Abstract
Friedreich ataxia (FA) is a progressive neurodegenerative disease caused by expansion of a trinucleotide repeat within the first intron of the gene that encodes frataxin. In our study, we investigated the regulation of frataxin expression by iron and demonstrated that frataxin mRNA levels decrease significantly in multiple human cell lines treated with the iron chelator, desferal (DFO). In addition, frataxin mRNA and protein levels decrease in fibroblast and lymphoblast cells derived from both normal controls and from patients with FA when treated with DFO. Lymphoblasts and fibroblasts of FA patients have evidence of cytosolic iron depletion, as indicated by increased levels of iron regulatory protein 2 (IRP2) and/or increased IRE-binding activity of IRP1. We postulate that this inferred cytosolic iron depletion occurs as frataxin-deficient cells overload their mitochondria with iron, a downstream regulatory effect that has been observed previously when mitochondrial iron–sulfur cluster assembly is disrupted. The mitochondrial iron overload and presumed cytosolic iron depletion potentially further compromise function in frataxin-deficient cells by decreasing frataxin expression. Thus, our results imply that therapeutic efforts should focus on an approach that combines iron removal from mitochondria with a treatment that increases cytosolic iron levels to maximize residual frataxin expression in FA patients.
INTRODUCTION
Friedreich ataxia (FA) is a progressive disease caused by expansion of a GAA trinucleotide repeat in the gene that encodes frataxin. Patients develop ataxia, sensory loss and cardiomyopathy, and symptoms are limited mainly to the central nervous system and heart (reviewed in 1,2). Frataxin is important in iron–sulfur cluster assembly where it likely functions as a chaperone that provides iron in a bioavailable form in the early steps of iron–sulfur cluster biosynthesis (reviewed in 3). Patients manifest mitochondrial iron–sulfur cluster assembly deficiency, with decreased activity of iron–sulfur proteins such as mitochondrial aconitase and succinate dehydrogenase. As disease progresses, mitochondrial iron overload develops in the heart and central nervous system (4–6). Because of the mitochondrial iron overload, it has been considered that iron chelation therapy should ameliorate disease by protecting mitochondria from the adverse effects of iron overload. Studies in tissue culture performed with the primarily cytosolic iron chelator, desferal (DFO) (7), or with lipophilic iron chelators that specifically target mitochondrial iron, including pyridoxal isonicotinoyl hydrazone and analogues of 2-pyridylcarboxaldehyde isonicotinoyl hydrazone (8,9), as well as studies in patients using deferiprone, a membrane-permeable chelator thought to be capable of shuttling chelated iron from cells to transferrin (10) have shown positive results.
Although many chelator studies have been performed on FA cells and models, none has evaluated the effects of iron chelation on levels of residual frataxin expression in patients. Low expression of FXN causes iron accumulation in mitochondria, a common phenotype observed when proteins involved in iron–sulfur cluster biogenesis are deficient (11). Previous studies have shown that yeast which lacks the mitochondrial ABC transporter, Atm1 (a homologue of human ABCB7), display features of both mitochondrial iron overload and cytosolic iron starvation (12), which is similar to the phenotype of frataxin-deficient yeast (13). Interestingly, levels of yfh1 (frataxin) mRNA level decreased 4-fold in Atm1-deficient yeast, whereas atm1 mRNA levels were unchanged in the yfh1 mutant (12). Although this study suggested that yfh1 levels were regulated by iron, the authors were unable to confirm iron-dependent regulation of frataxin when they manipulated iron levels in yeast.
Since we have previously observed that expression of the iron–sulfur scaffold protein, ISCU, decreases with iron chelation (14), we decided to evaluate the effects of iron depletion on frataxin expression. We treated human cell lines with the iron chelator, DFO, or the iron salt, ferric ammonium citrate (FAC), to determine whether frataxin expression changes in normal controls or in cell lines derived from FA patients. We discovered that frataxin mRNA levels decrease significantly in multiple human cell lines treated with DFO. In addition, frataxin levels decrease in fibroblasts and lymphoblast cells derived from both normal controls and from patients with FA when treated with DFO. Using activity of iron regulatory proteins as an indicator of cytosolic iron status, we find evidence to support that there is cytosolic iron depletion in fibroblast and lymphoblast cell lines from FA patients compared with normal control cell lines. Our results suggest that cytosolic iron depletion may worsen FA by further diminishing expression of frataxin.
RESULTS
To study whether frataxin expression was regulated by iron, we studied human cell lines, including HEK293. After adding either FAC or the iron chelator DFO (Sigma) to cells for 16 h, we observed that frataxin mRNA levels decreased in cells treated with DFO, as detected by northern blot (Fig. 1A) and confirmed by quantitative real-time PCR (qRT–PCR) (Fig. 1B). In addition to finding the major frataxin transcript of 1.24 kb as previously reported (15), we observed some similarly regulated bands (e.g. ∼1.7 and 2.3 kb), whereas some others did not appear to be regulated (e.g 3.2, 4.5 and 5.8 kb bands). All of these bands were detected using an antisense RNA probe to the sense strand of FXN, and we do not know whether the larger weak bands are non-specific, or whether they represent different isoforms or incompletely spliced intermediate forms. We also tested fibroblast cell lines (GM08399, Coriell Cell Repositories), and found increased mRNA levels of frataxin in cells treated with FAC versus DFO (Fig. 1C), which was confirmed by qRT–PCR (Fig. 1D). This decrease in frataxin expression with DFO treatment was also observed in other cell lines we tested, including the rhabdomyosarcoma cell line, RD4 (data not shown).
