Abstract
UL18 is a human cytomegalovirus class I MHC (MHCI) homolog that binds the host inhibitory receptor LIR-1 and the only known viral MHC homolog that presents peptides. The 2.2-Å structure of a LIR-1/UL18/peptide complex reveals increased contacts and optimal surface complementarity in the LIR-1/UL18 interface compared with LIR/MHCI interfaces, resulting in a >1,000-fold higher affinity. Despite sharing only ≈25% sequence identity, UL18's structure and peptide binding are surprisingly similar to host MHCI. The crystal structure suggests that most of the UL18 surface, except where LIR-1 and the host-derived light chain bind, is covered by carbohydrates attached to 13 potential N-glycosylation sites, thereby preventing access to bound peptide and association with most MHCI-binding proteins. The LIR-1/UL18 structure demonstrates how a viral protein evolves from its host ancestor to impede unwanted interactions while preserving and improving its receptor-binding site.
Keywords: cytomegalovirus, immune evasion, LIR-1, x-ray crystallography
Human cytomegalovirus (HCMV) is prevalent among all human populations, infecting 70–90% of adults (1). HCMV infection is usually asymptomatic in immunocompetent individuals, but the virus causes a latent infection that can be reactivated during immune suppression. To maintain persistence in the presence of a primed immune system, HCMV has developed mechanisms to subvert the host immune response (2). One strategy is to down-regulate cell surface expression of host class I MHC molecules, thereby allowing HCMV-infected cells to avoid recognition by virus-specific cytotoxic T cells. However, cells that lack surface class I MHC molecules are susceptible to lysis by natural killer (NK) cells (3). NK cells express both activating receptors and inhibitory receptors, many of which recognize class I MHC molecules (4). Stimulation of the activating receptors leads to lysis of target cells when expression of class I MHC proteins is down-regulated (5); however, clinical isolates of HCMV can evade NK lysis (6) by down-regulation (7) or inhibition (8) of the activating ligands for NK cells; up-regulation of HLA-E (9), which binds the NK cell inhibitory receptor NKG2A/CD94; and potentially through the expression of two virally encoded class I MHC homologs, UL142 (10) and UL18 (11).
UL18 is a heavily glycosylated transmembrane protein that shares ≈25% sequence identity with class I MHC molecules (11). It associates with the class I MHC light chain, β2-microglobulin (β2m) (12), and with endogenous peptides derived from cytoplasmic proteins that resemble those bound to host class I proteins (13). Peptide binding renders UL18 unique among viral MHC homologs and unusual among host MHC homologs. The role of UL18 in NK cell recognition is controversial; it has been reported to inhibit NK-cell-mediated lysis in some experimental conditions but not others (14).
The only host receptor identified for UL18 is LIR-1 (15) (also known as ILT2, CD85j, or LILRB1) (16). LIR-1 is expressed on most or all monocytes, dendritic and B cells, and subsets of T cells and NK cells, where it functions as an inhibitory receptor by virtue of inhibitory motifs in its cytoplasmic tail and association with SHP-1, a tyrosine phosphatase that inhibits activating signals (15). Unlike the related killer inhibitory receptors (KIRs) on NK cells, which show allele-specific binding to class I MHC molecules (17), LIR-1 and its relative LIR-2 bind a broad range of classical and nonclassical class I MHC proteins (18, 19). Structural and biochemical results suggest that LIR-1 uses a common binding interaction to recognize UL18 and class I MHC molecules, but it binds to UL18 with a >1,000-fold higher affinity (18). Crystal structures of LIR-1 (20), LIR-2 (21), and their complexes with classical (HLA-A2) (22) and nonclassical (HLA-G) (23) class I MHC molecules revealed that the LIR proteins bind to the relatively conserved α3 domain of the heavy chain and to the invariant β2m light chain, rationalizing the broad cross-reactivity of these receptors.
Structural studies involving UL18 have been hindered by its heavy glycosylation. Here we report the 2.2-Å crystal structure of a complex between LIR-1 and a glycosylation-deficient UL18 occupied with an actin-derived peptide. The structure reveals a surprising mimicry of classical class I MHC peptide, suggests that carbohydrates prevent interactions with many potential UL18 binding partners, and explains how UL18 binds more tightly than host MHC proteins to LIR-1, demonstrating how a viral protein can effectively compete with host proteins for a receptor to subvert the host immune response.
Results and Discussion
Production of Glycosylation-Deficient UL18 for Crystallization.
UL18 contains 13 potential N-glycosylation sites, compared with one in human class I MHC molecules (11). When expressed in mammalian cells, the ectodomain of UL18 was heavily (≈55% carbohydrate by weight) and heterogeneously glycosylated (18) and therefore unsuitable for forming well ordered crystals. We produced a more homogeneous form of UL18 by expressing soluble UL18/β2m heterodimers in insect cells, which attach relatively small N-linked oligomannose structures (24), and removing 3 of the 13 potential N-linked sites chosen as described in the supporting information (SI) Materials and Methods. An actin-derived peptide (ALPHAILRL) previously identified as associating with UL18 (13) was added during purification. The resulting mutant protein formed a peptide-binding heterodimer with β2m, showed a reduction in apparent molecular mass of its heavy chain consistent with removal of N-linked carbohydrate, and bound LIR-1 with an affinity similar to that of wild-type UL18 (data not shown).
