lightning bolt) four nucleotides downstream of the 32P label. (B) Splicing was carried out with the indicated precursors and aliquots of each mixture were analyzed by denaturing (7M urea) PAGE and autoradiography. (C) The samples were separated by SDS-PAGE after UV irradiation and RNase T1 digestion and 32P-labeled polypeptides were visualized by autoradiography. The positions (in kDa) of pre-stained marker proteins are indicated at the right. (D) Singly-labeled pre-mRNAs with (+) or without (−) s4U at position +17, were spliced in wild-type extract supplemented with wild-type Prp22 or the dominant-negative Prp22-T765A protein. The RNA species were analyzed by denaturing PAGE (left panel) and the 32P-labeled polypeptides were visualized by SDS-PAGE and autoradiography after cross-linking and RNase T1 digestion (right panel). (E) Pre-mRNA containing a single 32P, and s4U at position +17 was spliced in wild-type extract supplemented with Prp22, Prp22-T765A or Prp22-K512A-TAP proteins. After cross-linking and RNase T1 digestion, the samples were analyzed by SDS-PAGE and autoradiography. (F) s4U-containing +17 pre-mRNA was spliced in Δprp22 extract supplemented with Prp22-T765A or Prp22-N 260-T765A polypeptides. The reaction mixtures were exposed to UV, treated with RNase T1 and analyzed by SDS-PAGE. An autoradiogram of the dried gel is shown.
To gauge if the observed protein-RNA contacts were specific to spliceosome-associated mRNA, splicing and cross-linking experiments were performed using pre-mRNA with s4U at +17 and wild-type extract supplemented with wild-type Prp22 or the inactive Prp22-T765A protein (Fig. 6D). The 40 kDa and 65 kDa polypeptides were cross-linked to s4U irrespective of whether mRNA release was inhibited or not (lanes 2, 3, 5 and 6), indicating that they are not stage-specific. In contrast, the ~155 kDa and ≥250 kDa cross-linked species were specific to s4U contained in spliceosome-associated mRNA (lanes 5 and 6). A control experiment verified that formation of the ~155 kDa and ≥250 kDa RNA-protein adducts depended on a photoreactive s4U nucleobase in the RNA (lanes 7–12).
To ascertain whether the labeled ~155 kDa species was indeed Prp22, splicing and site-specific cross-linking experiments were carried out in the presence of dominant-negative Prp22 proteins, either fused to a 183-aa C-terminal TAP peptide, or lacking the 260-aa N-terminal segment (Fig. 6E and F). When wild-type extract was supplemented with Prp22-K512A-TAP protein, a new higher molecular weight labeled species was detected at the expense of the ~155 kDa species seen in reactions where spliceosome disassembly was inhibited by Prp22-T765A (compare lanes 8, 9 to lanes 5, 6). When Δprp22 extract was complemented with Prp22-NΔ260-T765A protein, a new cross-linked polypeptide was detected that migrated faster than the ~155 kDa species. I conclude that Prp22 is positioned on the mRNA downstream of the splice junction when spliceosome disassembly is inhibited. The Prp22 cross-link was centered at +17, indicating that Prp22 is responsible for, or significantly contributes to the observed RNase H protection in the mRNA prior to spliceosome disassembly.
DISCUSSION
The present study illuminates the mechanism of pre-mRNA splicing in the following ways: (i) it demonstrates the requirement for an RNA segment downstream of the exon-exon junction for the mRNA release step of splicing, (ii) it shows where Prp22 is positioned on the mRNA after exon ligation, and (iii) it describes a new intermediate in the splicing pathway that arises concurrent with the second chemical reaction and precedes mRNA release. The findings support a model for Prp22-catalyzed mRNA release wherein an ATP-independent rearrangement deposits Prp22 on the mRNA downstream of the splice junction, thus allowing the 3′→5′ helicase to liberate mRNA from the spliceosome by disrupting contacts that involve the U5 snRNP (Fig. 7).
Fig. 7.

