Abstract
Rotaviruses are the major etiological agents of diarrhea in children less than 5 years of age. Two unusual rotavirus strains not previously reported in India, G11P[25] (CRI 10795) and G3P[3] (CRI 33594) were isolated from faecal samples of asymptomatic children in India. The strains were characterized by sequence analysis of the genes encoding the VP7, VP4, VP6, and NSP4. The G11P[25] strain was closely related to the human G11P[25] strains from Bangladesh (with 98% identity at the nucleotide [nt] level and the amino acid [aa] level for the VP7 gene and 96% identity at the nt and 98% at the aa level for the VP4 gene). The G3P[3] strain was found to be related to a G3P[3] strain isolated in Thailand (CMH222; 88% identity at the nt level and 97% at aa level for the VP7 gene and 84% identity at the nt level and 90% at the aa level for the VP4 gene). Phylogenetic analysis of the VP6 and the NSP4 genes revealed that the Vellore G11P[25] strain was of VP6 subgroup II and NSP4 genotype B. The G3P[3] strain was identified as NSP4 genotype C and the VP6 gene showed 97% identity at the deduced amino acid level with strain CMH222 (Thailand) strain but did not cluster with sequences of SGI, SGII, SGI+II or SG-nonI/nonII. Both strains had gene segments of animal rotavirus origin suggesting inter-species transmission of rotavirus, and in the case of G11P[25] possibly underwent reassortment subsequently with human strains resulting in an animal-human hybrid strain.
Keywords: rotavirus, genotyping, phylogenetic analysis, zoonotic transmission
INTRODUCTION
Rotaviruses are a member of the Reoviridae family and are the most common cause of dehydrating gastroenteritis in children [Kapikian et al., 2001]. Worldwide, rotavirus kills an estimated 611,000 children every year, accounting for about 5% of all deaths among those younger than 5 years [Parashar et al., 2006].
To date, at least 15 different G types and 28 different P types have been found in human and animal infections, determined serologically or through the diversity of the genes encoding the VP7, and VP4, respectively [Estes, 1996; Kapikian et al., 2001; Martella et al., 2006; Chan-It et al., 2007; Khamrin et al., 2007]. Currently, the commonest human rotavirus strains worldwide belong to G1, G2, G3, G4, and G9 types in association with either P[4], P[6] or P[8] types [Santos and Hoshino, 2005]. Rotavirus strains diversity is generated through the accumulation of point mutations leading to antibody-escape mutants, or through reassortment of co-circulating strains upon dual infection of a single cell. Reassortment among human strains is unlikely to have a major public health impact as the antigenic properties remain unchanged. However, reassortment between animal and human strains may result in the introduction of strains and proteins to which there is no pre-existing immunity. From time to time, novel rotavirus types emerge in the human population, most likely through zoonotic transmission, and these are often localized temporally and/or geographically. Zoonotic transmission is likely to result in asymptomatic infection and no onward transmission as animal strains replicate poorly in the human host. However, there have been a few reports of symptomatic infection by rotavirus strains with distinct animal origin like the AU-1, and HCR3 strains, suggesting that sporadic disease by animal strains is also possible [Nakagomi et al., 1987; Nakagomi and Nakagomi, 2000]. Reassortment with a human strain provides the replicative advantage that may result in a strain to persist and spread within the human population. Occasionally these “unusual” strains are modified through reassortment and transmitted within the human population to become global strains of public health importance. This is exemplified by rotavirus G9 strains that may have emerged through reassortment between animal and human strains to spread globally in the mid-1990s, and become one of the most frequently identified genotypes circulating globally at present.
In this study, we report the first two cases in India of human infections identified as caused by rotavirus G11P[25] and G3P[3] strains and their molecular characterization through sequence analysis of the genes encoding VP7, VP4, VP6, and NSP4.
MATERIALS AND METHODS
Samples
A total of 452 children were recruited and monitored for rotavirus infection for a period of 3 years as part of a community based birth cohort study in Vellore, South India. The children were from an urban slum area, overcrowded with closely clustered houses with garbage dumps, open drains, inadequate sanitation, and with few residents owning their homes. Surveillance stool samples were collected from these children every 2 weeks, and daily during each episode of diarrhea. Voluntary, informed consent was obtained from parents of all children prior to enrollment, and the study was approved by the Research Committee of the Christian Medical College, Vellore. The epidemiological setting of this cohort, details of monitoring of the children, and the method of sample collection have been described previous [Banerjee et al., 2006; Banerjee et al., 2007]. All stool samples were screened for rotavirus using an enzyme-linked immunoassay (EIA) detecting the VP6 antigen as per the manufacturer’s instructions (Rota IDEIA, Thermo Fisher Scientific, United Kingdom).