Crystals were produced from a 1:1 complex between a mutant UL18 and the UL18-binding and class-I-MHC-binding domains of LIR-1, domains 1 and 2 (D1 and D2), respectively. Elimination of one of the N-glycosylation sites (Asn-147) in UL18 was critical for crystallization because this region of UL18 was in a crystal contact. A structure was refined to 2.6-Å resolution (Rcryst = 22.2%, Rfree = 25.1%; Table S1) by using data from crystals that had not been supplemented with peptide. Although both proteins were well ordered, the electron density for the actin peptide was poor, suggesting that a mixture of peptide-filled and empty UL18 had been crystallized. A subsequent UL18/LIR-1 structure with a fully loaded UL18 peptide-binding groove was refined to 2.2 Å (Rcryst = 24.1%, Rfree = 25.9%; Table S1) by using data from crystals that had been incubated with excess peptide. Although there were slight differences in crystal packing, the structures were similar (rmsd of 0.4 Å calculated for all Cα atoms), suggesting that peptide binding does not significantly alter the UL18 structure, which is consistent with studies showing that empty, partially filled, and fully peptide-loaded UL18 bind similarly to LIR-1 (18). Structural analyses were done by using the 2.2-Å structure involving fully peptide-loaded UL18.
Overview of the UL18/LIR-1 Complex Structure.
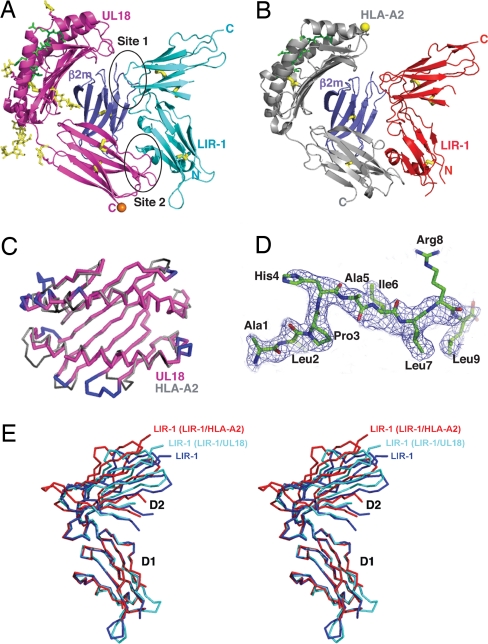
The structures of individual proteins in the LIR-1/UL18 complex resembled their counterparts in structures of the LIR-1/HLA-A2 complex (22) and of LIR-1 or LIR-2 proteins alone (20, 21) (Fig. 1). UL18 and host class I MHC molecules are composed of a heavy chain with an α1-α2 peptide-binding platform and a membrane-proximal α3 domain, a bound peptide, and a noncovalently associated β2m light chain. Despite sharing only ≈25% sequence identity, the UL18 heavy chain was remarkably similar to its MHC counterparts in overall structure and association with bound peptides and β2m. In both types of protein, the peptide-binding α1-α2 superdomain (Figs. 1 A–C and 2) is positioned on top of the Ig-like α3 and β2m domains.
Fig. 1.
Structure of UL18/LIR-1 and comparison with HLA-A2/LIR-1. (A and B) Ribbon diagrams of the UL18/LIR-1 (A) and HLA-A2/LIR-1 (B) complexes. Disulfide bonds are thick yellow sticks, and ordered carbohydrates on UL18 are thin yellow sticks. A predicted O-glycosylation site in the UL18 ectodomain is indicated by an orange sphere, and the single N-glycosylation site on HLA-A2 is indicated as a yellow sphere. (C) Cα superposition of the peptide-binding platforms of UL18 and HLA-A2. Insertions in UL18 are in blue on the UL18 structure, and deletions in UL18 are black on the HLA-A2 structure. (D) The ALPHAILRL peptide model superimposed on a 2.2-Å 2Fo − Fc annealed omit electron density map contoured at 1.0 σ. (E) Stereo superposition of Cα traces of LIR-1 D1-D2 structures after superposition of the D1 domains.
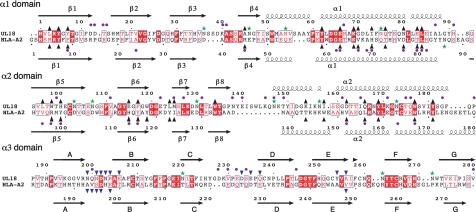
Fig. 2.
Structure-based sequence alignment of UL18 and HLA-A2. Secondary structures are indicated as arrows for β-strands and springs for α-helices. Conserved residues are shown as white letters in red boxes, and conservative substitutions are indicated as red letters. UL18 residues that vary between clinical and laboratory isolates of HCMV (identified by using 21 unique HCMV sequences available in the NCBI protein database, www.ncbi.nlm.nih.gov/) are indicated with a purple dot above the UL18 sequence. Highly variable residues, defined as described previously (46), are indicated by purple dots below the HLA-A2 sequence. The asparagines in potential N-glycosylation sites in both sequences are marked with green stars. Residues in each peptide binding groove are indicated as upward-pointing black triangles. Residues that interact with LIR-1 are marked with downward-pointing blue triangles.
LIR-1 D1-D2 consists of two nearly orthogonal Ig superfamily domains (20). Both LIR-1 domains in the UL18/LIR-1 complex were structurally similar to their counterparts in previous structures (rmsd <1 Å for individual domain superpositions), but their orientation with respect to each other was different in the structures of free LIR-1, UL18-bound LIR-1, and HLA-A2-bound LIR-1 (Fig. 1E). When the D1 domains of LIR-1 pairs were superimposed, the rmsd for the 87 Cα atoms in the D2 domain was 3.0 Å (free LIR-1 versus UL18-bound LIR-1), 4.8 Å (free LIR-1 versus HLA-A2-bound LIR-1), and 3.7 Å (UL18-bound LIR-1 versus HLA-A2-bound LIR-1).
