Abstract
Multiple-complete-digest mapping is a DNA mapping technique based on complete-restriction-digest fingerprints of a set of clones that provides highly redundant coverage of the mapping target. The maps assembled from these fingerprints order both the clones and the restriction fragments. Maps are coordinated across three enzymes in the examples presented. Starting with yeast artificial chromosome contigs from the 7q31.3 and 7p14 regions of the human genome, we have produced cosmid-based maps spanning more than one million base pairs. Each yeast artificial chromosome is first subcloned into cosmids at a redundancy of ×15–30. Complete-digest fragments are electrophoresed on agarose gels, poststained, and imaged on a fluorescent scanner. Aberrant clones that are not representative of the underlying genome are rejected in the map construction process. Almost every restriction fragment is ordered, allowing selection of minimal tiling paths with clone-to-clone overlaps of only a few thousand base pairs. These maps demonstrate the practicality of applying the experimental and software-based steps in multiple-complete-digest mapping to a target of significant size and complexity. We present evidence that the maps are sufficiently accurate to validate both the clones selected for sequencing and the sequence assemblies obtained once these clones have been sequenced by a “shotgun” method.
With the impressive progress that has been made in the sequencing of the genomes of model organisms such as Caenorhabditis elegans (1), Saccharomyces cerevisiae (2, 3), and others (4, 5), the Human Genome Project is approaching its final phase—large-scale sequencing of human genomic DNA (6, 7). The sequencing of the two largest genomes for which there is extensive experience, C. elegans (100 Mbp) and S. cerevisiae (15 Mbp), has been aided by the existence of high-quality physical maps that were constructed over a period of many years (8–10). Although a small proportion of the human genome has been mapped at high resolution by methods similar to those employed for model organisms (11–13), global physical mapping has proceeded at much lower resolution (14, 15) on the assumption that the final mapping of clones chosen as sequencing templates will be carried out on a “just-in-time” basis. Despite its importance in the overall logic of large-scale genome sequencing, this final phase of the human mapping has received little attention.
We describe here our early experience in analyzing human DNA by the multiple-complete-digest (MCD) restriction fragment mapping technique, which has been developed as a potential solution to the sequence-ready mapping problem. MCD mapping is an extension of the single-complete-digest method employed to produce a high-resolution physical map for S. cerevisiae (9, 10). In that project, a mixture of two restriction enzymes with 6 bp recognition sites, EcoRI and HindIII, was used to digest bacteriophage λ and cosmid clones. A single list of fragment sizes was obtained for each clone. Here we increase the number of enzymes to three and perform the digestions independently. This yields three fragment-size lists for each clone. Every fragment-size list is referred to as one “enzyme domain,” regardless of whether it results from a single-enzyme digest or a two-enzyme double digest. Reconstruction of the underlying fragment ordering and synchronization of this information across enzyme domains is accomplished with the software package dnam (16, 17). High sampling redundancies of ×15–30 are required, not only for closure of the maps, but also for ordering of the fragments and automatic detection of bad data. We require absolute consistency between the maps and the underlying data because, at the restriction fragment level, errors in the analysis of the gel images and subtle aberrations in the individual clones are virtually indistinguishable from errors in the map assembly. The strength of the multiple enzyme system is that it allows the detection of nearly all such problems, making the final maps highly reliable.
MCD mapping makes no detailed assumptions about the clones that are to be fingerprinted. Although our focus has been on cosmid and bacterial-artificial-chromosome (BAC) clones (18), this paper will describe the mapping of cosmids subcloned from yeast artificial chromosomes (YACs). The YACs come from a YAC-based sequence tagged site (STS)-content map (19) of human chromosome 7 (20). The density of STS markers in the chromosome 7 map (average spacing 100 kbp) is higher than that for typical large-scale STS maps, and a high proportion of the STSs are reliably ordered. Most of this map is based on a specialized YAC library that was derived from a monochromosomal hybrid cell line (20). This library has a lower chimerism rate, estimated at less than 15%, than most other YAC libraries. Over the same chromosome, Centre d’Etudes du Polymorphisone Humain “mega-YACs” (15) exhibited a chimerism rate of roughly 50%. In this paper, only hybrid-cell-line-derived YACs that are consistent with the consensus STS map are chosen for subcloning into cosmids, and any cosmid that is chosen for shotgun sequencing is first validated by demonstrating that its restriction pattern is consistent with two independently mapped YACs. This ×2 YAC redundancy provides protection against the risk of passing YAC aberrations down to the final sequence via a cosmid that is a faithful representation of the YAC from which it was subcloned, but not of the human genome.
MATERIALS AND METHODS
Chromosome 7 YACs.