Viral RNA Extraction and Genotyping
The EIA-positive specimens were processed for extraction of viral RNA using the guanidium isothiocyanate-silica method [Boom et al., 1990]. Complementary DNA was synthesized from the extracted viral RNA through reverse transcription in the presence of random hexamers. G and P typing was performed using VP7- and VP4-specific multiplex hemi-nested RT-PCRs as described previously [Gouvea et al., 1990; Gentsch et al., 1992; Iturriza-Gomara et al., 2004]. Amplification of the VP6 and NSP4 genes was performed using primers described previously [Ciarlet et al., 2000; Iturriza Gomara et al., 2002].
When strains failed to genotype through the amplification of genotype-specific fragments, the first round PCR products generated through the use of consensus primers were sequenced and the genotype determined through sequence and phylogenetic analysis.
Sequencing of VP7, VP4, VP6, and NSP4 Encoding Gene
Sequencing was done using the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA). For sequencing of the VP7, and VP4 genes, the same primer pairs as in the first round PCR reaction were used. Amplicons of the genes encoding the VP6 specificities were sequenced using the oligonucleotide primers VP6-F and VP6-R [Iturriza Gomara et al., 2002]. Sequencing was also carried out for the NSP4 gene using the primers NSP4-F’ and NSP4-R [Ciarlet et al., 2000].
The PCR amplicons obtained by the RT-PCR were purified and sequenced in both directions. Sequences were resolved in an automated DNA sequencer (ABI PRISM 310 Genetic Analyzer [Applied Biosystems, Foster City, CA]), and electropherograms were analyzed using the sequencing analysis software (Sequence Navigator, version 1.01, Applied Biosystems). The nt and deduced aa sequences of VP4, VP6, VP7, and NSP4 genes were compared with sequences available in the NCBI (National Center for Biotechnology Information) GenBank database using BLAST (Basic Local Alignment Search Tool) program.
Sequence Analysis
Multiple alignments and phylogenetic analysis were performed using Bioedit (version 7.0.5.3) (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and dendogram was constructed using MEGA version 3.1 software [Kumar et al., 2004] and/or Bionumerics software (Applied Maths, Kortrij, Belgium). Dendrograms were generated, and confirmed by at least two different methods, Neighbour Joining, Maximum Parsimony and/or Maximum Likelihood. Genetic clusters, indicated with parentheses, were defined when bootstrap values were ≥95% (values not shown). Partial nt sequences of VP7, VP4, VP6, and NSP4 of the G11P[25] and G3P[3] strains were submitted to the GenBank database (accession numbers; EF014906, EF202610, EF175922, EF175923, EF175919, EF202611, EF175920, EF175921).
RESULTS
Two strains that failed to genotype (CRI 10795 and CRI 33594) in the conventional multiplex PCRs were identified as G11P[25] and G3P[3]. These two infections were asymptomatic and were identified from routine surveillance stool samples collected on September 15, 2003 and August 8, 2005, respectively. Both the affected children were male and were 6 and 30 months old, respectively.