As observed in the LIR-1/HLA-A2 complex, the LIR-1/UL18-binding interaction involved two distinct surfaces on each binding partner, with the D1-D2-interdomain-hinge region contacting β2m (site 1) and the tip of the LIR-1 D1 domain contacting the UL18 α3 domain (site 2) (Fig. 1 A and B). Variable residues in the UL18 α1 domain, identified by sequence analysis of 21 clinical and laboratory isolates (Fig. 2 and Fig. S1), were predicted to contact LIR-1 (25, 26); however, there are no UL18-α1-domain residues within 4 Å of LIR-1 D1-D2. Although LIR-1 D3-D4, which is not present in the crystals, could contact this region of UL18, analytical ultracentrifugation data suggested an extended structure for LIR-1 D1-D4 (18), whereas a substantial bend would be required to achieve D3-D4 contacts with UL18 α1. In addition, the affinities of D1-D4 and D1-D2 for UL18 did not differ significantly (18), which is consistent with no critical contacts between LIR-1 D3-D4 and UL18.
The C termini of the UL18 ectodomain and the LIR-1 D1-D2 fragment were oriented in opposite directions, which is most consistent with a trans interaction on opposing membranes (Fig. 1A). A cis interaction on the same membrane between LIR-1 and class I MHC molecules has been proposed (27) but is hard to rationalize when analytical ultracentrifugation data suggest an extended LIR-1 structure (18), because a cis interaction would require LIR-1 D3, D4, and the C-terminal residues before the transmembrane region to nearly reverse directions compared with D1-D2.
Comparison of UL18 with Classical Class I MHC and MHC Homolog Structures.
In a comparison of UL18 with other class I MHC homologs that share similar or higher percent sequence identities with classical class I MHC proteins, including CD1, HFE, FcRn, ZAG, MIC-A, M10.5, T-22, and the viral class I homologs m144 and m157, UL18 was the most similar to host class I molecules by virtue of being the only homolog to contain an open and peptide-occupied groove (Fig. S2). Thus, unlike the structures of MHC homologs with closed grooves, the α-helices in the UL18 and HLA-A2 peptide-binding platforms were separated by a similar distance (Fig. 1C). The actin-derived ALPHAILRL peptide assumed an extended conformation in the UL18 groove, similar to the conformations of nonamer peptides bound to HLA-A2 and other class I MHC molecules (Fig. 1D and Fig. S3; see the SI Discussion for a detailed comparison). Despite several insertions and deletions in aligned sequences of the peptide-binding domains of UL18 and class I MHC molecules (Fig. 2), superposition of the UL18 and HLA-A2 α1-α2 domains (Fig. 1C) (rmsd = 0.5 Å for 50 Cα atoms) revealed that the changes were usually accommodated without generating the protruding loops that had been suggested by computational modeling of the UL18 structure (25, 26, 28).
Minor differences between the UL18 and HLA-A2 structures included the relative orientation between the α3 domain and α1–α2 platform (Fig. S2), which differed slightly due in part to a four-residue deletion in UL18 at a position corresponding to the short helix in class I MHC molecules that connects the α2 and α3 domains (class I residues 175–178) (Fig. 2). Due to differences in this region and in the loop connecting the α1 and α2 domains in the platform (Fig. 1C), the UL18 heavy chain made more contacts with β2m than did the HLA-A2 heavy chain, leading to a somewhat larger total buried surface area in the UL18/β2m interface (2,900 Å2) than in the HLA-A2/β2m interface (2,700 Å2). The UL18 heavy chain also differed from the class I MHC molecules in that three, rather than two, disulfide bonds were present. In addition to the conserved disulfide bonds in the α2 and α3 domains found in class I MHC molecules and homologs (UL18 residues Cys-102–Cys-174 and Cys-209–Cys-265), the UL18 α3 domain included a disulfide bond linking strands D and E (Cys-238–Cys-253) (Fig. 1A). Both α3 domain disulfide bonds are critical for UL18 stability because disruption of either of them abolished the association with β2m and the binding to LIR-1 (25, 28).
Comparison of LIR Binding to UL18 and Host Class I MHC Proteins.
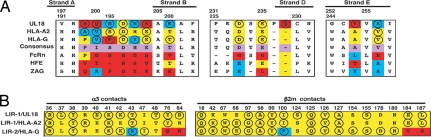
The residues in the site 1 interaction were very similar in the LIR-1/UL18 and LIR-1/HLA-A2 complexes (Fig. 3 and Table 1). In contrast, residues in the site 2 interaction differed, as expected given the sequence differences between the two α3 domains. The sequence differences, combined with slight conformational changes, created a more favorable binding interface at the UL18 site 2 interface with LIR-1. For example, the UL18 site 2 interface included salt bridges (UL18 Asp-202–LIR-1 Arg-36, UL18 Glu-235–LIR-1 Lys-41), hydrogen bonds (UL18 Asn-201–LIR-1 Leu-37, UL18 Asp-233 and Gln-237–LIR-1 Lys-41, UL18 Asn-199 and Glu-206–LIR-1 Thr-43), van der Waals contacts, and water-mediated hydrogen bonds (Table S2), whereas most of the HLA-A2 site 2 interface consisted of van der Waals interactions. Residues from three regions in UL18 α3 contacted LIR-1 (Fig. 3A): the loop between strand A and B (residues 199–206), the loop between strand C and D (residues 231–237), and residues from strand E (residues 254–256). Only residues from the first and third regions were at the LIR-1/HLA-A2 interface. Asp-202 of UL18 (HLA-A2 residue Asp-196), which formed a salt bridge with Arg-36 of LIR-1 and van der Waals contacts with Tyr-76 in both structures, was the only conserved residue in site 2. This residue is conserved throughout LIR-1-binding classical and nonclassical class I MHC molecules but is substituted nonconservatively in the MHC homologs HFE, FcRn, and ZAG, which do not bind LIR-1 (Fig. 3A) (18, 22).
Fig. 3.