YACs to be subcloned into cosmids are chosen from two target regions on the chromosome 7 STS-content map (20). All but one of the mapped YACs come from the 7q31.3 region. These are named yWSS771, yWSS1346, yWSS1434, yWSS1572, yWSS1613, yWSS1862, and yWSS1980. One other YAC, named yWSS1564, comes from the 7p14 region.
Cosmid Vector.
The MCD cosmid vector, s-Cos-DBI, was derived from the widely used cosmid vector s-Cos-1 (ref. 21; Stratagene). s-Cos-1 was first digested with Eco47III (Stratagene) and the 4376-bp fragment containing the dual cos sites and Ampr gene was gel purified and ligated to produce s-Cos-D. By replacing the polylinker with a synthetic oligodeoxynucleotide in which the two EcoRI sites had been mutated to GAATTT and CAATTC, we obtained s-Cos-DBI, which is suitable for cloning inserts prepared by MboI partial digestion into the single BamHI site; mutation of the flanking EcoRI sites preserves the utility of EcoRI as an MCD mapping enzyme.
Cosmid Libraries.
Total yeast DNA is prepared in agarose plugs (22), partially digested with MboI, and size-selected (35–45 kbp) by pulsed-field agarose gel electrophoresis. Fractionated DNA is electroeluted into dialysis tubing, extracted with phenol, precipitated with ethanol, and ligated to the linearized MCD cosmid vector. The ligation mixture is packaged with Gigapack III Gold (Stratagene); DH5α MCR (Life Technologies, Gaithersburg, MD) is employed as the host. Cosmids containing human DNA inserts are selected by screening with unfractionated human DNA (CLONTECH). Probes are nonradioactively labeled with the Genius System (Boehringer Mannheim).
Purification of Cosmid DNA.
Cosmid DNA mini-preparations are performed with a modified version of the alkaline–lysis protocol of Coulson and Sulston (23). Bacteria are harvested from 1.5-ml cultures and resuspended in 250 μl of buffer containing 50 mM glucose, 25 mM Tris (pH 8.0), 10 mM EDTA (pH 8.0), 0.1 mg/ml RNase A. Final DNA pellets are dissolved in 100 μl of TE buffer (pH 8.0) and supplemented with an RNase A/RNase T1 cocktail (final concentration 0.01 mg/ml RNase A and 10 units per ml RNase T1). DNA concentration is measured with a DyNA Quant 200 Fluorometer according to the manufacturer’s instructions (Hoefer).
Restriction Digestion and Agarose Gel Electrophoresis.
Aliquots containing 45 ng of cosmid DNA are completely and independently digested for 2 hr at 37°C with 5 units each of three different restriction enzymes (EcoRI, HindIII, and NsiI). Each gel lane is loaded with 15 ng of digested DNA. A mixture of 1 kbp ladder (Life Technologies), XbaI-digested λgt11 DNA (yielding three fragments at 43.7, 24.8, and 18.9 kbp), and two supplementary fragments of size 1,204 and 691 bp, is used as the size marker. Agarose gel electrophoresis (1%; Eastman Kodak; gel dimensions 19 cm × 19 cm × 0.5 cm) is carried out in ×2 GGB buffer (80 mM Tris base/40 mM sodium acetate/4 mM EDTA/52 mM glacial acetic acid, pH 8.0–8.3; Ref. 9) at 6 V/cm, in a custom-made chamber with circulating buffer thermostated at 14°C to 18°C. Total run time under these conditions is 4 hr. We also obtain good results with overnight runs at 2 V/cm.
Image Acquisition and Vector Band Identification.
The gel is stained for 1 hr in SYBR–green I solution (Molecular Probes), diluted 1:20,000 in ×2 GGB buffer. After the gel is scanned on a FluorImager 575 (Molecular Dynamics), the DNA is transferred by capillary action onto a nylon filter and fixed by UV crosslinking. The filter is probed with nonradioactively labeled vector DNA (Genius System, Boehringer Mannheim). Vector bands are identified by eye and entered into the computer manually.
Data Analysis.
All of the data analyses are performed with custom-designed software. The gel-image analysis system (G.K.-S.W., unpublished work) performs automatic lane finding, lane-profile generation, peak detection, size calibration, band quantitation, and fragment-count estimation. Results are written out as lists of fragment sizes, with the vector fragments flagged, and passed to the map assembly system, dnam (16, 17). The output from dnam is passed to a third software package, atlas (E.C.T., unpublished work), which performs stringent quality control checks and produces a graphical representation of the MCD map.