Figure 1.
Frataxin (FXN) mRNA expression is regulated by iron status in human cell lines. HEK293 cells were treated with desferal (DFO, 100 µM), normal unsupplemented medium (Ctr) or ferric ammonium citrate (FAC, 200 µM) for 16 h. (A) mRNA levels of FXN decreased in DFO treated cells compared with control (Ctr) and FAC treated cells. Equal loading was verified by GAPDH and L32 measurements. (B) Quantitative real-time PCR (qRT–PCR) was carried out for FXN and mRNA levels decreased significantly (P = 0.0013) after DFO treatment. (C) Northern analysis and (D) qRT–PCR were carried out for FXN on fibroblasts (GM08399) derived from a normal control, treated either with DFO (100 µM) or FAC (200 µM). Frataxin levels in qRT–PCR were significantly reduced (P = 0.028). Ribosomal RNA levels (28S and 18S) were used as loading controls for (C) and GAPDH was used as an internal control for (D). (E) Luciferase activity driven by FXN promoter decreased significantly (P = 0.0025) when the cells were treated with DFO compared with growth in normal iron-replete media. The empty plasmid was used as a negative control. FXN-luc1, 2 and 3 contained different fragments of genomic DNA, and FXN-GAA-luc contained the frataxin promoter, first exon and a portion of the first intron as described in Materials and Methods. Statistical analysis was done with the Student's t-test: *P < 0.05, **P < 0.01.
To evaluate the mechanisms of regulation of FXN mRNA levels by iron status, we cloned the previously defined human frataxin promoter region (16,17), and generated constructs that incorporated 1.2 (luc 1), 1.8 (luc 2) or 3.2 kb (luc 3) upstream of the first AUG of exon 1 into a luciferase reporter plasmid, as described previously (17). In another construct (pGL-GAA-luc), we fused a DNA fragment including the promoter region (1.3 kb upstream of the first AUG in exon 1), the entire first exon, and 1.2 kb of the 5′ end of the first intron extending to the position of the GAA triple repeat expansion with luciferase. Each of the four constructs showed that DFO treatment significantly reduced the expression of luciferase compared with untreated controls (Fig. 1E), although the pGL4-GAA-luc construct showed lower promoter activity.
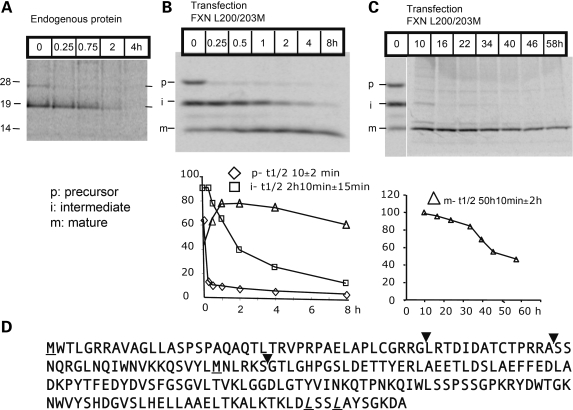
To study whether frataxin protein levels were regulated by iron, as expected based on mRNA results, we analyzed the half-life of frataxin in metabolic labeling experiments to define the optimal length of iron or iron chelator treatments. After immunoprecipitation (IP) of frataxin radiolabeled with 35S-methionine, we observed the frataxin precursor and the frataxin intermediate band generated during mitochondrial import (Fig. 2A), but we were unable to observe the mature fully processed product (Fig. 2D), as the mature form does not contain methionine (18). There are two methionines in the unprocessed frataxin precursor, both of which are removed during mitochondrial import. The inferred cleavage sites are indicated in Figure 2D with inverted arrowheads (18,19), and methionines are underlined. In order to measure the half-life of mature frataxin in cells, we replaced two leucines (L) (amino acids 200th and 203rd) of frataxin, with methionine (M) (described in Materials and Methods). Metabolic labeling results showed that the half-life of the initial precursor was 10 min, whereas that of the intermediate was 2 h 10 min, and that of the mature form was 50 h (Fig. 2B and C).
Figure 2.
Half-life determination of human FXN in HEK293 cells with metabolic labeling. (A) Endogenous FXN of HEK293 cells was chased and only the precursor (p) and intermediate (i) forms were identified. (B) and (C) A mutagenized form of frataxin (FXN L200/203M- described in Materials and Methods) was expressed in HEK293 cells to allow visualization of the mature isoform (m). The half life of human FXN was calculated from three independent experiments, and precursor (p), intermediate (i) and mature (m) forms are represented in lower panels of (B and C) with the half-life of the mature form calculated as ∼50 h. (D) The amino acid sequence of the human FXN open reading frame. The initiator methionine and an in-frame methionine (M) are underlined. The inverted arrowheads indicate proposed sites of cleavage associated with mitochondrial uptake. The first two cleavage sites were initially proposed according to molecular weight in a number of in vitro studies (19,43), whereas the third cleavage site of human endogenous FXN was identified by mass spectrometry (18). The last two leucines (L) were replaced with methionine (M), underlined and in italics.