Sequence comparisons of residues at LIR-contacting positions. Red, blue, and yellow indicate nonconservatively substituted, conservatively substituted, and conserved positions, respectively. (A) Comparison of the LIR-1-binding (LIR-2-binding for HLA-G) epitope on the α3 domains of UL18, HLA-A2, and HLA-G. Residues contacting LIR-1 or LIR-2 (≤ 4.0 Å) are circled. Corresponding residues on class I MHC homologs that do not bind LIR-1 or LIR-2 are also listed. (B) Comparison of α3- and β2m-contacting residues on LIR-1 (LIR-2 for HLA-G) in the LIR-1/UL18, LIR-1/HLA-A2, and LIR-2/HLA-G complexes. Interacting (≤ 4.0 Å) residues are circled.
Table 1.
Comparison of amino acid contacts at the LIR-1/UL18 and LIR-1/HLA-A2 interfaces
| LIR-1 | UL18 | HLA-A2 |
|---|---|---|
| D1 | UL18 | HLA-A2 |
| Arg-36 | Asp-202* | Asp-196* |
| Leu-37 | Asn-201† | None |
| Tyr-38 | Asn-201, Arg-204, Asp-233 | Val-194, Ser-195 |
| Arg-39 | None | Val-194 |
| Glu-40 | Asp-233 | None |
| Lys-41 | Asp-233†, Glu-235*, Gln-237†, Tyr-254, Ala-256 | Val-194, Glu-198, Val-248 |
| Lys-42 | Tyr-254 | None |
| Thr-43 | Asn-199†, Glu-206† | Ala-193 |
| Thr-48 | Gln-200, Asn-201 | None |
| Tyr-76 | Asp-202, Asn-203 | Asp-196 |
| Arg-84 | Asp-202, Asn-203 | None |
| D1 | β2m in LIR-1/UL18 | β2m in LIR-1/HLA-A2 |
| Gln-18 | Gln-89† | Gln-89† |
| Lys-42 | None | Asp-96* |
| Trp-67 | None | Lys-91, Ile-92†, Val-93 |
| Glu-68 | None | Lys-94 |
| Gly-97 | Ser-88 | Ser-88 |
| Ala-98 | Leu-87, Ser-88† | Leu-87, Ser-88 |
| D2 | β2m in LIR-1/UL18 | β2m in LIR-1/HLA-A2 |
| Tyr-99 | Thr-4, Thr-86, Lys-91 | Thr-4, Thr-86, Lys-91† |
| Ile-100 | Gln-2, Val-85, Thr-86† | Val-85, Thr-86† |
| Ser-124 | Ile-1 | None |
| Gln-125 | Ile-1, Gln-2† | Ile-1, Gln-2† |
| Val-126 | Ile-1, Gln-2, Thr-4, Thr-86 | Gln-2, Arg-3, Thr-4, Thr-86 |
| Ala-127 | Ile-1, Gln-2†, Arg-3 | Gln-2†, Arg-3 |
| Ser-155 | Ile-1 | None |
| Glu-184 | Lys-91† | Lys-91 |
| Leu-187 | Ser-88 | Ser-88 |
*Salt bridge.
†Hydrogen bond.
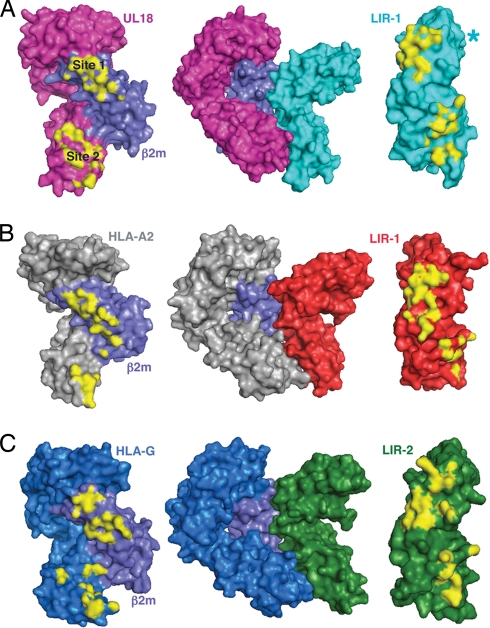
The total solvent-accessible surface area buried at the LIR-1/UL18 interface (≈2,200 Å2) was higher than the areas of typical protein–protein recognition interfaces (1560–1700 Å2) (29) and the LIR-1/HLA-A2 interface (≈1700 Å2) (Fig. 4 A and B). In the heavy-chain-specific binding surface (site 2), LIR-1 made more contacts with UL18 α3 than with HLA-A2 α3 (Table 1 and Fig. 3), and consequently the total buried surface area at this site increased from ≈500 Å2 in the LIR-1/HLA-A2 complex to ≈1200 Å2 for LIR-1/UL18. In contrast, there was slightly less buried surface area at site 1 in the LIR-1/UL18 complex (≈1,000 Å2) than in the LIR-1/HLA-A2 complex (≈1,200 Å2). Thus the majority of the contacts between LIR-1 and UL18 involved the UL18-specific portion of the UL18/β2m heterodimer, whereas the LIR-1 interaction with HLA-A2 involved a majority of contacts to the invariant β2m light chain.
Fig. 4.
Surface representations of LIR proteins and their binding partners. Contact surfaces (≤4.0 Å) are highlighted in yellow. (A) UL18, UL18/LIR-1, and LIR-1. An asterisk marks the position of a LIR-1 loop (residues 148–154) that is disordered in the UL18/LIR-1 complex structure. (B) HLA-A2, HLA-A2/LIR-1, and LIR-1 (PDB ID code 1P7Q). (C) HLA-G, HLA-G/LIR-2, and LIR-2 (PDB ID code 2DYP).