RESULTS
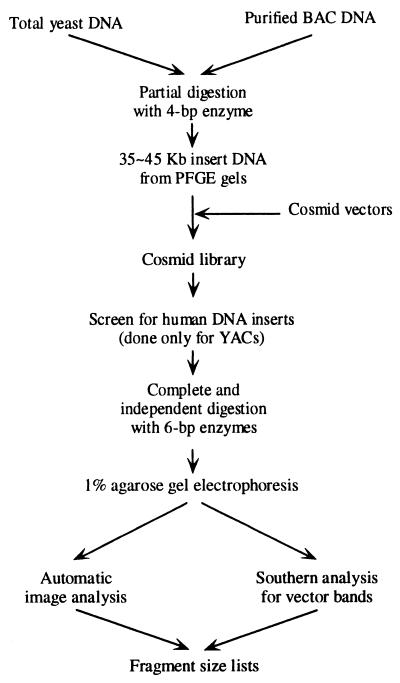
The experimental procedures behind MCD mapping are shown in Fig. 1, and a conceptual overview of this process is shown in Fig. 2. Standard molecular biology protocols are employed throughout. However, a number of adaptations have been made to produce data of adequate quality for MCD mapping. Very high-quality gel images are essential because the precision of the fragment size measurements determines the information content of the fingerprint data and hence the frequency at which different fragments of similar size are confused with one another. Furthermore, large-scale mapping is only practical when the gel images can be analyzed automatically with few errors. This goal is only achievable with consistent, high-quality images.
Figure 1.
Flow chart of wet bench procedures for YAC → cosmid and BAC → cosmid MCD mapping. The main difference is that, while BAC DNA can readily be purified from bacterial chromosomal DNA, there is no good preparative method to separate YAC DNA from yeast chromosomal DNA. In the YAC case, the few percent of the cosmids that are derived from the YAC are identified by a hybridization-based colony-screening protocol. With BAC-derived cosmids, this step is unnecessary because the mapping software can readily eliminate the small number of cosmids that do not originate from the BAC.
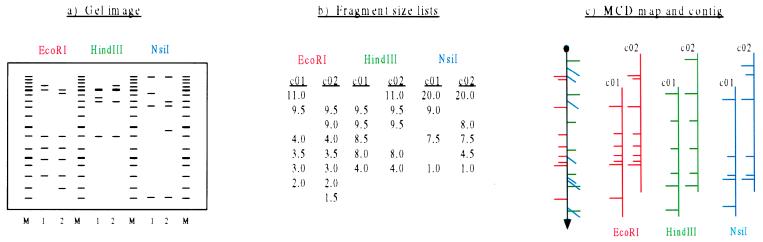
Figure 2.
Schematic representation of MCD mapping process. (a) Gel image. (b) List of fragment sizes for each enzyme domain in each clone. Lanes labeled with a number identify the clone as c01 or c02. Lanes labeled with the letter M identify size markers. (c) Three single-enzyme maps are independently constructed (Right). Synchronization across enzyme domains results in a composite map (Left). Long tick marks indicate boundaries between ordered groups of fragments; short tick marks demarcate unordered fragments within a group, arbitrarily drawn in order of decreasing size.
The successful implementation of MCD mapping has required a co-evolution of the experimental process and the data analysis software. One example of this interaction is the design of the cosmid vector. For shotgun sequencing, the vector should be as small as possible to minimize the overhead associated with repeated sequencing of the vector. For MCD mapping, the vector should contain no sites for the mapping enzymes and allow no possibility for creation of an artifactual site at the vector-insert junction (e.g., when an MboI partial-digest fragment is ligated into a BamHI cloning site, there is a chance that an artifactual BamHI site will be created at the junction). When the vector s-Cos-DBI is used to clone MboI partial-digest fragments, a single vector-containing fragment of known minimum size (3205 bp) is produced in each of our three enzyme domains. Because this vector-containing fragment is not representative of any complete-digest fragment in the underlying genome, it is identified by gel-transfer hybridization and eliminated from the list of fragments used for map assembly.
A major improvement in image quality was achieved by switching to the intercalating dye SYBR–green I. At the excitation wavelength of 488 nm used by our gel scanner, we find that SYBR–green I is five times more sensitive than thiazole orange, which is in turn three times more sensitive than ethidium bromide. We typically load only 15 ng of cosmid DNA per gel lane when using SYBR–green I to stain gels of ordinary dimensions. Band distortion due to local overloading is never a problem because the largest bands contain only 5–10 ng of DNA. Furthermore, when employing DNA of only moderate purity, as we do, the cleanliness of the restriction digests is inversely related to the volume of bacterial culture from which the DNA is extracted. SYBR–green I has greatly reduced the number of gel lanes that are unusable because of poor or failed digestions. The only serious complication is that, for unknown reasons, SYBR–green I displays a narrow and variable range over which integrated fluorescence increases linearly with the amount of DNA in the band.