A model for Prp22-catalyzed mRNA release. Prp22 interacts with intron RNA adjacent to the 3′ splice site in spliceosomes that are poised for the 2nd transesterification reaction. The U5 snRNP contributes RNA and protein contacts to configure the active site for step 2 catalysis. Concurrent with exon ligation, Prp22 is deposited on the mRNA downstream of the exon-exon junction. Bound to its target RNA, Prp22 helicase hydrolyzes ATP and moves in the 3′→5′ direction to disrupt contacts with the U5 snRNP, thereby liberating spliced mRNA from the residual spliceosome that still contains excised lariat-intron RNA.
Although not required for the 1st and 2nd transesterification reactions, an RNA segment downstream of the 3′ splice site is necessary for mRNA release in vitro. With respect to length and composition, the 3′ exon requirements for spliceosome disassembly mirror those of the RNA cofactor requirements for Prp22. Specifically, previous studies showed that RNA, but not DNA can serve as a cofactor for Prp22’s enzymatic activities. Those studies also demonstrated a direct correlation between the length of the RNA cofactor and (i) the stimulatory effect on the ATPase activity of Prp22, (ii) RNA binding by Prp22, and (iii) helicase activity of Prp22, whereby longer single-stranded RNAs were more effective than shorter RNAs. For example, increasing the RNA length from 10 to 20 or 30 nt lead to a 13- or 40-fold increase, respectively, in the catalytic efficiency of the Prp22 ATPase (Tanaka and Schwer, 2005). The findings that >13 ribonucleotides downstream of the splice junction are necessary for mRNA release, and that Prp22 interacts with this segment in the mRNA, are consistent with the idea that the region downstream of the exon-exon junction is the cofactor for Prp22 action within the spliceosome.
The ATPase activities of DExD/H splicing factors are either stimulated by or dependent on an RNA cofactor (Schwer and Guthrie, 1991; Kim et al., 1992; O’Day et al., 1996; Schwer and Gross, 1998; Kim and Rossi, 1999; Martin et al., 2002). Because none of the enzymes exhibits overt sequence-specificity in vitro, it has been difficult to pinpoint the stimulatory RNA in the spliceosome. By comparing the effect of five spliceosomal snRNAs on Prp5’s ATPase activity, O’Day et al. (1996) found that Prp5 exhibits a slight preference for U2 snRNA. Thus, Prp5, which facilitates stable binding of U2 snRNP to the branchpoint region and which is thought to modulate fidelity of spliceosome assembly, might be stimulated by the U2 snRNA within the spliceosome (Ruby et al., 1993; Xu and Query, 2007). Site-specific cross-linking studies have documented interactions between hPrp28 and the 5′ splice site and Prp16 with sequence elements in the 3′ splice site region in the course of the splicing reaction (Ismaili et al., 2001; Umen and Guthrie, 1995; McPheeters et al., 2000; McPheeters and Muhlenkamp, 2003), but the functional relevance of these interactions for splicing has not been explored in detail.
In addition to its essential function for mRNA release, Prp22 plays an ATP-independent role during the second step of splicing (Schwer and Gross, 1998). Prp22 associates with spliceosomes before exon ligation (James et al., 2002). At this stage, Prp22 does not interact with the 3′ exon, but rather with intronic RNA. Specifically, Prp22 can be cross-linked to s4U in an ~8 nt segment within the intron immediately upstream of the 3′ splice site (McPheeters et al. 2000; McPheeters and Muhlenkamp, 2003). This interaction represents an authentic intermediate (McPheeters and Muhlenkamp, 2003), and it does not persist after exon ligation, insofar as the ATPase-deficient Prp22-K512A mutant that supports step 2, but prevents mRNA release, was not cross-linked to the 3′ end of the excised intron (McPheeters et al., 2000). In previous studies, the fate of Prp22-RNA interactions after exon ligation was not determined, but it was clear that Prp22 remains associated with splicing complexes until mRNA is released (James et al., 2002). I show here that Prp22 is situated on the mRNA downstream of the splice junction after step 2.