Strain Characterization of G11P[25]
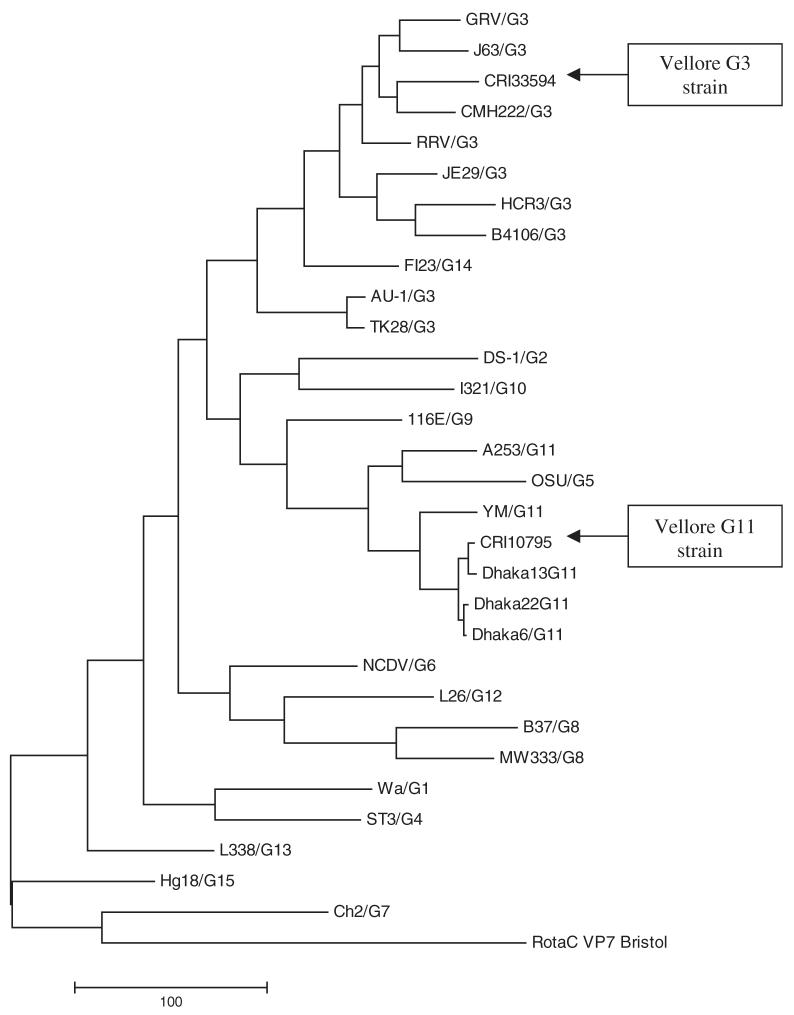
The surveillance stool sample CRI 10795 was positive for rotavirus antigen by ELISA and VP6 PCR. RNA-PAGE showed a long electropherotype and no evidence of mixed infection. However, G and P typing was not possible after the second round of multiplex PCR using type specific primers [Iturriza-Gomara et al., 2004]. A fragment of 881 bp of the VP7 gene amplified in the first round PCR using VP7 consensus primers was sequenced. The partial nucleotide sequence of the VP7 gene and the deduced amino acid sequence were determined and compared with the VP7 gene sequences of prototype strains belonging to G1-G15 types (data not shown). Phylogenetic analysis identified the VP7 gene sequence of CRI 10795 as a G11 strain that closely resembled the previously reported Dhaka6 strain (98% identity at nucleotide level and 98% at the deduced amino acid level). A phylogenetic tree including all the known 15 G prototype sequences was constructed (Fig. 1).
Fig. 1.
Phylogenetic tree constructed from sequences of the VP7 gene of the G11 and G3 rotavirus strains and with other representative G-types using the Maximum Parsimony method. The tree was rooted using VP7 sequence of human Group C rotavirus Bristol strain. The bar indicates the variation scale.
Similarly, partial sequence analysis of the VP4 gene of CRI 10795 identified it as a P[25] strain with maximum identity to the prototype P[25] strain, Dhaka6 (96% identity at nucleotide level and 98% at the deduced amino acid level).
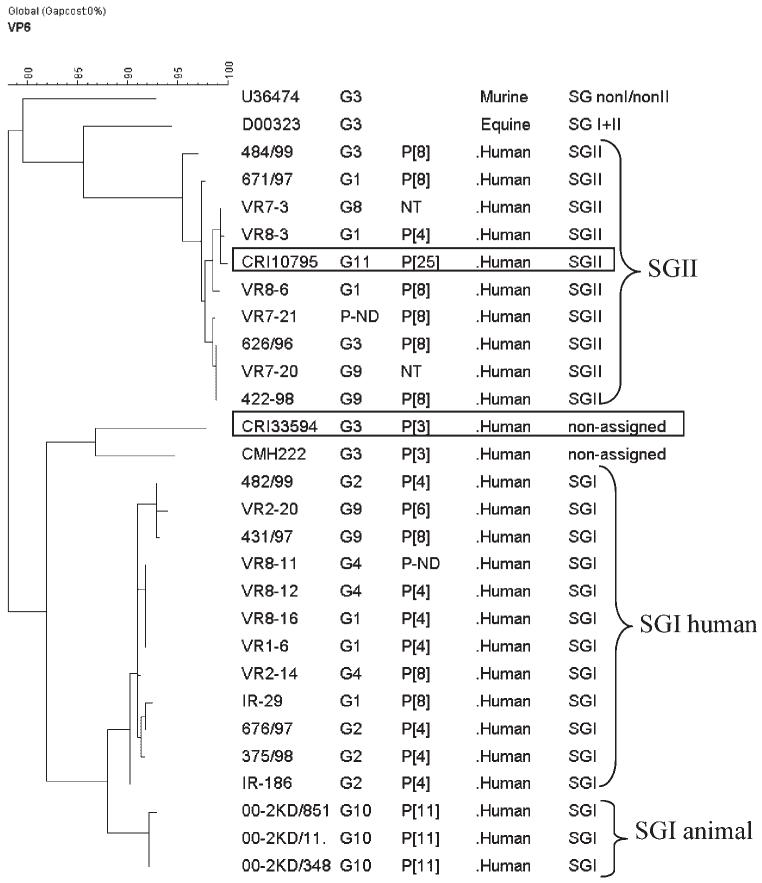
Analysis of the gene encoding VP6 indicated that the G11P[25] strain was of subgroup II and segregated with human strains on phylogenetic analysis (Fig. 2). The gene encoding NSP4 clustered with NSP4 genogroup B and with those of animal origin (Fig. 3).