Automatic, robust, and accurate determination of fragment sizes requires carefully designed DNA size markers. Ideally, the marker bands should be uniformly spaced along the arc length of the size mobility curve. There must be an increasing number of marker bands as the fragment size approaches the threshold at which mobilities become size independent. Attention to curve-fitting stability in this region allows excellent fragment sizing precision up to 15 kbp (SD ± 1%) and adequate fragment sizing precision up to 40 kbp (SD ± 5%). A second requirement is that there must be three bands that are easily recognized as local intensity maxima. Recognition of these conspicuous bands nucleates the automatic pattern-match procedure by which the image analysis software identifies the marker bands. In our standard gel format (Fig. 3), sets of six digest lanes are flanked by two marker lanes. All of the five marker lanes on the gel are used in the two-dimensional interpolation algorithm that assigns sizes to the digest bands.
Figure 3.

Gray scale image of a typical mapping gel poststained with SYBR–green I. There are five marker lanes, at positions 1, 8, 15, 22, and 29. Two clones, each independently digested with EcoRI, HindIII, and NsiI (and loaded in that order) are placed between every pair of marker lanes.
The image analysis problem associated with a restriction digest pattern is quite different from the “base calling” problem associated with a sequencing ladder. Base calling software needs only to identify the dominant band at every ladder position. In contrast, software designed to analyze restriction patterns must determine the number of fragments in each band, since any number of fragments of similar size may comigrate at any position in a lane. Under normal electrophoretic conditions, band multiplicities of two or three are common. Band multiplicities must be computed in spite of diminishing signal-to-noise ratios at small fragment sizes and nonlinearities in the relationship between integrated fluorescence intensity and DNA quantity per band. These image characteristics can vary from lane to lane even on the same gel. Effective image analysis software must account for all such experimental realities. The analysis of a typical gel lane is shown Fig. 4. We have now successfully analyzed over 1,000 gels with our software and, on balance, it is almost as good as an expert interpreter. It makes some mistakes that a human expert would not make, but it also correctly analyzes many bands that an expert would miscount.
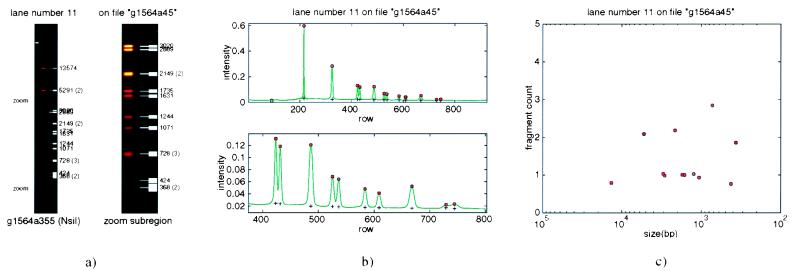
Figure 4.
Processing of agarose gel images. (a) False-color image of digest from lane 11 of the gel shown in Fig. 3. The full-lane image is shown (Left), and an intensity-rescaled image of the region demarcated by “zoom” is shown (Right). White bars point to bands that are automatically identified by the image-analysis software. Fragment sizes in base pairs are indicated, and any band multiplicities greater than one are given in parentheses. (b) One-dimensional representation of the full lane (Upper) and the zoom region (Lower). The collapse to one dimension is done with a median-biased averaging scheme. Each row is analyzed separately. Pixels are first sorted by intensity, and a fixed number of the lowest intensity pixels are eliminated to account for the gap between gel lanes. From the remainder, an average of the middle quartile is computed. (c) Fragment counts for the lane, which contains eight singlets, three doublets, and one triplet. Fragment count estimates are based on the trend in integrated band intensity versus fragment size. This trend is variable from gel to gel and is highly nonlinear. Every digest lane on the gel that has not been rejected because of bad data is analyzed simultaneously to build a composite trend line for the relationship between integrated intensity and DNA quantity.
A key feature of the system is the automatic rejection of low quality data. No attempt is made to identify the source of the problem. The software has an internal model of what a good data lane should look like, and it rejects any lane that does not satisfy this model. A partial list of the types of problems that are detected includes deleted clones, mixed clones, partial digestions, failed digestions, cleavage at secondary sites, overloaded lanes, underloaded lanes, and dirt on the gel. In current practice, 80–90% of the gel lanes are usable. However, even good lanes can be misinterpreted. A powerful tool for detecting misinterpretations is the cross enzyme sum-of-fragments consistency test. Except for contributions from a few missing small fragments of size less than 500 bp, which are on average expected to be less than 1% of the total cosmid length, the sum of fragments should be consistent across enzyme domains. It can vary between 40 and 50 kbp from clone to clone, but from enzyme to enzyme on a given clone total deviations of more than 1 or 2 kbp are almost certain indication that something is wrong with the image analysis. By using this test to detect misanalyzed lanes, and manually correcting the fragment counts, we have essentially eliminated fragment miscounts on all bands larger than 2 kbp.