The placement of Prp22 on the mRNA concomitant with exon joining is reflected in a conformational change that was detected by comparing the RNase H cleavage patterns of 3′ exon RNA in complexes arrested before and immediately after the second transesterification step. A region of ~23 nt downstream of the exon-exon junction was impervious to RNase H in mRNA, when mRNA release was blocked by inactivating the ATP-dependent function of Prp22. Protection was seen for several spliced RNAs, arguing that it is not transcript-specific and attesting to the generality of the conformational change. Furthermore, protection was seen in spliceosomes that were stalled in the presence of wild-type Prp22 by depletion of ATP, indicating that the observed change reflects a natural intermediate step in the splicing pathway.
The RNA-dependent ATPase/helicase function of Prp22 is required after exon ligation, yet Prp22 interacts with the 3′ end of the intron before step 2 (Fig. 7). This raises the question of whether Prp22 engages with RNA similarly at both stages. Prp22 may associate with intronic RNA in a fashion that does not trigger NTP hydrolysis, and the enzyme may use a different RNA binding site to interact productively with mRNA. Alternatively, Prp22’s helicase action may be repressed specifically prior to mRNA release. There is precedent for a scenario in which a DEAD-box protein is inhibited when bound to RNA. eIF4AIII is a constituent of the metazoan exon junction complex (EJC), which is deposited on spliced mRNA and escorts the mRNP to the cytoplasm (Le Hir et al., 2000). eIF4AIII is locked in the ATP-bound state on RNA by specific EJC components, thereby ensuring the stable association of the multiprotein complex ~24 nt upstream of the exon-exon junction (Ballut et al., 2005; Bono et al., 2006). Whether and how Prp22’s helicase activity is controlled prior to exon ligation remains to be determined. Regulating Prp22 activity at this stage could provide a mechanism to ensure fidelity of exon ligation, a role for Prp22 that was proposed by Mayas et al. (2006). Whereas the Prp22 helicase is normally active when bound to the mRNA downstream of the exon-exon junction, it is conceivable that in spliceosomes assembled on aberrant pre-mRNAs, Prp22’s activity is unleashed/deregulated before the 2nd transesterification reaction, thus leading to the disassembly of aberrant spliceosomes (Mayas et al., 2006).
Once bound to mRNA downstream of the splice junction, Prp22 acts to liberate the mRNA from the splicing complex (Fig. 7). To accomplish this, Prp22 harnesses the energy of ATP hydrolysis to perform mechanical work that can be measured as directional unwinding of a nucleic acid duplex in vitro (Schwer and Meszaros, 2000). It is conceivable that Prp22 moves along the mRNA in the 3′→5′ direction to peel the mRNA off the residual spliceosome that still contains excised lariat-intron RNA. This proposal is based on current models for the mechanism of directional helicases, which suggest that helicases translocate on the loading strand, thereby dissociating the second strand in the duplex substrate (reviewed in Patel and Donmez, 2006). For example, the 3′→5′ DExH-box helicase NPH-II depends on a continuous ribose-phosphate backbone for unwinding, while the enzyme is insensitive to gaps or deoxynucleotides in the displaced strand (Kawaoka et al., 2004; Kawaoka and Pyle, 2005). By tracking on RNA, NPH-II can unwind nucleic acid duplexes, but NPH-II was also shown to dislodge proteins (Jankowsky et al., 2001; Fairman et al., 2004). Genetic studies revealed that Prp22 helicase disrupts contacts within the splicing complex that involve the U5 snRNP. Specifically, Arg-1753 in the U5 snRNP component Prp8, and bases in the conserved loop 1 of the U5 snRNA that pair with exon bases adjacent to the splice sites, were implicated in interactions that are broken by Prp22 helicase (Schneider et al., 2004; Aronova et al., 2007).