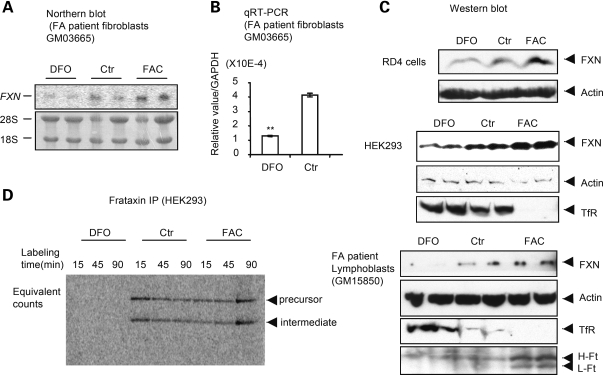
We then analyzed regulation of frataxin mRNA and protein levels in fibroblast cell lines of controls and FA patients after treatment with DFO or FAC for 48 h. We found that frataxin mRNA levels decreased in fibroblasts (GM03665) of patients treated with DFO (Fig. 3A) and we verified this result with qRT–PCR (Fig. 3B). We then verified that protein levels changed in RD4 (top), HEK293 (middle), fibroblasts (GM04078 and GM03816, not shown) and lymphoblasts (GM15850) from affected patients (bottom, Fig. 3C). The increased levels of transferrin receptor and decreased levels of ferritin H and L chains after DFO treatment in lymphoblasts confirmed that the iron treatment was efficacious. Instantaneous biosynthetic rate measurements showed markedly diminished biosynthesis of endogenous frataxin after treatment with DFO (Fig. 3D), further confirming regulation of frataxin expression by iron.
Figure 3.
Iron deprivation reduces FXN protein levels. Northern analysis (A) and qRT–PCR (B) were performed for FXN in fibroblasts GM03665 derived from an FA patient. Ribosomal RNAs (28S and 18S) were used as loading controls in (A) and GAPDH was used to normalize in (B), where frataxin levels were significantly reduced by treatment with desferal (**P = 0.0006). (C) Immunoblots showed that FXN protein levels diminished with iron deprivation, observed with DFO treatment in the rhabdomyosarcoma cell line RD4 (top), HEK293 cells (middle) and lymphoblasts from an FA patient (GM15850) (bottom). Transferrin receptor (TfR) and ferritin (H- and L subunits, H-Ft and L-Ft) were used to verify that iron treatments were efficacious. (D) Biosynthetic labeling of FXN was performed after cellular iron status was manipulated. HEK293 cells were treated with DFO (30 µM) or FAC (200 µM) for 48 h prior to labeling with [35S]-methionine at the indicated time points. Immunoprecipitation was performed using equal counts of radioactively labeled material.
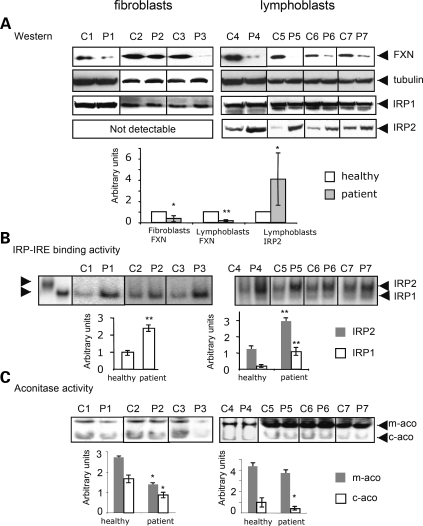
To more fully characterize cellular iron status in cell lines from FA patients, we compared fibroblast and lymphoblast cell lines derived from either normal controls or patients that are either age and sex-matched, or proband and parent-paired, for levels of frataxin, iron regulatory proteins 1 (IRP1) and 2 (IRP2), IRE-binding activity and aconitase activities. Comparing lymphoblasts from patients (P) to control (C) cell lines, we found that frataxin expression levels varied from 8.5% [e.g. the pair of GM16200 (C5) and GM16197 (P5) in Fig. 4A, right column] to 25% [e.g. the pairs of GM16215 (C6) and GM16214 (P6), AG15799 (C7) and GM16209 (P7), Fig. 4A, right column] of control levels, whereas in fibroblasts from three different patients, we found that frataxin levels varied from 10% [e.g. the pair of GM08399 (C3) and GM03665 (P3), Fig. 4A, left column] to 66% [e.g. the pair of AG09429 (C2) and GM03816 (P2), Fig. 4A, left column] of normal control levels, quantified as described in Materials and Methods. We found that without any iron manipulations, lymphoblast cell lines of FA patients showed increased levels of IRP2 in western blots (Fig. 4A) and RNA gel-shift studies (Fig. 4B) compared with normal controls. In the presence of high iron and oxygen, IRP2 undergoes iron-dependent degradation (20,21). Although low oxygen levels and increased cobalt levels can stabilize IRP2, high IRP2 levels are in general an excellent indicator of cytosolic iron status in mammalian cells (reviewed in 22). IRP2 was not detectable in fibroblasts, but gel-shift studies indicated that the IRE-binding ability of IRP1 was markedly increased, as has been previously observed in FA patient cells (23) (Fig. 4B), as well as in RNAi knockdown studies in HeLa cells (24) and in mouse models (25). In correlation, both mitochondrial and cytosolic aconitase activities were decreased in patient fibroblasts, and cytosolic aconitase activity was significantly decreased in lymphoblasts (Fig. 4C). Thus, IRP1 converted from the cytosolic aconitase form to the IRE-binding form without change of IRP1 protein levels in cell lines of patients with FA.
Figure 4.