The automatic phase of the MCD map assembly proceeds as a series of steps during which the order of the clone ends and restriction fragments are progressively refined (16, 17). Fragment sizing outliers are handled by the “gray zone” concept. A fragment pairing that is more precise than the lower gray zone threshold is automatically accepted unless it violates a topological constraint of the map. Within the gray zone, fragment pairings are only made if they are required for topological consistency; otherwise, they are deferred. Pairings that are less precise than the upper gray zone threshold are rejected outright. We currently set the gray zone thresholds at 2.0 and 4.0% over most of the usable size range. These thresholds are increased both for large fragments (because of the severe loss of electrophoretic resolution) and for small fragments (because of the moderate loss of electrophoretic resolution and the increased band broadening). Statistical outliers generally fall below the gray zone. Valid pairings end up in the gray zone primarily as a result of a multiplet band that is not properly decomposed by the image-analysis software into its component fragments.
Ultimately, the key to obtaining accurate maps lies in a “fix it as you grow” strategy. The basic premise is that errors are rare, because of the high quality of the input data. When errors do occur, and regardless of whether they are due to cloning aberrations, image-analysis errors, or map-assembly errors, the problem is usually limited to just one of the three enzyme domains. Often, the problem is limited to a single clone. Removal of the suspect clone allows the map to grow. Once the map extends beyond the end of the suspect clone, it is generally quite easy to determine why that clone originally interfered with map growth. If the problem is an obvious mistake in the image-analysis or vector-band identification, we fix the data set and put the clone back into the map. At our high sampling depths, these constraints on the map construction are sufficiently strong everywhere but at the ends to allow nearly all errors to be detected and fixed. Any undetected errors are either within a clone length of the end of the map or in a region of exceptionally low coverage.
Table 1 is a summary of the YAC → cosmid maps that we have built on human chromosome 7. Not every fragment is ordered, and locally unordered fragments are placed into “fragment groups.” In most cases, there is an average of 1.2–1.3 unordered fragments per fragment group, meaning that we closely approach the goal of ordering all the restriction fragments. A typical MCD map, which combines the results of four independently constructed YAC → cosmid maps, is shown in Fig. 5. The high sampling depths allow the selection of a truly minimal tiling path, with overlaps of only a few kilobase pairs. YAC fidelity is validated by comparing the overlapping regions between these independently constructed maps. To date, no discrepancies have been found. As an even more rigorous test of YAC fidelity, we fingerprinted a small collection of cosmids from a library that was directly subcloned from the same hybrid cell line used to construct the YACs (E. D. Green, unpublished results). No discrepancies were found between these cosmids and the ones that were derived from YAC clones. Popular perceptions about YAC instability are based largely on experience with a relatively small number of libraries. What these results establish is that stable YAC libraries can be built, and that YACs can be used as the starting clones for systematic sequencing.
Table 1.
Summary of YAC → cosmid MCD maps for portions of human chromosome 7
| Chromosome 7 YACs | Coverage* | Nf† (EcoRI) | Nf† (HindIII) | Nf† (NsiI) | Coligations,‡ % | Map size,§ kbp |
|---|---|---|---|---|---|---|
| yWSS771 | 30.3 | 9.8/1.2 | 8.4/1.2 | 11.4/1.2 | 2.8 | 44 + 170 |
| yWSS1346 | 29.2 | 10.5/1.2 | 12.4/1.3 | 10.0/1.3 | 3.0 | 281 |
| yWSS1434 | 20.5 | 7.4/1.3 | 6.8/1.4 | 7.4/1.6 | 7.8 | 156 |
| yWSS1564 | 16.7 | 9.2/1.3 | 10.4/1.5 | 9.8/1.3 | 7.9 | 640 |
| yWSS1572 | 31.5 | 8.0/1.2 | 9.1/1.2 | 9.0/1.3 | 4.5 | 292 |
| yWSS1613 | 26.3 | 10.6/1.2 | 10.6/1.1 | 11.5/1.3 | 3.5 | 136 + 56 |
| yWSS1862 | 23.4 | 8.4/1.2 | 11.0/1.2 | 11.6/1.3 | 3.4 | 261 |
| yWSS1980 | 20.7 | 8.3/1.1 | 8.5/1.1 | 10.8/1.1 | 5.7 | 278 |
Coverage is calculated assuming a 40-kbp insert size. Clones left out of the map because they could not be uniquely placed are included in this calculation; coligations and yeast impurities are not.
Nf refers to the average number of fragments observed in a clone, which is the first number given in each row. The second number indicates the average number of fragments per fragment group, an indication of how well ordered the restriction fragments are in the maps. Contigs smaller than 100 kbp are not included when summarizing fragments per fragment group.
Coligations are cosmids that contain a human insert from the targeted region and an unrelated piece of DNA that is inserted between the end of the human insert and the cosmid vector.