The UL18-specific portion of the LIR-1/UL18 interface was also more complementary than the LIR-1/HLA-A2 interface, as assessed by the shape complementarity index (SC) (30), an index that ranges from 0 (not complementary) to 1 (a perfect fit). The site 1 interaction, although rather large in the LIR-1/HLA-A2 complex, was not particularly complementary compared with its LIR-1/UL18 counterpart (SC values of 0.76 and 0.61 for the LIR-1/UL18 and LIR-1/HLA-A2 complexes, respectively). By comparison, SC values are 0.64–0.68 for typical antibody/antigen interfaces (30). The SC value was 0.59 for the site 2 interaction in the LIR-1/UL18 complex and 0.57 in the LIR-1/HLA-A2 complex. The LIR-2 interaction with HLA-G (23) also involved a large (≈2,200 Å2) (Fig. 4C) but not particularly complementary interface (SC values of 0.61 and 0.51 for sites 1 and 2, respectively). The combination of increased contacts, a large buried surface area, and optimal surface complementarity explains why LIR-1 binds >1,000-fold more tightly to UL18 (Kd = ≈4.0 nM) than it binds to HLA-A2 and other class I molecules (18) and >500-fold more tightly than LIR-2 binds to HLA-G (19).
Comparisons of the UL18/LIR-1, LIR-2, and HLA-G/LIR-2 structures (21, 23) explain why UL18 binds >3,000-fold more tightly to LIR-1 than to LIR-2 (18). An overall similarity between the D2 domains of LIR-1 and LIR-2 suggests that interactions with β2m at site 1 would be preserved in the UL18/LIR-2 interaction. However, a nonconservative substitution in LIR-2 (LIR-1 Ile-100 for LIR-2 Pro-99) might weaken the binding to LIR-2 (Fig. 3B). In addition, one of the loops in LIR-1 D1 that makes contacts with UL18 α3 in site 1 (LIR-1 residues 77–86; LIR-2 residues 77–85) showed an altered conformation in the LIR-2 structures (21, 23), such that a loop with this conformation would not contact UL18 in a LIR-2/UL18 complex. A LIR-2 mutant in which LIR-1 residues were introduced into this loop (Q76Y/R80D/W83R LIR-2) bound UL18 10- to 30-fold more tightly than did wild-type LIR-2 (20), suggesting the potential for flexibility and demonstrating that the wild-type LIR-2 sequence is not optimal for UL18 binding. Thus decreased interactions in both the site 1 and site 2 interfaces in a UL18/LIR-2 complex are predicted to be responsible for the lower affinity of LIR-2 versus LIR-1 for UL18.
UL18 Carbohydrates Prevent Interactions with Potential Binding Partners.
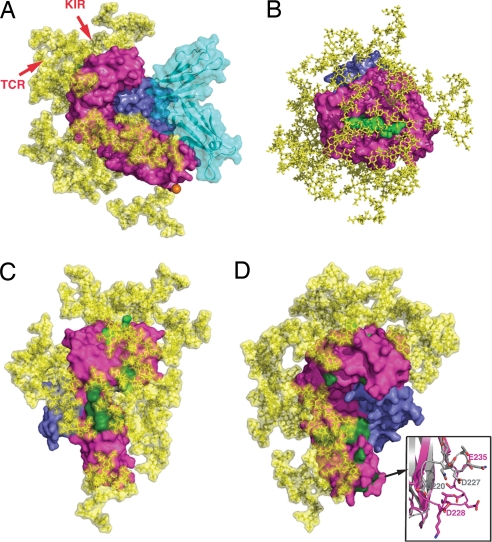
One of the major differences between UL18 and host class I MHC molecules is the large amount of carbohydrate resulting from UL18's 13 potential N-glycosylation sites as compared to the single N-glycan attached to human class I MHC molecules. Ordered carbohydrate was observed at 5 of the 10 potential N-linked sites in the UL18 mutant that was crystallized but not at the single predicted O-glycosylation site (Fig. 1A). Although much of the UL18 carbohydrate was disordered in the LIR-1/UL18 crystal structure, the molecular mass of purified UL18 protein suggests that most, if not all, of its N-linked sites are glycosylated (18). In addition, the 13 potential N-linked sites are conserved in 21 unique UL18 sequences from sequenced HCMV genomes (Fig. S1). Using the coordinates of a typical mammalian N-linked glycan, which can extend up to 30 Å from the asparagine in an Asn-Xaa-Ser/Thr N-glycosylation site (31), we modeled the structure of fully glycosylated UL18 bound to LIR-1 (Fig. 5A). Most of the available surface area on the UL18 heavy chain was predicted to be covered with N-linked carbohydrate, particularly the α1–α2 peptide binding platform of UL18, which includes 10 of 13 potential glycosylation sites (Fig. 5B). Two notable regions lacked carbohydrate: the binding site for LIR-1 and the interaction interface with β2m, suggesting that UL18 evolved to hide other potential binding surfaces, thereby preventing interactions with binding partners of host class I MHC molecules.
Fig. 5.
Interaction sites for potential binding partners highlighted on a fully glycosylated UL18 model. (A) Space-filling representation of the UL18/LIR-1 complex (UL18 in magenta, β2m in slate, peptide in light green, LIR-1 in cyan) with a complex carbohydrate model (yellow) attached to each of the 13 potential N-glycosylation sites. The single predicted O-glycosylation site (residue 281) within the UL18 ectodomain is indicated by an orange sphere. Two to three additional O-glycosylation sites are predicted in the region C-terminal to the UL18 ectodomain fragment that was crystallized, but these sites would be distant from the binding sites for all potential UL18 binding partners. The UL18 counterparts of the approximate binding sites on a class I MHC molecule for a TCR or KIR are indicated by arrows. (B) Top view of the fully glycosylated UL18 peptide binding platform. (C) Potential US2 binding site (dark green) on fully glycosylated UL18. (D) Potential CD8 binding site (dark green) on fully glycosylated UL18. An enlarged view of the CD8-binding loop in the class I MHC α3 domain (gray) is shown with the counterpart UL18 loop (magenta).