Cell lines derived from FA patients show evidence of cytosolic iron depletion when grown under normal conditions. Either fibroblasts (C1: GM08402 and P1:GM04078; C2:AG09429 and P2:GM03816; C3: GM08399 and P3: GM03665, left column) or lymphoblasts (C4: GM15849 and P4:GM15850, C5: GM16200 and P5:GM16197, C6: GM16215 and P6:GM16214, C7: AG15799 and P7:GM16209, right column) derived from both healthy controls (C) and FA patients (P) were compared. Representative results from multiple repetitions are shown here. Numbers 1–7 for controls and patients represent either age and sex-matched (C1/P1, C2/P2, C3/P3, C4/P4, C7/P7) pairs or proband and parent-paired (C5/P5, C6/P6) pairs. (A) The relative protein levels of FXN, IRP1 and IRP2 were determined by western blot, quantified as described in Materials and Methods. Both IRE-binding activities of IRP1 (white bar) and IRP2 (grey bar) (B) and aconitase activities (C) (mitochondrial, m-aco, grey bar; cytosolic, c-aco, white bar) were assessed and quantified, showing significant decreases in aconitase activities (*P < 0.05), and increases in IRE binding activities (**P < 0.01).
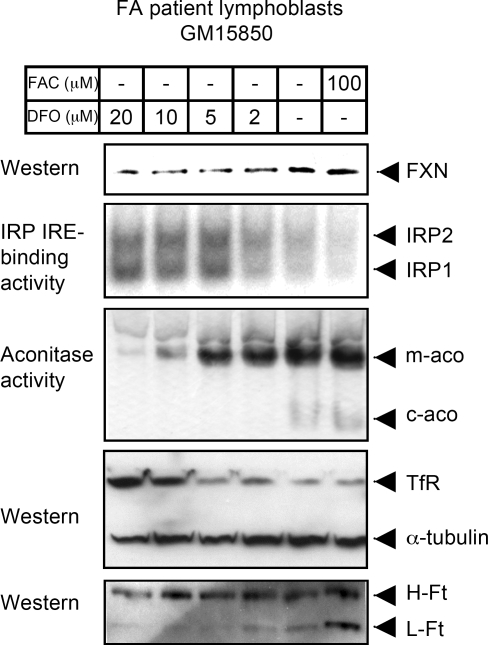
To further verify the effects of iron on FXN expression, we challenged the FA patient cells with different concentrations of DFO or FAC. We observed regulation of FXN, as well as sensitivity of FA fibroblasts to iron depletion, where as little as 1 or 2 µM DFO abolished cytosolic aconitase activity of IRP1, and 10 µM DFO was sufficient to dramatically decrease mitochondrial aconitase activity (Fig. 5). In addition, the IRE-binding activity of IRP1 and IRP2 were maximal at 5 µM of DFO concentration, suggesting that the cells of the FA patients were relatively iron-starved prior to treatment.
Figure 5.
A titration curve of FA patient cells reveals that FA cells are markedly affected by iron depletion, which further reduces FXN expression. Patient lymphoblasts GM15850 were treated either with indicated concentrations of DFO or with FAC for 60 h, then harvested, lysed and analyzed by the in-gel aconitase assay, bandshift assay and western blot. The abbreviations are the same as those used in Fig. 4.
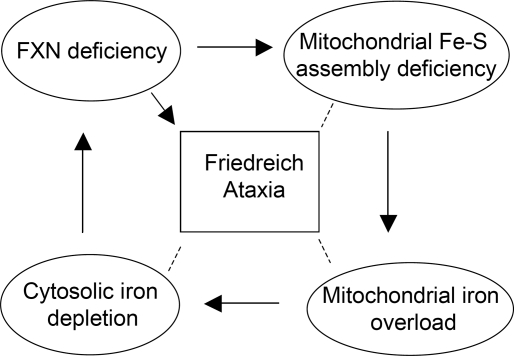
In summary, our results demonstrated that cytosolic iron depletion may contribute to decreased expression of frataxin in FA patients and may thereby worsen disease (modeled in Fig. 6).
Figure 6.
A model of how cytosolic iron depletion decreases expression of frataxin in FA patients and may thereby exacerbate disease. Frataxin deficiency mainly caused by the triplet GAA repeat expansion in the first intron of frataxin gene leads to decreased frataxin expression and impairment of mitochondrial iron–sulfur cluster [Fe–S] assembly. Mitochondrial iron overload develops as a consequence of deficient [Fe–S] assembly in mitochondria which leads to misregulation of mitochondrial iron homeostasis. Excess mitochondrial iron uptake and sequestration causes cytosolic iron depletion, which can further diminish frataxin expression by decreasing frataxin transcription. Thus, the decrease in frataxin levels caused by the trinucleotide repeat may be worsened as cells accumulate mitochondrial iron and deplete cytosolic iron stores. This negative feedback loop may contribute to progression of disease in long-lived cells, particularly neurons and cardiomyocytes.