The footprint of the spliceosome on mRNA spans ~38 nt, including 15 nt upstream of the exon-exon junction. It is conceivable that the U5 snRNP, via Prp8, contributes to the RNA-protein interactions across the splice junction, insofar as Prp8 was shown to interact with at least 8 and 13 nt in the 5′ and 3′ exons, respectively in the course of the splicing reaction (Teigelkamp et al., 1995a,b; McPheeters et al., 2000; McPheeters and Muhlenkamp, 2003). The RNP structure of the 5′ exon has been analyzed extensively in mammalian splicing systems. RNase protection and cross-linking studies demonstrated that the last 25–27 nt of the 5′ exon are engaged in numerous dynamic protein-RNA interactions in fully assembled spliceosomes (Reichert et al., 2002). Upon exon ligation, the interactions at the 3′ end of the 5′ exon disappeared and the EJC, centered ~24 nt upstream of the splice junction was detected (Le Hir et al., 2000; Reichert et al., 2002). EJC components are not present in the yeast proteome. Therefore it is not surprising that the RNase H protection analyses presented here did not reveal significant RNA-protein interactions after the mRNA was released from the spliceosome.
EXPERIMENTAL PROCEDURES
Splicing substrates
All precursor RNAs were generated as capped run-off transcripts using T7 RNA polymerase in the presence of cap analog (GpppG). The ACT1 pre-mRNAs contained a 90-nt 5′ exon and the ACT1- 6 intron (302 nt) that lacks a cryptic branch site (Vijayraghavan et al., 1986). The 3′ exons were 6, 13, 18, 31 or 126 nt in length. The 5′ and 3′ exons of the RPS17A pre-mRNA were 120 and 153 nt, respectively, and the 5′ and 3′ exons of the U3 snoRNA precursor were 78 and 318 nt long. Body-labeled transcripts were synthesized in the presence of [α–32P]GTP and purified as described (Ansari et al., 1995). Singly-labeled pre-mRNAs were generated by ligating 5′[32P]-oligonucleotides to unlabeled run-off transcripts using T4 Rnl2 (Nandakumar et al., 2004; Nandakumar and Shuman, 2004). The synthetic oligos (shown in the respective figures) were labeled with T4 polynucleotide kinase and [γ-32P]ATP and purified by electrophoresis through denaturing 15% polyacrylamide gels. To form the nicked substrate for ligation by Rnl2, 10 pmol of the transcript (5′ exon-intron and 3′ exon of various lengths) and the 5′[32P]-oligo were annealed to a complementary 26–30 nt bridging DNA oligo (1:2:5 molar ratio) in 150 mM NaCl, 10 mM Tris-HCl (pH8.0), 1 mM EDTA, by incubation for 15 min at 65°C, followed by incubation for 15 min at 37°C and then 30 min at 22°C. Reaction mixtures (100 μl) containing 50 mM Tris-acetate, pH 6.5, 40 mM NaCl, 5 mM DTT, 1 mM ATP, 1 mM MgCl2, nicked substrate and 20 μg Rnl2 were incubated at 22°C for 30 min. The full-length ligated products were purified by electrophoresis through a denaturing 5% polyacrylamide gel. Typically, the yield of labeled pre-mRNA was 15–25% of the input transcript.
UV cross-linking
Pre-mRNAs containing a single 32P and a single s4U, incorporated during synthesis of the oligonucleotide [Dharmacon] were spliced during incubation at 28°C for 20 min. The mixtures were then placed on ice and exposed to UV (312 nM) at a distance of ~8 cm using a FOTO/UV® transilluminator (Fotodyne). Aliquots (10 μl) were withdrawn after 0, 5 and 10 min. T1 ribonuclease (150 U) was added in 6 μl T1 buffer (13 mM Tris-HCl pH 7.5, 20 mM EDTA, 2x Complete™ protease inhibitor cocktail [Roche]) to each mixture and incubated at 37°C for 30 min. An equal volume of 2x SDS-loading buffer was added and the polypeptides were analyzed by electrophoresis in 6% SDS-polyacrylamide gels.
