Abstract
Nonsyndromic clefting of the lip and palate in humans has a highly complex etiology, with both multiple genetic loci and exposure to teratogens influencing susceptibility. Previous studies using mouse models have examined only very small portions of the genome. Here we report the findings of a genome-wide search for susceptibility genes for teratogen-induced clefting in the AXB and BXA set of recombinant inbred mouse strains. We compare results obtained using phenytoin (which induces cleft lip) and 6-aminonicotinamide (which induces cleft palate). We use a new statistical approach based on logistic regression suitable for these categorical data to identify several chromosomal regions as possible locations of clefting susceptibility loci, and we review candidate genes located within each region. Because cleft lip and cleft palate do not frequently co-aggregate in human families and because these structures arise semi-independently during development, these disorders are usually considered to be distinct in etiology. Our data, however, implicate several of the same chromosomal regions for both forms of clefting when teratogen-induced. Furthermore, different parental strain alleles are usually associated with clefting of the lip versus that of the palate (i.e., allelic heterogeneity). Because several other chromosomal regions are associated with only one form of clefting, locus heterogeneity also appears to be involved. Our findings in this mouse model suggest several priority areas for evaluation in human epidemiological studies.
Human cleft palate only (CPO) is usually thought to be multifactorially inherited and genetically distinct from cleft lip with or without cleft palate [CL(P)] (1, 2). However, a major gene effect (3, 4) and a relationship of the two birth defects (5, 6) have also been claimed. Genetic epidemiological analyses have yielded different results in different populations including major gene effects with recessive (7, 8) dominant or codominant (9–11) transmission or oligogenic models (12, 13). Molecular genetic analyses also suggest multiple loci in humans (reviewed in ref. 14). Association at the transforming growth factor-α (TGFA) locus was found (15) and confirmed by several (but not all) groups but no evidence of linkage of CL(P) to this region was found in multiplex families using tests assuming a monogenic major locus (16). It has also recently been shown that maternal smoking during pregnancy interacts with TGFA to alter susceptibility to clefting (17, 18). Contrary to the commonly held view that CL(P) and CPO are distinct in etiology, an influence of the TGFA locus on CPO has also been suggested (19). Association of CL(P) with the retinoic acid receptor α locus (17q21) has been reported (20). Chromosomal translocations and linkage studies suggest 6p23–24 as a candidate region (21, 22), some families exhibit evidence of linkage with BCL3 on 19q13.1 (23), and linkage disequilibrium between markers at 4q25–4q31.3 and CL(P) has been found (24, 25). Van der Woude syndrome with both CL(P) and CPO and lower lip pits maps to 1q32–41 (26), CPO with tongue tie maps to Xq21.3-q22 (27), Stickler syndrome of CPO with retinal detachments maps to chromosome 12 (28), and Velocardiofacial syndrome with an abnormal facies, CPO, or velopharyngeal incompetence, and cardiac defects is associated with deletions of 22q (29). A number of syndromes with oral facial clefting occurring at low frequencies have also been mapped (14).
Our work focuses on the genetics of susceptibility to teratogen-induced oral facial clefting because a high percentage of the xenobiotics that are proven teratogens for humans include oral facial clefting as a manifestation (30). Ethanol is the most common human teratogen, and fetuses exposed in utero to ethanol have an increased incidence of CL(P). The fetal hydantoin syndrome is also well known and includes CL(P). Trimethadione involves a much lower frequency of fetal exposure but it, too, includes CL(P). CPO can result from aminopterin and retinoic acid exposure in utero. Effects of exposure to smoking were noted above. Xenobiotics are not the only teratogens known to cause oral facial clefting in humans, as hyperthermia also increases incidence of CL(P).
Genetic studies reported to date for animal models of clefting are limited to evaluations of only very small portions of the mouse genome. Many reports consist of observations of clefting in animals carrying mutant copies of genes including the Msx1 gene on chromosome 5 (31), several Hox genes (32, 33), the retinoic acid receptor α locus (34), the β3 subunit of the A γ-aminobutyric acid receptor (35), transforming growth factor-β3 (36, 37), aggrecan (38), activin receptor IIA (39), and the Twirler (40) and Dancer (41) mutants. A number of studies also implicate major histocompatibility genes in susceptibility to teratogen-induced clefting (42–44).