DISCUSSION
In this study, we have demonstrated that frataxin expression levels are regulated by iron in various human tissue culture cell lines, and in lymphoblasts and fibroblasts from controls and FA patients. Through use of promoter-luciferase constructs, we have further observed that transcriptional regulation likely accounts for much of the iron-dependent regulation of frataxin in controls. Importantly, significant iron-dependent regulation was also observed in the lymphoblast and fibroblast cell lines derived from FA patients. To further evaluate the importance of iron-dependent regulation of frataxin in patients, we analyzed cytosolic iron status by assaying activity of IRP1 and IRP2. IRP1 and IRP2 are mammalian proteins that register cytosolic iron concentrations and post-transcriptionally regulate expression of numerous iron metabolism genes to optimize cellular iron availability. IRP1 registers cytosolic iron status mainly through an iron–sulfur cluster at its active site; when the iron–sulfur cluster is present, IRP1 is an active cytosolic aconitase, whereas when the cluster is absent, IRP1 becomes an apoprotein that undergoes a significant conformational change, enabling it to bind to IREs, RNA stem-loop structures found within transcripts (reviewed in 22,26,27). Although IRP2 is homologous to IRP1, IRP2 activity is regulated primarily by iron-dependent degradation through the ubiquitin-proteasomal system in iron-replete cells (22,26,27). Targeted deletions of IRP1 and IRP2 in animals have demonstrated that IRP2 is the chief physiologic iron sensor in animals (28,29). Gel-shifts in this study showed that the IRE-binding activities of both IRP1 and IRP2 were increased in FA patient lymphoblasts, and IRP2 levels were increased relative to controls (Fig. 4), suggesting that there is cytosolic iron depletion in FA cells. Our measurements were not performed on the patient tissues that are very adversely affected in FA, including the heart (6) and cerebellum (5). Thus, even though frataxin levels were markedly reduced, mitochondrial aconitase activity was normal in lymphoblasts from a patient with only 10% of normal frataxin levels, although cytosolic aconitase levels were significantly reduced (Fig. 4C, lanes C4 and P4, C5 and P5). Levels of both cytosolic aconitase and mitochondrial aconitase were significantly reduced in FA fibroblast cells in which frataxin levels were ∼35% of normal (Fig. 4C, 30% for lane C1 and P1, 66% for lane C2 and P2, 10% for lane C3 and P3), but not as significantly as has been reported in heart samples of patients (30), consistent with the observation that disease affects primarily heart and central nervous system, whereas other tissues are less adversely affected.
Since we found evidence for cytosolic iron depletion in cell types that do not develop phenotypes in FA, we postulate that cytosolic iron depletion may be even more pronounced in areas of the brain and heart that are functionally compromised in FA patients. Interestingly, expression of ISCU was decreased in the heart of frataxin-deficient mice (31), and cytosolic iron deficiency perhaps could explain the decrease in ISCU expression (14). In the brain and heart of FA patients, mitochondrial iron overload is prominent and is readily detected with tissue iron stains. In numerous model systems, mutations or deficiencies of iron–sulfur cluster biogenesis proteins result not only in diminished activities of iron–sulfur enzymes such as aconitase and succinate dehydrogenase, but also in mitochondrial iron overload, which appears to be a secondary effect that results from the compromise of iron–sulfur cluster biogenesis (32) (reviewed in 11). Thus, the cytosolic iron depletion that we observe in FA cells conforms to a pattern of compartmental iron misregulation observed when iron–sulfur cluster biogenesis is impaired, characterized by mitochondrial iron overload and cytosolic iron depletion.
Therapeutic approaches to treatment of FA presently include three broad areas: methods to increase frataxin expression by decreasing heterochromatin formation at the GAA repeat (33–35), methods to chelate iron and thereby reduce associated oxidative stress (9) (10) and methods to directly reduce oxidative stress (36), which have recently been shown to be effective in a fly model for FA (37).
Since evidence in favor of cytosolic iron depletion has not been previously reported in FA patients of cell lines, there have been no efforts to increase cytosolic iron stores in patients. Interestingly, several studies have reported that adverse phenotypes can be ameliorated by treatment with hemin, although the mechanism is unknown (38,39). Perhaps hemin treatment increases frataxin expression by increasing cytosolic iron levels, as we have observed in this study when cells were treated with the iron salt, FAC (Figs 3C and 5).
Our results imply that the proposed cytosolic iron depletion that develops with frataxin deficiency may exacerbate disease by further reducing frataxin transcription. Since we observed that frataxin levels varied over at least a 2.5-fold range with manipulations of iron, we suggest that frataxin expression could potentially be increased significantly in patients by judicious management of iron status. We conclude that addition of an iron supplementation regime that would replenish cytosolic iron might be of value in conjunction with other therapeutic approaches such as HDAC inhibitors (35), iron chelators that would reduce mitochondrial iron stores (9,40), and antioxidants that would mitigate the effects of mitochondrial iron overload (36).
MATERIALS AND METHODS
Chemicals and cell cultures
FAC and deferoxamine (DFO, commercial name ‘desferal’) were purchase from Sigma (St Louis, MO). HEK293 cell line was obtained from ATCC (Rockville, MD), lymphoblasts GM15851 and AG15799 (healthy), GM15849, GM16215 and GM16200 (unaffected carriers), GM15850, GM16209, GM16197, GM16214 and GM04079 (FA patients), apparently healthy fibroblasts, GM08399, AG09429 and GM08402, and FA patient fibroblasts GM03665, GM03816 and GM04078 from the Coriell Cell Repostories (Camden, NJ). All the medium and antibiotics (when necessary) were bought from Invitrogen (Carlsbad, CA). The condition for cultivation of the cells was followed according to the instruction of the manufactories. Cells were incubated in a humidified atmosphere of 5% CO2 at 37°C.
Northern blot and qRT–PCR
Total RNA was isolated from 80% confluence of cells after treatment as needs with TRizol (Invitrogen). polyA RNA for northern blot was isolated with PolyATract mRNA isolation kit (cat# Z5210, Promega, Madison, WI). Northern hybridization was carried out and the blot was exposed to PhosphoImage screen and detected with Typhoon 9200 (GE Healthcare). For qRT–PCR, 1 µg total RNA was used in the reverse transcription reaction with random primers (Applied Biosystems, cat# 4368814). Quantitative PCR was carried out with a kit (Invitrogen, cat#11744). Primers for FXN gene are: 5′-CCTTGCAGACAAGCCATACA-3′ and 5′-CCACTGGATGGAGAAGATAG-3′, for control GAPDH gene are: 5′-TGCACCACCAACTGCTTAGC-3′ and 5′-GGCATGGACTGTGGTCATGAG-3′.