In vitro splicing
Extracts were prepared and reactions were performed essentially as described (Ansari and Schwer, 1995) and the methods are summarized in the Supplement.
Supplementary Material
Acknowledgments
I am grateful to Jayakrishnan Nandakumar for T4 Rnl2 protein, to Stewart Shuman for stimulating discussions and to David Horowitz for comments on the manuscript. This work was supported by NIH GM050288 and funds from the William Randolph Hearst Foundation to the Department of Microbiology and Immunology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson K, Moore M. Bimolecular exon ligation by the human spliceosome. Science. 1997;276:1712–1716. doi: 10.1126/science.276.5319.1712. [DOI] [PubMed] [Google Scholar]
- Ansari A, Schwer B. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J. 1995;14:4001–4009. doi: 10.1002/j.1460-2075.1995.tb00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronova A, Bacikova D, Crotti LB, Horowitz DS, Schwer B. Functional interactions between Prp8, Prp18, Slu7, and U5 snRNA during the second step of pre-mRNA splicing. RNA. 2007;13:1437–1444. doi: 10.1261/rna.572807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Sérahin B, Le Hir H. The exon junction core is locked onto RNA by inhibition of eIF4III ATPase activity. Nat Struct Mol Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Burgess SM, Guthrie C. Beat the clock: paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem Sci. 1993;18:381–384. doi: 10.1016/0968-0004(93)90094-4. [DOI] [PubMed] [Google Scholar]
- Campodonico E, Schwer B. ATP-dependent remodeling of the spliceosome: Intragenic suppressors of release-defective mutants of Saccharomyces cerevisiae Prp22. Genetics. 2002;160:407–415. doi: 10.1093/genetics/160.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- Crotti LB, Bacikova D, Horowitz DS. The Prp18 protein stabilizes the interaction of both exons with the U5 snRNA during the second step of pre-mRNA splicing. Genes Dev. 2007;21:1204–1216. doi: 10.1101/gad.1538207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. Protein displacement by DExD/H "RNA helicases" without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- Horowitz DS, Abelson J. Stages in the second reaction of pre-mRNA splicing: the final step is ATP independent. Genes Dev. 1993;7:320–329. doi: 10.1101/gad.7.2.320. [DOI] [PubMed] [Google Scholar]
- Ismaïli N, Sha M, Gustafson EH, Konarska MM. The 100-kda U5 snRNP protein (hPrp28p) contacts the 5′ splice site through its ATPase site. RNA. 2001;7:182–193. doi: 10.1017/s1355838201001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SA, Turner W, Schwer B. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA. 2002;8:1068–1077. doi: 10.1017/s1355838202022033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNA-protein interaction by a DExD/H RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExD/H RNA helicases. Nucleic Acids Res. 2006;34:4181–4188. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka J, Jankowsky E, Pyle AM. Backbone tracking by the SF2 helicase NPH-II. Nat Struct Mol Biol. 2004;11:526–530. doi: 10.1038/nsmb771. [DOI] [PubMed] [Google Scholar]
- Kawaoka J, Pyle AM. Choosing between DNA and RNA: the polymer specificity of RNA helicase NPH-II. Nucleic Acids Res. 2005;33:644–649. doi: 10.1093/nar/gki208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Smith J, Claude A, Lin RJ. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 1992;11:2319–2326. doi: 10.1002/j.1460-2075.1992.tb05291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Rossi JJ. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA. 1999;5:959–971. doi: 10.1017/s135583829999012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Schneider S, Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J Biol Chem. 2002;277:17743–17750. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- Mayas RM, Maita H, Staley JP. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol. 2006;13:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters DS, Schwer B, Muhlenkamp P. Interaction of the yeast DExH-box RNA helicase Prp22p with the 3′ splice site during the second step of nuclear pre-mRNA splicing. Nucleic Acids Res. 2000;28:1313–1321. doi: 10.1093/nar/28.