Fig. 2.
Phylogenetic tree constructed using Maximum Parsimony, and partial sequences of the gene encoding the VP6 of SGI, SGII SGI + II, and SG-nonI/nonII strains of human, and animal origin. Strain identifier, G and P type, and host from which isolates were obtained are indicates in columns 1-4. The strains described in this study are boxed.
Fig. 3.
Phylogenetic tree constructed using Maximum Parsimony and sequences of the gene encoding the NSP4 of strains of human and animal origin. Strain identifier, G, and P type, and host from which isolates were obtained are indicated in columns 1-4. The strains described in this study are boxed.
Strain Characterization of G3P[3]
The CRI 33594 was not genotyped using the type specific primers described earlier [Iturriza-Gomara et al., 2004] but first round products were obtained and were sequenced. Sequence analysis of the genes encoding VP7 and VP4 of this strain identified it as G3P[3] strain. The G3 sequence did not cluster with currently circulating human G3 strains and showed <90% homology with previously identified animal strains. It showed maximum identity with the CMH222 strain with a simian—like VP7 gene (88% identity at nucleotide level and 97% at deduced amino acid level) (Fig. 1). RNA-PAGE showed a long electropherotype and no evidence of mixed infection.
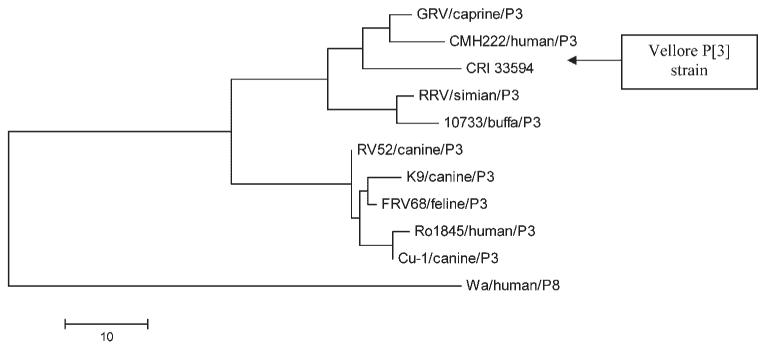
The analysis of the P[3] sequence of CRI 33594 strain failed to reveal strains of >90% homology at the nt level. The closest identity was with the GRV strain which was the first reported G3P[3] caprine rotavirus strain (84% identity at nucleotide level and 90% at the deduced amino acid level) (Fig. 4).
Fig. 4.
Phylogenetic tree constructed from amino acid sequences of the VP8* gene of the Vellore P[3] rotavirus strain and other P[3] strains using the Maximum Parsimony method. The bar indicates the variation scale.
The VP6 sequence of the CRI33594 strain showed 97% identity at the deduced amino acid level with CMH222 strain but did not cluster with SGI, SGII, SGI+II or SG-nonI/nonII sequences and again failed to identify any closely related strains within the recognized subgroups (Fig. 2). The NSP4 genotype was identified as genotype C, previously identified associated only with animal strains (Fig. 3).