Plasmid construction and transfection
Wild-type full length of human frataxin gene coding for 210 amino acids was amplified with primers 5′-AATCTCGAGATGGAAACTCTCGGGCGCCGCGCAGTAG-3′ and 5′-GGGGGAAGCTTTCAAGCATCTTTTCCGGAATAGGCC-3′, then cloned into plasmid pcDNA3.1(−) with restriction sites (XhoI and HindIII) to generate plasmid pcDNA3.1-mito-hFRDA. Site-directed mutagenesis was carried out with QuikChange® II Site-Directed Mutagenesis Kits (Stratagene, La Jolla, CA) to mutate 200th and 203rd amino acid leucine (L) into methionine (M) to produce plasmid pcDNA3.1-mito-hFRDA L200/203M with primers 5′-CCAAACTGGACATGTCTTCCATGGCCTATTCCGG-3′ and 5′-CCGGAATAGGCCATGGAAGACATGTCCAGTTTGG-3′. Plasmid transfections were performed using Fugene6 (Roche Applied Science, Indianapolis, IN) according to the manufactur′s instructions. Stable cell line formation was confirmed by western and metabolic labeling.
Reporter gene assays
A template DNA construct FXN81658 (16) or a BAC construct RP11-265B8 (BACPAC Resources Center, Oakland, CA) was used to generate different promoter constructs for cloning by PCR. Promoter fragments that extended 5′ of the initiation methionine of FXN exon 1 were cloned by adding KpnI and NcoI sites to the PCR primers, and were then inserted into pGL4.10[luc2] (Promega) (17). Cloned fragments contained the 1.2 kb promoter region (16) for construct FXN-luc1, a larger 1.8 kb fragment for FXN-luc2, or a 3.2 kb fragment in FXN-luc3. In addition, a fragment that contained the promoter (1.3 kb), the entire first exon, and the first 1241 bases of intron 1 of human FXN was included in the FXN-GAA-luc plasmid. For the FXN-GAA-luc plasmid, a reverse primer containing the splice acceptor site from the 3′ end of intron 1 and an NcoI site was designed so that the FXN coding sequence in exon 1 would be translated in frame with the luciferase open reading frame from pGL4.10[luc2] (16). The four plasmids, generated as FXN-luc1, FXN-luc2, FXN-luc3 and FXN-GAA-luc, were, respectively, co-transfected with pGL4.75 into HEK293, HeLa and RD4 cells. The luciferase assays were conducted with Veritas microplate luminometer (Turner Biosystems, Sunnyville, CA) according to the manufacturer's manual (Promega, cat#TM040). To control for transfection efficiency in each well, firefly luciferase activity was normalized to Rellina luciferase activity.
Metabolic labeling
HEK293 cells and stably transfected HEK293 with pcDNA3.1-mito-hFRDA L200/203M were cultivated to reach confluence 80%, then labeled with [35S]-methionine (Perkin Elmer, Waltham, MA) for 1 h. After radiolabeling, the cells were chased. Harvested cells were lysed, followed by IP with antibody against human frataxin (MitoScience, Eugene, OR). After the first IP, a second IP (recapture) was performed as previously described (41).
Western blot
Western blot analysis was performed as previously described (42), using the above-described frataxin antibody. IRP1 and IRP2 antibodies were polyclonal, raised from rabbit (29,14). H- and L-ferritin antibodies against human spleen ferritin were purchased from Santa Cruz Biotech Inc. (Santa Cruz, CA), transferrin receptor antibody from Zymed (San Francisco, CA), α-tubulin and actin antibodies from Sigma. Quantification of the density of the western bands was done with program ImageJ (http://rsb.info.nih.gov/ij/).
RNA mobility shift and in-gel aconitase activity assays
Gel retardation assays that separate human IRP1 and IRP2 and in-gel aconitase activity assays were performed as described previously (14).
Statistical analysis
All statistical analysis in this study was done with the Student's t-test (http://www.danielsoper.com/statcalc/).
FUNDING
This work was supported by the intramural program of the NICHD, National Institutes of Health, and in part by Friedreich ataxia research association.