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters DS, Muhlenkamp P. Spatial organization of protein-RNA interactions in the branch site-3′ splice site region during pre-mRNA splicing in yeast. Mol Cell Biol. 2003;23:4174–4186. doi: 10.1128/MCB.23.12.4174-4186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Ho CK, Lima CD, Shuman S. RNA substrate specificity and structure-guided mutational analysis of bacteriophage T4 RNA ligase 2. J Biol Chem. 2004;279:31337–31347. doi: 10.1074/jbc.M402394200. [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Shuman S. How an RNA ligase discriminates RNA versus DNA damage. Mol Cell. 2004;16:211–221. doi: 10.1016/j.molcel.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Newman AJ. The role of U5 snRNP in pre-mRNA splicing. EMBO J. 1997;16:5797–5800. doi: 10.1093/emboj/16.19.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Day CL, Dalbadie-McFarland G, Abelson J. The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J Biol Chem. 1996;271:33261–33267. doi: 10.1074/jbc.271.52.33261. [DOI] [PubMed] [Google Scholar]
- Pandit S, Lynn B, Rymond BC. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc Natl Acad Sci USA. 2006;103:13700–13705. doi: 10.1073/pnas.0603188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Donmez I. Mechanisms of helicases. J Biol Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- Reichert VL, Le Hir H, Jurica MS, Moore MJ. 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes Dev. 2002;16:2778–2791. doi: 10.1101/gad.1030602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby SW, Chang TH, Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- Rymond BC, Rosbash M. Cleavage of 5′ splice site and lariat formation are independent of 3′ splice site in yeast mRNA splicing. Nature. 1985;317:735–737. doi: 10.1038/317735a0. [DOI] [PubMed] [Google Scholar]
- Schneider S, Hotz HR, Schwer B. Characterization of dominant-negative mutants of the DEAH-box splicing factors Prp22 and Prp16. J Biol Chem. 2002;277:15452–15458. doi: 10.1074/jbc.M112473200. [DOI] [PubMed] [Google Scholar]
- Schneider S, Campodonico E, Schwer B. Motifs IV and V in the DEAH box splicing factor Prp22 are important for RNA unwinding, and helicase-defective Prp22 mutants are suppressed by Prp8. J Biol Chem. 2004;279:8617–8626. doi: 10.1074/jbc.M312715200. [DOI] [PubMed] [Google Scholar]
- Schwer B, Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;349:494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- Schwer B, Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Gross CH. Prp22, an RNA-dependent ATPase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998;17:2086–2094. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Meszaros T. RNA helicase dynamics in pre-mRNA splicing. EMBO J. 2000;19:6582–6591. doi: 10.1093/emboj/19.23.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Schwer B. Characterization of the NTPase, RNA-Binding, and RNA Helicase Activities of the DEAH-Box Splicing Factor Prp22. Biochemistry. 2005;44:9795–9803. doi: 10.1021/bi050407m. [DOI] [PubMed] [Google Scholar]
- Teigelkamp S, Newman AJ, Beggs JD. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 1995a;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp S, Whittaker E, Beggs JD. Interaction of the yeast splicing factor PRP8 with substrate RNA during both steps of splicing. Nucleic Acids Res. 1995b;23:320–326. doi: 10.1093/nar/23.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan U, Parker R, Tamm J, Iimura Y, Rossi J, Abelson J, Guthrie C. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986;5:1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Guthrie C. Prp16p, Slu7p, and Prp8p interact with the 3′ splice site in two distinct stages during the second catalytic step of pre-mRNA splicing. RNA. 1995;1:584–597. [PMC free article] [PubMed] [Google Scholar]
- Will CL, Lührmann R. Spliceosome structure and function. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2006. pp. 369–400. [Google Scholar]
- Valadkhan S. snRNAs as the catalysts of pre-mRNA splicing. Curr Opin Chem Biol. 2005;9:603–608. doi: 10.1016/j.cbpa.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Xu YZ, Query C. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell. 2007;28:838–849. doi: 10.1016/j.molcel.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.