DISCUSSION
The genetic analysis of these two rare strains raises interesting insights into their origin, and may suggest that the strain G3P[3] (CRI33594) is the result of direct zoonotic infection, whereas the G11P[25] strain (CRI10795) has arisen through reassortment between animal and human strains.
G11 genotypes are predominantly associated with porcine or bovine strains, except for the two recent reported human strains from Nepal, and Bangladesh [Rahman et al., 2005, 2007; Uchida et al., 2006]. The exact origin of the VP[4] gene of G11P[25] remains unclear as no animal reservoir has yet been identified. Analysis of the prototype P[25] strains (Dhaka6; accession number AY773004) revealed a single aa insertion at position 135 [Rahman et al., 2005]. This extra aa in the length of the VP4 has been demonstrated previously for animal rotavirus strains, suggesting the origin of Dhaka6 may have been zoonotic [Rahman et al., 2005]. Interestingly, the Vellore G11P[25] strain also contained the aa insertion in the gene encoding VP4. Similarly, the analysis of the genes encoding the VP7, and NSP4 would suggest a strain of animal origin, but it is unlikely that this strain is the result of a recent zoonotic introduction as the VP6 clusters with subgroup II sequences more closely related to human rotavirus strains, and not with porcine strains of subgroup II (RU172 and Gottfried strains). This suggests reassortment between animal and human strains.
Characterization of the gene encoding the VP7 of the G3P[3] strain (CRI 33594) revealed a strain more closely related to G3 strains of animal origin than to human G3 strains. The VP7 sequence showed greatest homology with the CMH222 strain from a 2 years child from Thailand, the Korean caprine GRV strain, and the simian RRV strain [Malherbe and Harwin, 1963; Lee et al., 2003; Khamrin et al., 2006]. There are two other reports of human G3 strains showing homology to animal strains, the first was the Ro1845 strain from Israel, and the second, the HCR3 strain from a healthy infant in the USA, both closely related to canine, and feline rotavirus strains [Nakagomi et al., 1990; Nakagomi and Nakagomi, 2000]. Additionally, the P[3] and NSP4-C specificities of the CRI 33594 strain strongly suggest a zoonotic origin. The VP4 sequence of CRI 33594 was closely linked to the GRV strain and the NSP4 sequence clustered with animal rotavirus strains (Figs. 3 and 4). Analysis of four rotavirus genes of the CRI 33594 indicates an animal origin but it is possible that one or more of its other segments were derived from human rotavirus strains making it a human-animal reassortant. The most compelling evidence of interspecies transmission of animal rotaviruses to humans as whole virions is the finding of a high level of homology of a human rotavirus isolate in all its eleven genome segments with rotaviruses commonly found in animals, as exemplified by the HCR3 strain [Nakagomi and Nakagomi, 2000]. CRI 33594 has exactly the same G3P[3] specificity as the HCR3 strain and it is likely that it also traces its origin from animal rotavirus strains.
Analysis of partial VP6 sequences derived from the Vellore and Thailand G3P [3] strains suggested that these were neither SGI nor SGII, and did not cluster with animal strains of SGI + II or SGnonI/nonII, sub-grouped both serologically, and molecularly. This would explain why sub-grouping of the Thailand strain, CMH222 failed when it was previously analyzed by ELISA using a SGI-specific Mab [Khamrin et al., 2006]. Previously, a close relationship between VP6 subgroup, and NSP4 genotype had been established [Iturriza-Gomara et al., 2003]. Additionally segregation by species of genes encoding NSP4 and VP6 was identified [Ciarlet et al., 2000]. To date, common human rotavirus strains possess either SGI/NSP4-A or SGII/NSP4-B specificities. However, the SGI, NSP4-A, and SGII, NSP4-B specificities are also shared by rotaviruses of animal origin. A total of five genotypes of NSP4 have been identified (A-E), but only four different SG specificities or VP6 genogroups have been described (SGI, SGII, SGI + II, and SG-nonI/nonII). The functioning of NSP4, a transmembrane protein, as a receptor for VP6 during morphogenesis supports the observed phylogenetic linkage between VP6 subgroups and NSP4 genotypes, indicating possible constraints to reassortment [Iturriza-Gomara et al., 2003]. Given the link that exists between these two proteins, it is plausible that the same number of SG specificities/VP6 genogroups as of NSP4-genotypes should exist. The “non-assigned” subgroup identified in the Vellore and Thailand G3P[3] strains (CRI33594 and CMH222) may represent a subgroup/VP6 genogroup linked to NSP4-C, suggesting that these two strains are of zoonotic origin. However, the VP6 sequence of the G11P[25] strain (CRI10795) clusters with SGII strains of human origin, whilst its corresponding NSP4 sequence clustered with genotype B sequences of animal strains. This may indicate reassortment between rotaviruses of animal, and human origin, whilst maintaining the VP6-NSP4 link, and suggests that this genetic link is not host-restricted, but VP6 genogroup/NSP4genotype restricted.