Gene mapping approaches in the mouse have focused on the set of closely related inbred “A” strains which exhibit increased susceptibility to clefting. Creation of a congenic strain pair by backcrossing the A/WySn strain to a strain not susceptible to clefting led to the recent identification of a region of mouse chromosome 11 influencing susceptibility to spontaneous (i.e., without induction by teratogens) CL(P) (45). This region contains a number of candidate genes, including that for the retinoic acid receptor α. Statistical analyses suggest that clefting susceptibility in this model may also involve interaction with a second recessive gene, as yet unmapped (46).
Experiments were performed using the AXB and BXA recombinant inbred (RI) strains to evaluate both spontaneous and teratogen-induced clefting [both CL(P) and CPO] (47–53). These studies examined only a few regions of the genome but suggested possible involvement of genes on several chromosomes. One region on chromosome 8 representing <0.7% of the genome containing the N-acetyl transferase gene was further supported by a congenic strain analysis. However, our re-evaluation of the statistical methods used in these previous studies has led us to conclude that the tests performed only indirectly relate to hypotheses regarding gene mapping of clefting susceptibility and the P values reported were highly anticonservative.
Here we combine the extensive data on teratogen-induced clefting in the AXB and BXA RI lines collected previously with a genome wide collection of marker typings for these RI lines. We first refined and evaluated the marker map to be used for these comparisons. We then developed and applied statistical tests appropriate for evaluating gene mapping hypotheses for categorical data such as clefting using RI lines. Our analyses identified several genome regions that may contain susceptibility genes for phenytoin-induced CL(P) and 6-aminonicotinamide (6-AN)-induced CPO and we review candidate genes located in these regions. Phenytoin was chosen as an inducer of CL(P) with known effects in humans while 6-AN is a potent experimental inducer of CPO which may mimic teratogenesis related to maternal smoking. When we compared the chromosomal regions identified for CL(P) and CPO, we found that many (but not all) are in common, but that they usually differ in which parental allele predisposes to clefting. This implies that both allelic and locus heterogeneity may distinguish these two forms of clefting. We consider how these findings may relate to the etiology of clefting in humans.
MATERIALS AND METHODS
Mouse Maintenance and Drug Administration.
RI lines derived from crosses between A (A/J) and B (C57BL6/J) strains were supplied by M. Nesbitt (54). Mice were then bred and maintained in a colony at the University of Michigan. Our teratogen exposure protocols were based on methods established in many previous studies identifying developmental stages and doses appropriate for induction of cleft lip or cleft palate. We recognize that other schedules of administration may produce different frequencies of clefting and some might more closely mimic exposure in human subjects (e.g., multiple, sequential exposures), but investigation of these alternatives was beyond the scope and available resources of our study. For phenytoin (Dilantin; Parke-Davis), RI strains were injected i.p. on day 10 of pregnancy. The day the plug was found was designated as day 0 (52). The dose was 60 mg/kg. Control injections of 40% propylene glycol in 10% ethanol (the drug solvent) were administered in a similar manner. Fetuses were examined on day 17 of pregnancy and scored for cleft lip and/or cleft palate (53). For 6-AN, RI strains were injected i.p. on day 13 of pregnancy. The dose was 9 mg/kg. Control injections of sterile distilled water (the drug solvent) were administered similarly. Fetuses were examined on day 17 and scored for facial clefting. We did not attempt to sex fetuses, as previous pilot studies (unpublished data) suggested a misclassification rate of 10–20%. Most cases of cleft lip observed were bilateral, but this information was not recorded for this study. Numbers of resorbed fetuses were recorded but these data were not included in the present analyses.
Marker Typings and Genetic Map.