Map sizes are based on the sum of the restriction fragment sizes. The gap in the overlap region between YACs yWSS771 and yWSS1613 has not yet been closed. These two maps agree perfectly on either side of the gap and stop abruptly at the same fragments at the gap.
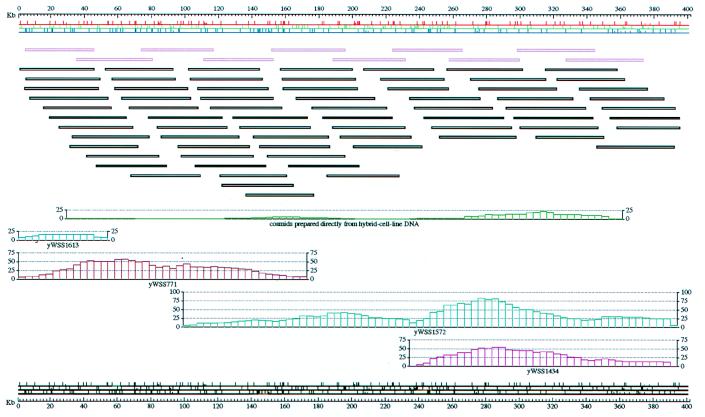
Figure 5.
Representative MCD map from chromosome 7. Four hybrid cell-line-derived YACs were subcloned into cosmids to map this 400 kbp region. In addition, a special cosmid library derived directly from the hybrid cell line (i.e., not derived from a YAC clone) was also placed on this map, with no inconsistencies. The map is depicted just below the upper scale bar. Enzyme domains EcoRI, HindIII, and NsiI are depicted, from top to bottom, in red, green, and blue. Ordered groups of fragments are separated by tall tick marks and unordered fragments within a group are separated by short tick marks. The minimal-tiling-path clones are displayed in purple just below the map. Below the tiling path clones, a larger set of clones is shown: this set includes all clones except those whose fragment content is identical to, or a subset of, that of a displayed clone. Next is a series of five histograms. From top to bottom, they reflect cosmid coverage derived from the following sources: the cosmid library prepared directly from hybrid cell line DNA, yWSS1613, yWSS771, yWSS1572, and yWSS1434. Below the histograms is a map quality assessment based on atlas (E. Thayer, unpublished work).
We have now sequenced cosmids from nearly 1 Mbp of the DNA whose mapping is summarized in Table 1. The shotgun sequencing data were analyzed with the Phred/Phrap sequence-assembly system (P. Green, unpublished results). No mapping errors were detected when the sequence derived maps were compared with the MCD maps. Not only were the fragments correctly ordered, but the accuracy of the intersite spacings was less than 1%, albeit with a systematic error somewhat more than 1% for the larger fragments. The maps involved in this test contained more than 700 different restriction fragments. In an independent MCD mapping/shotgun sequencing project of comparable size in the HLA class I region on human chromosome 6, similar results were obtained (D. E. Geraghty, T. Guillaudeux, and M. Janer, unpublished results). In the HLA project, a single mapping error was detected at the end of one map, which was traced to the miscounting of a 600 bp multiplet band in a single cosmid. Up to date maps, sequences, and software documentation can be found on our Web site at http://www.genome.washington.edu.
DISCUSSION
We have presented evidence that it is practical to construct detailed restriction maps of megabase pair-sized regions of human DNA that are sufficiently accurate to guide, and provide powerful cross-checks on, long-range DNA sequencing. MCD mapping is therefore a high-end solution to several major challenges confronting all large-scale sequencing projects directed at the genomes of higher organisms: sequence accuracy, clone validation, and map contiguity.
A commonly stated goal of the Human Genome Project is an error rate of less than 1 per 104 bp. Although the Phred/Phrap sequence-assembly system provides estimates of the single base pair sequence-error rate by analyzing the chromatogram traces produced by four-color fluorescence-based sequencing instruments (P. Green, unpublished results), it cannot resolve misassemblies due to exact repeats on the ≈500 bp length scale of the individual sequencing tracts. The performance of such sequence-assembly software can only be validated by independent experimental data. MCD maps allow strong validation of sequence assemblies because they rely on different input data and their construction is insensitive to the short interspersed repeats that cause most of the sequence-assembly difficulties. This insensitivity relates to the large size of the restriction fragments (average length 4096 bp) relative to the small size of the troublesome repeats (300 bp in the case of Alu). Our experience has been that the MCD maps also help to lower the cost of sequence finishing. Partially assembled cosmid sequences, which typically contain only one or two gaps at the stage where finishing commences, are readily aligned with the MCD maps. This allows us to establish the order and relative orientations of the assembled segments, thereby providing us with an estimate of the gap sizes.