ACKNOWLEDGEMENTS
We thank Dr Manik Ghosh for preparation of the IRE probe, and Dr Karen Usdin for the generous gift of plasmid FXN81658. Funding to pay the Open Access publication charges for this article was provided by the intramural program of NICHD.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Pandolfo M. Iron and Friedreich ataxia. J. Neural. Transm. Suppl. 2006:143–146. doi: 10.1007/978-3-211-45295-0_22. [DOI] [PubMed] [Google Scholar]
- 2.Babady N.E., Carelle N., Wells R.D., Rouault T.A., Hirano M., Lynch D.R., Delatycki M.B., Wilson R.B., Isaya G., Puccio H. Advancements in the pathophysiology of Friedreich's Ataxia and new prospects for treatments. Mol. Genet. Metab. 2007;92:23–35. doi: 10.1016/j.ymgme.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bencze K.Z., Kondapalli K.C., Cook J.D., McMahon S., Millan-Pacheco C., Pastor N., Stemmler T.L. The structure and function of frataxin. Crit. Rev. Biochem. Mol. Biol. 2006;41:269–291. doi: 10.1080/10409230600846058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puccio H., Simon D., Cossee M., Criqui-Filipe P., Tiziano F., Melki J., Hindelang C., Matyas R., Rustin P., Koenig M. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat. Genet. 2001;27:181–186. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- 5.Koeppen A.H., Michael S.C., Knutson M.D., Haile D.J., Qian J., Levi S., Santambrogio P., Garrick M.D., Lamarche J.B. The dentate nucleus in Friedreich's ataxia: the role of iron-responsive proteins. Acta. Neuropathol. 2007;114:163–173. doi: 10.1007/s00401-007-0220-y. [DOI] [PubMed] [Google Scholar]
- 6.Michael S., Petrocine S.V., Qian J., Lamarche J.B., Knutson M.D., Garrick M.D., Koeppen A.H. Iron and iron-responsive proteins in the cardiomyopathy of Friedreich's ataxia. Cerebellum. 2006;5:257–267. doi: 10.1080/14734220600913246. [DOI] [PubMed] [Google Scholar]
- 7.Wong A., Yang J., Cavadini P., Gellera C., Lonnerdal B., Taroni F., Cortopassi G. The Friedreich's ataxia mutation confers cellular sensitivity to oxidant stress which is rescued by chelators of iron and calcium and inhibitors of apoptosis. Hum. Mol. Genet. 1999;8:425–430. doi: 10.1093/hmg/8.3.425. [DOI] [PubMed] [Google Scholar]
- 8.Richardson D.R., Mouralian C., Ponka P., Becker E. Development of potential iron chelators for the treatment of Friedreich's ataxia: ligands that mobilize mitochondrial iron. Biochim. Biophys. Acta. 2001;1536:133–140. doi: 10.1016/s0925-4439(01)00041-2. [DOI] [PubMed] [Google Scholar]
- 9.Richardson D.R. Friedreich's ataxia: iron chelators that target the mitochondrion as a therapeutic strategy? Expert Opin. Investig. Drugs. 2003;12:235–245. doi: 10.1517/13543784.12.2.235. [DOI] [PubMed] [Google Scholar]
- 10.Boddaert N., Le Quan Sang K.H., Rotig A., Leroy-Willig A., Gallet S., Brunelle F., Sidi D., Thalabard J.C., Munnich A., Cabantchik Z.I. Selective iron chelation in Friedreich ataxia: biologic and clinical implications. Blood. 2007;110:401–408. doi: 10.1182/blood-2006-12-065433. [DOI] [PubMed] [Google Scholar]
- 11.Rouault T.A., Tong W.H. Opinion: Iron-sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat. Rev. Mol. Cell Biol. 2005;6:345–351. doi: 10.1038/nrm1620. [DOI] [PubMed] [Google Scholar]
- 12.Schueck N.D., Woontner M., Koeller D.M. The role of the mitochondrion in cellular iron homeostasis. Mitochondrion. 2001;1:51–60. doi: 10.1016/s1567-7249(01)00004-6. [DOI] [PubMed] [Google Scholar]
- 13.Knight S.A., Sepuri N.B., Pain D., Dancis A. Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J. Biol. Chem. 1998;273:18389–18393. doi: 10.1074/jbc.273.29.18389. [DOI] [PubMed] [Google Scholar]
- 14.Tong W.H., Rouault T.A. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab. 2006;3:199–210. doi: 10.1016/j.cmet.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Campuzano V., Montermini L., Molto M.D., Pianese L., Cossee M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., et al. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 16.Greene E., Entezam A., Kumari D., Usdin K. Ancient repeated DNA elements and the regulation of the human frataxin promoter. Genomics. 2005;85:221–230. doi: 10.1016/j.ygeno.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 17.Greene E., Mahishi L., Entezam A., Kumari D., Usdin K. Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res. 2007;35:3383–3390. doi: 10.1093/nar/gkm271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Condo I., Ventura N., Malisan F., Tomassini B., Testi R. A pool of extramitochondrial frataxin that promotes cell survival. J. Biol. Chem. 2006;281:16750–16756. doi: 10.1074/jbc.M511960200. [DOI] [PubMed] [Google Scholar]
- 19.Cavadini P., Adamec J., Taroni F., Gakh O., Isaya G. Two-step processing of human frataxin by mitochondrial processing peptidase. Precursor and intermediate forms are cleaved at different rates. J. Biol. Chem. 2000;275:41469–41475. doi: 10.1074/jbc.M006539200. [DOI] [PubMed] [Google Scholar]
- 20.Iwai K., Drake S.K., Wehr N.B., Weissman A.M., LaVaute T., Minato N., Klausner R.D., Levine R.L., Rouault T.A. Iron-dependent oxidation, ubiquitination, and degradation of iron regulatory protein 2: implications for degradation of oxidized proteins. Proc. Natl. Acad. Sci. USA. 1998;95:4924–4928. doi: 10.1073/pnas.95.9.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson E.S., Rawlins M.L., Leibold E.A. Oxygen and iron regulation of iron regulatory protein 2. J. Biol. Chem. 2003;278:40337–40342. doi: 10.1074/jbc.M302798200. [DOI] [PubMed] [Google Scholar]