To assess the effects of UL18's glycosylation on potential binding interactions, we evaluated the binding sites for class I MHC binding partners by using the fully glycosylated UL18 model. T cell receptors (TCRs) bind across the α1–α2 domain helices, contacting the bound peptide (32). Although the UL18 α1-α2 helices and associated peptide are structurally very similar to a class I MHC/peptide complex, it is unlikely that a TCR could contact the UL18-bound peptide because four predicted N-linked glycans (attached to Asn-75, Asn-147, Asn-157, and Asn-173) were located on the UL18 α-helices surrounding and effectively shielding the bound peptide (Fig. 5A). The footprint of NK cell KIRs involves a similar region on class I MHC molecules (33), thus carbohydrates also likely prevent KIR binding to UL18 (Fig. 5A). In addition, none of the residues critical for class I binding to KIRs (33) are conserved in UL18. Carbohydrates would also block binding of US2, one of several HCMV-encoded proteins that bind to newly synthesized class I MHC molecules in the endoplasmic reticulum, resulting in down-regulation of surface expression (34). US2 binds at the junction between the class I α1–α2 platform and the α3 domain (35); however, two class I MHC residues that interact with US2 (Asp-106 and Pro-267) are substituted by asparagines involved in N-glycosylation sites in UL18 (Asn-107 and Asn-271) (Fig. 5C), and UL18 has no equivalent of class I residue Arg-181, which plays an critical role in binding to US2 (36). Thus in an HCMV-infected cell, UL18 should escape from US2-mediated down-regulation, whereas host class I MHC molecules will not. A similar mechanism may account for why UL18 is not susceptible to down-regulation by either US3 or US11 (34).
A combination of carbohydrate shielding as well as sequence and conformational changes accounts for UL18's inability to interact with CD8, a T cell coreceptor that recognizes the α3 domain of host class I MHC molecules (37). Two predicted N-glycosylation sites (Asn-103 and Asn-220) were near the UL18 counterpart of the CD8 binding surface (Fig. 5D). In addition, most of the residues involved in the class I MHC interaction with CD8 were nonconservatively substituted in UL18 (Fig. 5D), and the α3 domain loop critical for CD8 binding to class I MHC molecules (37) was differently positioned in UL18 (Fig. 5D).
Significance of Peptide Binding by UL18.
Bound peptide is distant from the LIR-1 binding site (Fig. 1A) and not required for UL18 to bind to LIR-1 (18). Thus although the UL18/LIR-1 structure provided a clear picture of how UL18 preferentially binds short peptides with P2 (Leu or Met), P3 (Pro), and P9 (small hydrophobic residue) anchor positions (Fig. S3 and Table S3), it did not address the function of peptide binding. Of possible relevance is whether UL18 proteins from different HCMV strains show the same or different peptide-binding preferences. Although allelic polymorphism in classical class I MHC molecules mapped mainly to the MHC peptide-binding groove, thereby creating different allele-specific peptide-binding preferences (38), variability in UL18 sequences from different laboratory and clinical strains of HCMV systematically mapped to regions other than the UL18 peptide-binding groove (Fig. 2). Because UL18 variability does not create an opportunity for different strain-specific peptide-binding preferences, most or all UL18 proteins are likely to bind a similar class of peptides.
To address whether UL18 might sequester HCMV peptide epitopes from host class I MHC molecules, we investigated whether peptides conforming to the UL18-specific peptide-binding motif were present in the HCMV genome. We found >30 nonameric sequences in the genome of HCMV AD169 that conform to the UL18 binding motif (Fig. S4), thus UL18 could associate with viral as well as host peptides. However, a peptide-sequestering function seems unlikely given that only low levels of UL18 are expressed on virally infected cells (39) and UL18's strict peptide-binding preferences would allow sequestering from only a few class I MHC alleles, such as HLA-A2, whose peptide-binding motif is similar to that of UL18. Alternatively, peptide binding could be required to stabilize the UL18 fold (18) or for binding to an as yet undiscovered immune receptor, as previously hypothesized (40). However, direct contacts to the bound peptide are unlikely due to carbohydrate shielding (Fig. 5B).
Further studies will be required to understand why UL18, alone among viral class I MHC homologs, associates with endogenous peptides and what effects peptide binding has on the HCMV lifecycle and the host anti-viral immune response. The high-resolution UL18/LIR-1 structure allows the rational design of mutants that cannot bind peptides or bind a different class of peptides, which could be tested in viral and/or immunological assays.
Materials and Methods
For further experimental details, see the SI Materials and Methods.
Mutant UL18 (N36Q/N147Q/N220Q/C259S)/ALPHAILRL peptide complexes and LIR-1 D1-D2 were produced as described in the SI Materials and Methods. Two native data sets were collected at 100 K from LIR-1/UL18 complex crystals at the Stanford Synchrotron Radiation Laboratory. The first data set was collected from untreated crystals, and the second was collected from crystals grown from protein solutions supplemented with 2.5 mM ALPHAILRL peptide. Data were processed by using the HKL2000 program (41). The structure was determined by molecular replacement (42) using a LIR-1/HLA-A2 (22) search model and the first data set (Table S1). After rigid-body refinement (42), rebuilding was done by using the program O and 2Fo − Fc annealed omit maps, alternating with refinement using the CNS program (43). Noncrystallographic symmetry restraints (300 kcal/mol·Å2) and individual B factor refinement were used during refinement. A final model at 2.6-Å resolution (Rcrys = 22.2%, Rfree = 25.1%) consisted of residues 1–280 of the UL18 heavy chain; β2m residues 1–99; and LIR-1 residues 4–77, 83–136, 143–147, 155–166, and 170–198. Density for the ALPHAILRL peptide was poor.