The importance of clone validation by redundant analysis of overlapping clones should not be underestimated in evaluating sequencing strategies. With the high redundancy of cosmid coverage, and the requirement that the maps be independently determined from two different YACs, MCD mapping can detect all common instances of cloning artifacts. Numerous cosmid rearrangements are routinely detected. The most common class of artifacts is the large deletion. These events are easily detected without detailed mapping because they produce clones that are much smaller than the expected 40–50 kbp size of authentic cosmids. The next most common class of artifacts is the coligation: the juxtaposition of a normal human insert with a small extraneous fragment. Typically, 3–8% of our cosmid libraries consist of coligations, which are rejected in the MCD mapping process. It is conceivable that some region of the genome may undergo a reproducible rearrangement—such as the deletion of a segment between two repeats—in multiple clones. However, practical experience with both YACs and cosmids indicates that this problem is rare compared with the high incidence of idiosyncratic cloning artifacts that affect individual clones.
Map contiguity depends on the depth of cloned coverage and the intrinsic characteristics of the cloning system. Our experience indicates that genome sampling in cosmids is so strongly nonrandom that long-range contiguity can only be achieved by using much higher redundancies than those normally employed. There is no strong reason to expect other cloning systems to provide sampling that is any more random. Our clone depth histograms exhibit a quasi-periodic fluctuation whose variance greatly exceeds random expectations. When extrapolated down to the roughly ×5 coverage used in other mapping projects, these fluctuations would lead to a gap every 100 to 200 kbp, as is commonly observed (11–13). In contrast, we have mapped about 1.5 Mbp of nonredundant DNA on human chromosome 7, and a comparable amount in the HLA class I region on human chromosome 6 (D. E. Geraghty, T. Guillaudeux, and M. Janer, unpublished results), with only one gap. This gap was encountered in two different but overlapping YACs, yWSS771 and yWSS1613. The two maps agreed exactly on both sides of the gap, and stopped abruptly at the same fragments at the gap. Combined coverage in the vicinity of the gap was ×60.
Increasingly, all candidate steps in large-scale sequencing will need close assessment of cost and scalability. Even as currently implemented, the number of experimental steps required to position MCD mapping upstream from DNA shotgun sequencing does not appear to be prohibitive. One relevant measure is the number of gel lanes that must be produced. Assuming ×2 YAC validation, ×20 cosmid coverage, and 3 enzymes per cosmid, 120 gel lanes are required for each sequenced cosmid—a number that compares favorably with the 600 gel lanes that are required to sequence a cosmid by the shotgun method. The intrinsic labor, supply, and instrumentation costs per mapping gel lane all appear to be lower than the comparable costs for sequencing gel lanes. If the starting material from the low-resolution mapping stage were BACs rather than YACs, the clone validation could be carried out by comparing the fingerprints for the complete set of BACs with cosmid-based MCD maps obtained by subcloning a minimal tiling path of BACs. Subcloning is necessary for any of three reasons: (i) restriction patterns for the largest BACs are essentially impossible to analyze because there are too many fragments, (ii) existing BAC libraries are not deep enough to allow us to order all the restriction fragments, and (iii) haplotype differences are easier to resolve when the composite diploid map is “anchored” by these cosmid-based MCD maps (each of which is of a single haplotype). We have built a few such BAC → cosmid maps and found that they cost significantly less than comparably sized YAC → cosmid maps. The cost savings result from the elimination of the most expensive step in the current protocol—the screening of the cosmid libraries prepared from total yeast DNA for the small fraction of clones that are derived from the YAC. Screening is unnecessary with BAC-derived cosmids because BAC DNA is readily purified from host chromosomal DNA.
The major challenges associated with increasing the scale of MCD mapping involve software improvements. Whereas the complexity of the data analysis, starting with a raw gel image, greatly exceeds anything that could be carried out manually for even a single YAC, considerable manual intervention remains essential to produce maps of the quality reported here. Our long-term goal is to develop MCD map assembly software that can automatically detect and remove the small but significant percentage of bad data that require expert attention. In this sense, MCD mapping—with its high level of abstract definition, its reliance on experimental overdetermination, and its dependence on high-quality but imperfect data—provides an appealing context within which to confront one of the great challenges in practical computation: how to impose an ideal model on real world data without reliance on human intelligence to resolve the occasional clashes between the ideal and the real.
Acknowledgments
We gratefully acknowledge the help of the following individuals: Eric Green for providing YAC and cosmid clones, as well as unpublished mapping data for chromosome 7; Will Gillett and Elizabeth Hanks for a continuing collaboration on dnam; Richard Karp for collaborating on the development of atlas; Daniel Geraghty and Thierry Guillaudeux for sharing their unpublished experiences with many of the protocols described here; Phil Green and Shawn Iadonato for collaborating on the sequencing of chromosome 7 cosmids; Charles Magness for assistance with segmap; Regina Lim, Ying Ge, Roulan Qiu, Kim Erickson, Channakhone Saenphimmachak, Ellen Knebel, and Joseph Manakkil for technical assistance with cosmid subcloning, fingerprinting, and data analysis. This work was supported by Department of Energy Grants DE-FG06-92ER61487 and DE-FG02396ER62173 and National Institutes of Health Grant 1 R01 HG01475.