- 22.Rouault T.A. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 23.Lobmayr L., Brooks D.G., Wilson R.B. Increased IRP1 activity in Friedreich ataxia. Gene. 2005;354:157–161. doi: 10.1016/j.gene.2005.04.040. [DOI] [PubMed] [Google Scholar]
- 24.Stehling O., Elsasser H.P., Bruckel B., Muhlenhoff U., Lill R. Iron-sulfur protein maturation in human cells: evidence for a function of frataxin. Hum. Mol. Genet. 2004;13:3007–3015. doi: 10.1093/hmg/ddh324. [DOI] [PubMed] [Google Scholar]
- 25.Seznec H., Simon D., Bouton C., Reutenauer L., Hertzog A., Golik P., Procaccio V., Patel M., Drapier J.C., Koenig M., et al. Friedreich ataxia: the oxidative stress paradox. Hum. Mol. Genet. 2005;14:463–474. doi: 10.1093/hmg/ddi042. [DOI] [PubMed] [Google Scholar]
- 26.Wallander M.L., Leibold E.A., Eisenstein R.S. Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim. Biophys. Acta. 2006;1763:668–689. doi: 10.1016/j.bbamcr.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hentze M.W., Muckenthaler M.U., Andrews N.C. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 28.Meyron-Holtz E.G., Ghosh M.C., Iwai K., LaVaute T., Brazzolotto X., Berger U.V., Land W., Ollivierre-Wilson H., Grinberg A., Love P., et al. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 2004;23:386–395. doi: 10.1038/sj.emboj.7600041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyron-Holtz E.G., Ghosh M.C., Rouault T.A. Mammalian tissue oxygen levels modulate iron-regulatory protein activities in vivo. Science. 2004;306:2087–2090. doi: 10.1126/science.1103786. [DOI] [PubMed] [Google Scholar]
- 30.Rotig A., de Lonlay P., Chretien D., Foury F., Koenig M., Sidi D., Munnich A., Rustin P. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat. Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 31.Schoenfeld R.A., Napoli E., Wong A., Zhan S., Reutenauer L., Morin D., Buckpitt A.R., Taroni F., Lonnerdal B., Ristow M., et al. Frataxin deficiency alters heme pathway transcripts and decreases mitochondrial heme metabolites in mammalian cells. Hum. Mol. Genet. 2005;14:3787–3799. doi: 10.1093/hmg/ddi393. [DOI] [PubMed] [Google Scholar]
- 32.Seznec H., Simon D., Monassier L., Criqui-Filipe P., Gansmuller A., Rustin P., Koenig M., Puccio H. Idebenone delays the onset of cardiac functional alteration without correction of Fe-S enzymes deficit in a mouse model for Friedreich ataxia. Hum. Mol. Genet. 2004;13:1017–1024. doi: 10.1093/hmg/ddh114. [DOI] [PubMed] [Google Scholar]
- 33.Burnett R., Melander C., Puckett J.W., Son L.S., Wells R.D., Dervan P.B., Gottesfeld J.M. DNA sequence-specific polyamides alleviate transcription inhibition associated with long GAA.TTC repeats in Friedreich's ataxia. Proc. Natl. Acad. Sci. USA. 2006;103:11497–11502. doi: 10.1073/pnas.0604939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herman D., Jenssen K., Burnett R., Soragni E., Perlman S.L., Gottesfeld J.M. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat, Chem, Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 35.Gottesfeld J.M. Small molecules affecting transcription in Friedreich ataxia. Pharmacol. Ther. 2007;116:236–248. doi: 10.1016/j.pharmthera.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myers L., Farmer J.M., Wilson R.B., Friedman L., Perlman S.L., Subramony S.H., Gomez C.M., Ashizawa T., Wilmot G.R., Mathews K.D., et al. Antioxidant use in Friedreich ataxia. J. Neurol. Sci. 2008;267:174–176. doi: 10.1016/j.jns.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson P.R., Kirby K., Orr W.C., Hilliker A.J., Phillips J.P. Hydrogen peroxide scavenging rescues frataxin deficiency in a Drosophila model of Friedreich's ataxia. Proc. Natl. Acad. Sci. USA. 2008;105:611–616. doi: 10.1073/pnas.0709691105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarsero J.P., Li L., Wardan H., Sitte K., Williamson R., Ioannou P.A. Upregulation of expression from the FRDA genomic locus for the therapy of Friedreich ataxia. J. Gene Med. 2003;5:72–81. doi: 10.1002/jgm.320. [DOI] [PubMed] [Google Scholar]
- 39.Napoli E., Morin D., Bernhardt R., Buckpitt A., Cortopassi G. Hemin rescues adrenodoxin, heme a and cytochrome oxidase activity in frataxin-deficient oligodendroglioma cells. Biochim. Biophys. Acta. 2007;1772:773–780. doi: 10.1016/j.bbadis.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Sohn Y.S., Breuer W., Munnich A., Cabantchik Z.I. Redistribution of accumulated cell iron: a modality of chelation with therapeutic implications. Blood. 2008;111:1690–1699. doi: 10.1182/blood-2007-07-102335. [DOI] [PubMed] [Google Scholar]
- 41.Tong W.H., Rouault T. Distinct iron-sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells. EMBO J. 2000;19:5692–5700. doi: 10.1093/emboj/19.21.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li K., Tong W.H., Hughes R.M., Rouault T.A. Roles of the mammalian cytosolic cysteine desulfurase, ISCS, and scaffold protein, ISCU, in iron-sulfur cluster assembly. J. Biol. Chem. 2006;281:12344–12351. doi: 10.1074/jbc.M600582200. [DOI] [PubMed] [Google Scholar]
- 43.Branda S.S., Cavadini P., Adamec J., Kalousek F., Taroni F., Isaya G. Yeast and human frataxin are processed to mature form in two sequential steps by the mitochondrial processing peptidase. J. Biol. Chem. 1999;274:22763–22769. doi: 10.1074/jbc.274.32.22763. [DOI] [PubMed] [Google Scholar]