The structure was solved again by using data from the second data set (Table S1) (collected from crystals treated with excess peptide) by molecular replacement using the refined 2.6-Å UL18/LIR-1 structure. Clear density was visible for all nine residues of the peptide, UL18, β2m, and most of LIR-1. The model was refined to 2.2-Å resolution (Rcryst = 24.1%, Rfree = 25.9%). The final model consisted of residues 1–281 of UL18; peptide residues 1–9; β2m residues 1–99; LIR-1 residues 4–136, 143–147, 155–198; 362 water molecules, and 214 atoms of carbohydrate (ordered carbohydrate attached to UL18 residues Asn-54, Asn-103, Asn-157, Asn-173, and Asn-271). The side chains of UL18 residues 140 and 229; β2m 58; and LIR-1 31, 156, and 168 were modeled as alanines. The following cysteine pairs were disulfide bonded: LIR-1 26–75, 122–174, 134–144, UL18 102–174, 209–265, 238–253, and β2m 25–80.
For analysis of contacts and buried surface areas, D1 was defined as residues 1–98 and D2 was defined as residues 99–198. The CCP4 program Areaimol (44) was used to calculate buried surface areas using a 1.4-Å probe and to identify interacting residues using the following criteria: a distance of <3.5 Å and a hydrogen bond angle of >90° for hydrogen bonds and a maximum distance of 4.0 Å for a van der Waals interaction. Shape complementarity indices were calculated as described previously (30) by using the Sc program in CCP4 (44). Figures were prepared with PyMOL (45).
Supplementary Material
Acknowledgments.
We thank Inder Nangiana and Peter Snow at the Caltech Protein Expression Center for assistance with expression of UL18, Yan Qi for setting up wild-type UL18/LIR-1 crystallization trials, David Mathog in the Caltech Sequence Analysis Facility for identifying potential UL18-binding peptides encoded by HCMV, and Tara Chapman-Arvedson and members of the P.J.B. laboratory for helpful discussions and critical reading of the manuscript. Z.Y. was supported by a Life Sciences Research Foundation Postdoctoral Fellowship (Howard Hughes Medical Institute Fellow).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3D2U).
This article contains supporting information online at www.pnas.org/cgi/content/full/0804551105/DCSupplemental.
References
- 1.Britt WJ, Alford CA. Cytomegalovirus. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. Vol 2. Philadelphia: Lippincott-Raven; 1996. pp. 2493–2523. [Google Scholar]
- 2.Ploegh HL. Viral strategies of immune evasion. Science. 1998;280:248–253. doi: 10.1126/science.280.5361.248. [DOI] [PubMed] [Google Scholar]
- 3.Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 4.Yokoyama WM, Kim S, French AR. The dynamic life of natural killer cells. Annu Rev Immunol. 2004;22:405–429. doi: 10.1146/annurev.immunol.22.012703.104711. [DOI] [PubMed] [Google Scholar]
- 5.Moser JM, Byers AM, Lukacher AE. NK cell receptors in antiviral immunity. Curr Opin Immunol. 2002;14:509–516. doi: 10.1016/s0952-7915(02)00357-6. [DOI] [PubMed] [Google Scholar]
- 6.Cerboni C, et al. Human cytomegalovirus strain-dependent changes in NK cell recognition of infected fibroblasts. J Immunol. 2000;164:4775–4782. doi: 10.4049/jimmunol.164.9.4775. [DOI] [PubMed] [Google Scholar]
- 7.Tomasec P, et al. Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat Immunol. 2005;6:181–188. doi: 10.1038/ni1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnon TI, et al. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat Immunol. 2005;6:515–523. doi: 10.1038/ni1190. [DOI] [PubMed] [Google Scholar]
- 9.Tomasec P, et al. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. 2000;287:1031. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- 10.Wills MR, et al. Human cytomegalovirus encodes an MHC class I-like molecule (UL142) that functions to inhibit NK cell lysis. J Immunol. 2005;175:7457–7465. doi: 10.4049/jimmunol.175.11.7457. [DOI] [PubMed] [Google Scholar]
- 11.Beck S, Barrell BG. Human cytomegalovirus encodes a glycoprotein homologous to MHC class-I antigens. Nature. 1988;331:269–272. doi: 10.1038/331269a0. [DOI] [PubMed] [Google Scholar]
- 12.Browne H, Smith G, Beck S, Minson T. A complex between the MHC class I homologue encoded by human cytomegalovirus and beta 2 microglobulin. Nature. 1990;347:770–772. doi: 10.1038/347770a0. [DOI] [PubMed] [Google Scholar]
- 13.Fahnestock ML, et al. The MHC class I homolog encoded by human cytomegalovirus binds endogenous peptides. Immunity. 1995;3:583–590. doi: 10.1016/1074-7613(95)90129-9. [DOI] [PubMed] [Google Scholar]
- 14.Wagner CS, Ljunggren HG, Achour A. Immune modulation by the human cytomegalovirus-encoded molecule UL18, a mystery yet to be solved. J Immunol. 2008;180:19–24. doi: 10.4049/jimmunol.180.1.19. [DOI] [PubMed] [Google Scholar]
- 15.Cosman D, et al. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity. 1997;7:273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- 16.Colonna M, et al. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J Exp Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Natarajan K, Dimasi N, Wang J, Mariuzza RA, Margulies DH. Structure and function of natural killer cell receptors: Multiple molecular solutions to self, nonself discrimination. Annu Rev Immunol. 2002;20:853–885. doi: 10.1146/annurev.immunol.20.100301.064812. [DOI] [PubMed] [Google Scholar]