ABBREVIATIONS
- BAC
bacterial artificial chromosome
- MCD
multiple-complete-digest
- STS
sequence-tagged site
- YAC
yeast artificial chromosome
References
- 1.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, et al. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 2.Johnston M, Andrews S, Brinkman R, Cooper J, Ding H, et al. Science. 1994;265:2077–2082. doi: 10.1126/science.8091229. [DOI] [PubMed] [Google Scholar]
- 3.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, et al. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 4.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 5.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 6.Olson M V. Science. 1995;270:394–396. doi: 10.1126/science.270.5235.394. [DOI] [PubMed] [Google Scholar]
- 7.Gibbs R A. Nat Genet. 1995;11:121–125. doi: 10.1038/ng1095-121. [DOI] [PubMed] [Google Scholar]
- 8.Coulson A, Sulston J, Brenner S, Karn J. Proc Natl Acad Sci USA. 1986;83:7821–7825. doi: 10.1073/pnas.83.20.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson M V, Dutchik J E, Graham M Y, Brodeur G M, Helms C, Frank M, MacCollin M, Scheinman R, Frank T. Proc Natl Acad Sci USA. 1986;83:7826–7830. doi: 10.1073/pnas.83.20.7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riles L, Dutchik J E, Baktha A, McCauley B K, Thayer E C, Leckie M P, Braden V V, Depke J E, Olson M V. Genetics. 1993;134:81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stallings R L, Doggett N A, Callen D, Apostolou S, Chen L Z, et al. Genomics. 1992;13:1031–1039. doi: 10.1016/0888-7543(92)90016-l. [DOI] [PubMed] [Google Scholar]
- 12.Doggett N A, Goodwin L A, Tesmer J G, Meincke L J, Bruce D C, et al. Nature (London) 1995;377:335S–365S. doi: 10.1038/377335a0. [DOI] [PubMed] [Google Scholar]
- 13.Ashworth L K, Batzer M A, Brandriff B, Branscom E, de Jong P, Garcia E, Garnes J A, Gordon L A, Lamerdin J E, Lennon G, Mohrenweiser H, Olson A S, Slezak T, Carrano A V. Nat Genet. 1995;11:422–427. doi: 10.1038/ng1295-422. [DOI] [PubMed] [Google Scholar]
- 14.Hudson T J, Stein L D, Gerety D S, Ma J, Castle A B, et al. Science. 1995;270:1945–1954. doi: 10.1126/science.270.5244.1945. [DOI] [PubMed] [Google Scholar]
- 15.Chumakov I M, Rigault P, LeGall I, Bellann’e-Chantelot C, Billault A, et al. Nature (London) 1995;377:175S–197S. doi: 10.1038/377175a0. [DOI] [PubMed] [Google Scholar]
- 16.Gillett W, Hanks L, Wong G K-S, Yu J, Lim R, Olson M V. Genomics. 1996;33:389–408. doi: 10.1006/geno.1996.0215. [DOI] [PubMed] [Google Scholar]
- 17.Gillett W, Daues J, Hanks L, Capra R. J Comput Biol. 1995;2:185–205. doi: 10.1089/cmb.1995.2.185. [DOI] [PubMed] [Google Scholar]
- 18.Shizuya H, Birren B, Kim U J, Mancino V, Slepak T, Tachiiri Y, Simon M I. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green E D, Olson M V. Science. 1990;250:94–98. doi: 10.1126/science.2218515. [DOI] [PubMed] [Google Scholar]
- 20.Green E D, Braden V V, Fulton R S, Lim R, Ueltzen M S, Peluso D C, Mohr-Tidwell R M, Idol J R, Smith L M, Chumakov I, Le Paslier D, Cohen D, Featherstone T, Green P. Genomics. 1995;25:170–183. doi: 10.1016/0888-7543(95)80123-4. [DOI] [PubMed] [Google Scholar]
- 21.Wahl G M, Lewis K A, Ruiz J C, Rothenberg B, Zhao J, Evans G A. Proc Natl Acad Sci USA. 1987;84:2160–2164. doi: 10.1073/pnas.84.8.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Southern E M, Anand R, Brown W R A, Flethcher D S. Nucleic Acids Res. 1987;15:5925–5943. doi: 10.1093/nar/15.15.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coulson A, Sulston J. In: Genome Analysis: A Practical Approach. Davis K E, editor. Oxford: IRL; 1988. pp. 19–39. [Google Scholar]