A collection of 448 marker typings for AXB and BXA RI lines was established by Beverly Paigen (The Jackson Laboratory) from reports in the literature and unpublished information. These data were kindly supplied to us in 1994 and updated in 1995 and most or all are now available via the internet in the Mouse Genome Database (55). Recent genetic control testing of these RI lines indicated that several strains were contaminated at an undetermined time in the past (56), and so we discarded our data obtained from these lines. These data were included in the analyses reported previously (47–53). Marker data files were carefully cleaned (removing duplicates, marker name pseudonyms, or other erroneous typings) and a total of 361 markers were retained for our analyses. We located 293 markers on the Mouse Chromosome Committee Reports and we used the map positions assigned in the reports. Sixty-eight markers were not in the reports, and we mapped these using the program map manager (57) based on the strain distribution patterns in the full set of 41 AXB and BXA RI lines. Our final map was consistent with expectations when we compared strain distribution pattern similarity among pairs of markers as a function of map distance (for syntenic markers) and for markers mapped to different chromosomes (data not shown).
We created a data set that combined the RI strain distribution patterns for all 361 markers, our final marker map, and the observed frequencies of clefting in 9 RI lines exposed to phenytoin and 10 RI lines exposed to 6-AN. It was necessary to pool replicate litters within each RI line as there were insufficient data to estimate the many parameters required to incorporate this variable into the model. Markers with less than 100 pups evaluated for clefting from both the A and B parental marker allele groups (due to missing typing data from some RI lines) were not included in the analysis, leaving a total of 342 markers assessed for the phenytoin-exposed lines and 334 for the 6-AN-exposed lines. Using these loci, the average distance between markers for the phenytoin data set was 4.4 centimorgans (cM) with a maximum gap of 26 cM and only 10 intervals greater than 15 cM. The average distance between markers for the 6-AN data set was 4.5 cM with a maximum gap of 26 cM and 11 intervals greater than 15 cM.
Statistical Analyses.
Previously reported analyses using clefting data from these lines classified litters as “clefting positive” if one or more pups had a cleft, and thus did not distinguish between litters with just one pup affected versus litters with more than one or all pups affected. A 2 × 2 table was then constructed contrasting the frequency of litters with some clefting versus litters with no clefting pooled for all litters for all RI lines which retained the A versus the B parental allele. Thus, this analyses did not take into account variation among RI lines within each parental allele group when evaluating the statistical significance of the comparison between parental allele groups. The relative magnitude of variation among RI lines within the same parental allele group in comparison to that observed between different parental allele groups is at the heart of the strategy of gene mapping using RI lines. Furthermore, the P value obtained from the Fisher’s exact test of the 2 × 2 table is not the type I error of a false positive gene mapping finding, but is only an evaluation of the null hypothesis that the entire group of A-allele litters does not differ in clefting frequency from the entire group of B-allele litters. As shown below, RI lines differ very dramatically in frequency of clefting and so it is quite likely that by chance alone a random splitting of the lines into A versus B groups will produce a highly significant contrast between the groups. In fact, a highly significant finding using the 2 × 2 table approach is a necessary but not sufficient condition for inferring the location of a clefting susceptibility gene, and the P values obtained from this analysis strategy are highly anticonservative. For these reasons, and also because of the problem of contamination of some of the RI lines, we suggest that the previously reported clefting studies using these AXB lines should be interpreted with caution.