- 18.Chapman TL, Heikema AP, Bjorkman PJ. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral MHC homolog UL18. Immunity. 1999;11:603–613. doi: 10.1016/s1074-7613(00)80135-1. [DOI] [PubMed] [Google Scholar]
- 19.Shiroishi M, et al. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc Natl Acad Sci USA. 2003;100:8856–8861. doi: 10.1073/pnas.1431057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chapman TL, Heikema AP, West AP, Jr, Bjorkman PJ. Crystal structure and ligand binding properties of the D1D2 region of the inhibitory receptor LIR-1 (ILT2) Immunity. 2000;13:727–736. doi: 10.1016/s1074-7613(00)00071-6. [DOI] [PubMed] [Google Scholar]
- 21.Willcox BE, et al. Crystal structure of LIR-2 (ILT4) at 1.8 A: Differences from LIR-1 (ILT2) in regions implicated in the binding of the human cytomegalovirus class I MHC homolog UL18. BMC Struct Biol. 2002;2:6. doi: 10.1186/1472-6807-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willcox BE, Thomas LE, Bjorkman PJ. Crystal structure of HLA-A2 bound to LIR-1, a host and viral MHC receptor. Nat Immunol. 2003;4:913–919. doi: 10.1038/ni961. [DOI] [PubMed] [Google Scholar]
- 23.Shiroishi M, et al. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d) Proc Natl Acad Sci USA. 2006;103:16412–16417. doi: 10.1073/pnas.0605228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomiya N, Betenbaugh MJ, Lee YC. Humanization of lepidopteran insect-cell-produced glycoproteins. Acc Chem Res. 2003;36:613–620. doi: 10.1021/ar020202v. [DOI] [PubMed] [Google Scholar]
- 25.Wagner CS, et al. Structural elements underlying the high binding affinity of human cytomegalovirus UL18 to leukocyte immunoglobulin-like receptor-1. J Mol Biol. 2007;373:695–705. doi: 10.1016/j.jmb.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 26.Cerboni C, et al. Spontaneous mutations in the human CMV HLA class I homologue UL18 affect its binding to the inhibitory receptor LIR-1/ILT2/CD85j. Eur J Immunol. 2006;36:732–741. doi: 10.1002/eji.200425220. [DOI] [PubMed] [Google Scholar]
- 27.Masuda A, Nakamura A, Maeda T, Sakamoto Y, Takai T. Cis binding between inhibitory receptors and MHC class I can regulate mast cell activation. J Exp Med. 2007;204:907–920. doi: 10.1084/jem.20060631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Occhino M, et al. Dissecting the structural determinants of the interaction between the human cytomegalovirus UL18 protein and the CD85j immune receptor. J Immunol. 2008;180:957–968. doi: 10.4049/jimmunol.180.2.957. [DOI] [PubMed] [Google Scholar]
- 29.Jones S, Thornton JM. Principles of protein–protein interaction. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 31.Molinari M. N-glycan structure dictates extension of protein folding or onset of disposal. Nat Chem Biol. 2007;3:313–320. doi: 10.1038/nchembio880. [DOI] [PubMed] [Google Scholar]
- 32.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–66. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 33.Boyington JC, Brooks AG, Sun PD. Structure of killer cell immunoglobulin-like receptors and their recognition of the class I MHC molecules. Immunol Rev. 2001;181:66–78. doi: 10.1034/j.1600-065x.2001.1810105.x. [DOI] [PubMed] [Google Scholar]
- 34.Park B, et al. The MHC class I homolog of human cytomegalovirus is resistant to down-regulation mediated by the unique short region protein (US)2, US3, US6, and US11 gene products. J Immunol. 2002;168:3464–3469. doi: 10.4049/jimmunol.168.7.3464. [DOI] [PubMed] [Google Scholar]
- 35.Gewurz BE, et al. Antigen presentation subverted: Structure of the human cytomegalovirus protein US2 bound to the class I molecule HLA-A2. Proc Natl Acad Sci USA. 2001;98:6794–6799. doi: 10.1073/pnas.121172898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thilo C, et al. Dissection of the interaction of the human cytomegalovirus-derived US2 protein with major histocompatibility complex class I molecules: Prominent role of a single arginine residue in human leukocyte antigen-A2. J Biol Chem. 2006;281:8950–8957. doi: 10.1074/jbc.M507121200. [DOI] [PubMed] [Google Scholar]
- 37.Gao GF, et al. Crystal structure of the complex between human CD8 αα and HLA-A2. Nature. 1997;387:630–634. doi: 10.1038/42523. [DOI] [PubMed] [Google Scholar]
- 38.Madden DR. The three dimensional structure of peptide-MHC complexes. Ann Rev Immunol. 1995;13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- 39.Leong CC, et al. Modulation of natural killer cell cytotoxicity in human cytomegalovirus infection: The role of endogenous class I major histocompatibility complex and a viral class I homolog. J Exp Med. 1998;187:1681–1687. doi: 10.1084/jem.187.10.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prod'homme V, et al. The human cytomegalovirus MHC class I homolog UL18 inhibits LIR-1+ but activates LIR-1− NK cells. J Immunol. 2007;178:4473–4481. doi: 10.4049/jimmunol.178.7.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brünger AT, et al. Crystallography and NMR system: A new software system for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 44.CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 45.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, California: DeLano Scientific; 2002. [Google Scholar]
- 46.Bjorkman PJ, Parham P. Structure, function and diversity of class I major histocompatibility complex molecules. Annu Rev Biochem. 1990;90:253–288. doi: 10.1146/annurev.bi.59.070190.001345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.