The sequence data described here, the asymptomatic nature of the infection in children >6 months of age, strongly suggests zoonotic introduction of these two rotavirus strains. Animal rotaviruses replicate poorly in the human gut, leading to mild or asymptomatic infections, and excretion of the virus is limited, which is unlikely to favor human-to-human transmission of these strains. However, in settings such as Vellore, mixed infections with rotaviruses are frequent, increasing the probability of co-infection with human, and animal strains resulting in reassortment and the emergence of novel rotavirus strains which can spread globally. It is possible that a large group of asymptomatic children in the developing world comprise the “melting pot” for rotavirus reassortment. This report of G11P[25] and G3P[3] human rotavirus infections also highlights the genomic diversity of circulating rotavirus strains and underlines the need for frequent surveillance of domestic animals as they may be potential reservoirs for future rotavirus outbreaks in the human population.
ACKNOWLEDGMENTS
This work was supported by the Wellcome Trust under the Trilateral Cooperative Initiative for Research in Infectious Diseases in the Developing World (Grant number 063144). The authors thank Dr. Vito Martella for a helpful discussion on the possible origins of unusual strains in human infections.
Grant sponsor: Wellcome Trust; Grant number: 063144.
REFERENCES
- Banerjee I, Ramani S, Primrose B, Moses P, Iturriza-Gomara M, Gray JJ, Jaffar S, Monica B, Muliyil JP, Brown DW, Estes MK, Kang G. Comparative study of the epidemiology of rotavirus in children from a community-based birth cohort and a hospital in South India. J Clin Microbiol. 2006;44:2468–2474. doi: 10.1128/JCM.01882-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee I, Gladstone BP, Le Fevre AM, Ramani S, Iturriza-Gomara M, Gray JJ, Brown DW, Estes MK, Muliyil JP, Jaffar S, Kang G. Neonatal infection with G10P[11] rotavirus did not confer protection against subsequent rotavirus infection in a community cohort in Vellore, South India. J Infect Dis. 2007;195:625–632. doi: 10.1086/510853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-It W, Khamrin P, Saekhow P, Pantip C, Thongprachum A, Peerakome S, Ushijima H, Maneekarn N. Molecular characterization of VP4 and VP7 genes of nontypeable strains identifies a novel P[28] genotype in porcine rotaviruses. J Clin Microbiol. 2007 doi: 10.1128/JCM.01709-06. [DOI] [PubMed] [Google Scholar]
- Ciarlet M, Liprandi F, Conner ME, Estes MK. Species specificity and interspecies relatedness of NSP4 genetic groups by comparative NSP4 sequence analyses of animal rotaviruses. Arch Virol. 2000;145:371–383. doi: 10.1007/s007050050029. [DOI] [PubMed] [Google Scholar]
- Estes M. Rotaviruses and their replication. In: Knipe DM, Howley PM, editors. Fields virology. 3rd edition Philadelphia: Lippincott-Raven Publishers; 1996. pp. 1625–1655. [Google Scholar]
- Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, Das BK, Bhan MK. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992;30:1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, Fang ZY. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990;28:276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriza Gomara M, Wong C, Blome S, Desselberger U, Gray J. Molecular characterization of VP6 genes of human rotavirus isolates: Correlation of genogroups with subgroups and evidence of independent segregation. J Virol. 2002;76:6596–6601. doi: 10.1128/JVI.76.13.6596-6601.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriza-Gomara M, Anderton E, Kang G, Gallimore C, Phillips W, Desselberger U, Gray J. Evidence for genetic linkage between the gene segments encoding NSP4 and VP6 proteins in common and reassortant human rotavirus strains. J Clin Microbiol. 2003;41:3566–3573. doi: 10.1128/JCM.41.8.3566-3573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriza-Gomara M, Kang G, Gray J. Rotavirus genotyping: Keeping up with an evolving population of human rotaviruses. J Clin Virol. 2004;31:259–265. doi: 10.1016/j.jcv.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Kapikian AZ, Hoshino Y, Chanock RM. Rotaviruses. In: Knipe DM, editor. Fields virology. 4th edition Philadelphia: Lippincott-Raven Publishers; 2001. pp. 1787–1833. [Google Scholar]