We developed a new analytical strategy to perform a robust assessment of support for mapping of clefting susceptibility loci. First, we used logistic regression to model the proportions of pups affected by clefting in the RI lines. A logistic model was fit to the probability of clefting at the level of individual pups for each marker, with the RI lines as the independent variable [i.e., log (proportion affected/proportion unaffected) = intercept + RI]. This resulted in a completely saturated model—i.e., there were as many parameters in the model as RI lines. We used the SAS program genmod (58) to implement this logistic regression analysis using the statement: model (no. of affected pups/total no. of pups) = RI/dist=binomial type 3. To evaluate whether clefting susceptibility genes reside in the vicinity of each marker, a contrast was specified in the model comparing the probability of clefting in RI lines with the marker’s A allele versus RI lines with the B allele. This was done by weighting RI lines in a genmod contrast statement so that the logistic regression model parameters for RI lines with the same marker allele are weighted as equal. For example, consider a hypothetical marker typed for 11 RI lines where the A parental allele was present in the first, fifth, and eight through eleventh RI lines (corresponding to their order in the genmod data set) while the other 5 RI lines inherited the B parental allele. Here, we weight the six A allele RI lines as 1/6 and the five B allele RI lines as 1/5. Multiplying these weights by their common denominator 30 (which does not affect the magnitude of the χ2 statistic of the contrast) produces the following program statement which we use for the genmod analysis: contrast RI “marker name” 5 -6 -6 -6 5 -6 -6 5 5 5 5. Unlike previous studies of these data, this contrast explicitly takes into account variation in the proportion of clefting among the set of RI lines classified as belonging to each of the parental allele groups (A or B, defined by the allele observed at each marker position) when assessing the statistical significance of the contrast between the two parental allele groups.
The statistical significance of the χ2 statistic obtained from this contrast among parental allele groups via the genmod logistic regression analysis is still not an assessment of the type I error of the gene mapping hypothesis. However, we used this contrast statistic as a measure of the association between clefting frequency and marker alleles to calculate the appropriate gene mapping P value. Each marker sorts the RI lines into two different parental allele groups which are then contrasted in their frequency of clefting. A genome region that contains a clefting susceptibility locus is expected to produce an especially large contrast in the frequency of clefting between those lines that inherit the parental allele which causes high clefting susceptibility versus the other group of lines that inherit the low clefting susceptibility allele. The key question in addressing the gene mapping hypothesis is how often one would expect to see a contrast in clefting frequency between the parental allele groups as large or larger than that actually observed under the null hypothesis where no cleft predisposing gene actually resides in the vicinity of the marker locus. We calculated these type I error probabilities for each marker by assessing the distribution of χ2 statistics obtained by contrasting all possible combinations of different RI lines sorted into hypothetical parental allele groups using a computer program which generates the combinations and runs them through the same SAS genmod analysis as was used for the actual marker data. This approach is based on the fact that RI lines have an equal chance of retaining either the A or B parental allele at each position in the genome. Thus, all combinations of RI lines grouped according to their A or B alleles at each marker position are equally probable under the null hypothesis. The type I error for the gene mapping test was thus assigned as the proportion of RI combinations which produced an equal or greater χ2 statistic compared with the observed statistic obtained for a marker. An example of the application of this test is presented in the results.
Due to the way our logistic regression analysis accounts for variation in frequency of clefting among RI lines within each parental allele group, the distribution of χ2 statistics correlates closely (but not exactly) with the difference in frequency of clefting between the parental allele groups. Thus, a large χ2 statistic indicates a large difference in clefting frequency between the A versus B groups. However, our use of the contrast χ2 statistic as a measure of association between clefting frequency and marker alleles is superior to a simple statistic such as the difference in clefting frequency between RI lines (pooled for the A versus the B parental alleles), because it takes into account variation among RI lines within each of the parent allele groups in assessing this association. Because marker typings were missing for some RI lines for some markers, it was necessary to independently derive the distribution of χ2 statistics under the null hypothesis for each unique combination of data sets missing typings for particular RI lines to correctly determine the relevant type I error. Gene mapping type I errors were estimated separately for cleft lip induced by phenytoin and cleft palate induced by 6-AN. Combined P values, assessing the null hypothesis that a susceptibility locus for neither CL(P) nor CPO lies in the vicinity of a marker, were calculated using the method of Fisher (59) where 2 ∑ ln (P) is distributed as χ2 with 4 df [2 times the number of independent tests of clefting (phenytoin and 6-AN), each with P value P].
RESULTS
Distribution of the Gene Mapping Test Statistic.