- Khamrin P, Maneekarn N, Peerakome S, Yagyu F, Okitsu S, Ushijima H. Molecular characterization of a rare G3P[3] human rotavirus reassortant strain reveals evidence for multiple human-animal interspecies transmissions. J Med Virol. 2006;78:986–994. doi: 10.1002/jmv.20651. [DOI] [PubMed] [Google Scholar]
- Khamrin P, Maneekarn N, Peerakome S, Chan-It W, Yagyu F, Okitsu S, Ushijima H. Novel porcine rotavirus of genotype P[27] shares new phylogenetic lineage with G2 porcine rotavirus strain. Virology. 2007;361:243–252. doi: 10.1016/j.virol.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEG A3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lee JB, Youn SJ, Nakagomi T, Park SY, Kim TJ, Song CS, Jang HK, Kim BS, Nakagomi O. Isolation, serologic and molecular characterization of the first G3 caprine rotavirus. Arch Virol. 2003;148:643–657. doi: 10.1007/s00705-002-0963-7. [DOI] [PubMed] [Google Scholar]
- Malherbe H, Harwin R. The cytopathic effects of vervet monkey viruses. S Afr Med J. 1963;37:407–411. [PubMed] [Google Scholar]
- Martella V, Ciarlet M, Banyai K, Lorusso E, Cavalli A, Corrente M, Elia G, Arista S, Camero M, Desario C, Decaro N, Lavazza A, Buonavoglia C. Identification of a novel VP4 genotype carried by a serotype G5 porcine rotavirus strain. Virology. 2006;346:301–311. doi: 10.1016/j.virol.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Nakagomi T, Nakagomi O. Human rotavirus HCR3 possesses a genomic RNA constellation indistinguishable from that of feline and canine rotaviruses. Arch Virol. 2000;145:2403–2409. doi: 10.1007/s007050070029. [DOI] [PubMed] [Google Scholar]
- Nakagomi O, Nakagomi T, Hoshino Y, Flores J, Kapikian AZ. Genetic analysis of a human rotavirus that belongs to subgroup I but has an RNA pattern typical of subgroup II human rotaviruses. J Clin Microbiol. 1987;25:1159–1164. doi: 10.1128/jcm.25.7.1159-1164.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagomi O, Ohshima A, Aboudy Y, Shif I, Mochizuki M, Nakagomi T, Gotlieb-Stematsky T. Molecular identification by RNA-RNA hybridization of a human rotavirus that is closely related to rotaviruses of feline and canine origin. J Clin Microbiol. 1990;28:1198–1203. doi: 10.1128/jcm.28.6.1198-1203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar UD, Gibson CJ, Bresse JS, Glass RI. Rotavirus and severe childhood diarrhea. Emerg Infect Dis. 2006;12:304–306. doi: 10.3201/eid1202.050006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Matthijnssens J, Nahar S, Podder G, Sack DA, Azim T, Van Ranst M. Characterization of a novel P[25],G11 human group a rotavirus. J Clin Microbiol. 2005;43:3208–3212. doi: 10.1128/JCM.43.7.3208-3212.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Sultana R, Ahmed G, Nahar S, Hassan ZM, Saiada F, Podder G, Faruque AS, Siddique AK, Sack DA, Matthijnssens J, Van Ranst M, Azim T. Prevalence of G2P[4] and G12P[6] rotavirus, Bangladesh. Emerg Infect Dis. 2007;13:18–24. doi: 10.3201/eid1301.060910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos N, Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol. 2005;15:29–56. doi: 10.1002/rmv.448. [DOI] [PubMed] [Google Scholar]
- Uchida R, Pandey BD, Sherchand JB, Ahmed K, Yokoo M, Nakagomi T, Cuevas LE, Cunliffe NA, Hart CA, Nakagomi O. Molecular epidemiology of rotavirus diarrhea among children and adults in Nepal: Detection of G12 strains with P[6] or P[8] and a G11P[25] strain. J Clin Microbiol. 2006;44:3499–3505. doi: 10.1128/JCM.01089-06. [DOI] [PMC free article] [PubMed] [Google Scholar]