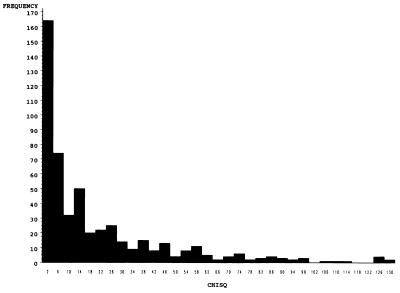
The frequencies of teratogen-induced clefting in the A and B parental lines and in the AXB and BXA RI lines assayed in this study are shown in Table 1. The very wide range of clefting observed, including clefting frequencies higher than those found in the susceptible A parental line (e.g., 86% 6-AN induced cleft palate in AXB9), rules out simple monogenic inheritance for this phenotype. An example of the application of our method for estimating type I error for the gene mapping hypothesis is illustrated in Fig. 1 for 6-AN induced cleft palate for markers with typings available for all 10 RI lines. Note that the random combination of RI lines into different hypothetical parental allele groups frequently results in many large χ2 statistics (1 df). This is because the RI lines differ so substantially among each other in frequencies of clefting. In fact, this is what we expect if genes are segregating among the RI lines that influence clefting. Thus, most of the random sortings of lines into two alternative groups yield a contrast that is assessed as quite unlikely to occur by chance alone if all of the RI lines actually had the same clefting frequency. But as noted above, this is not the hypothesis of interest for gene mapping studies. To calculate the type I error for the gene mapping null hypothesis, we determine the proportion of contrast statistics ≥ that observed for a marker. For example, 0.043 of the χ2 statistics were of size 82.3 or greater among all possible combinations of the RI lines used to generate the distribution in Fig. 1. Under the null hypothesis that no gene exists in the chromosomal region of interest that influences clefting susceptibility, we expect to obtain a statistic of this magnitude or greater 4.3% of the time. This illustrates how we use the contrast χ2 statistic as a standardized quantitative assessment of the difference in clefting frequency between the two parental allele groups defined for each marker.
Table 1.
Differences among RI lines in frequency of cleft lip (with exposure to phenytoin) or cleft palate (with exposure to 6-AN)
| Line | Phenytoin
|
6-AN
|
||
|---|---|---|---|---|
| No. of pups | % clefting | No. of pups | % clefting | |
| A parent | 153 | 43.1 | 109 | 54.1 |
| B parent | 180 | 1.1 | 146 | 8.9 |
| AXB2 | 108 | 12.0 | 55 | 5.5 |
| AXB5 | 51 | 5.9 | 24 | 50.0 |
| AXB6 | 105 | 34.3 | 123 | 22.0 |
| AXB7 | 99 | 4.0 | — | — |
| AXB9 | — | — | 59 | 86.4 |
| AXB12 | — | — | 71 | 62.0 |
| AXB13 | 61 | 16.4 | — | — |
| AXB15 | 122 | 0.0 | 107 | 32.7 |
| AXB17 | 109 | 1.8 | 102 | 21.6 |
| BXA2 | — | — | 41 | 39.0 |
| BXA4 | — | — | 37 | 62.2 |
| BXA8 | 58 | 1.7 | — | — |
| BXA14 | 130 | 0.0 | 99 | 85.9 |
Figure 1.
Distribution of χ2 statistics obtained from a logistic regression analysis contrasting the frequency of clefting between all possible combinations of RI lines exposed to 6-AN. Type I error for the gene mapping hypothesis estimated as the proportion of χ2 statistics ≥ the statistic observed for each marker.
Genome Regions Associated with Clefting Susceptibility.
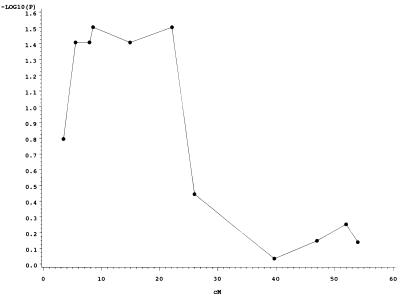
Genome regions with P values < 0.05 for phenytoin-induced cleft lip, for 6-AN induced cleft palate or for the combination of both are presented in Table 2. Regions where several adjacent markers had P values < 0.05 are shown as a range of map locations. In some cases, P values for a region peaked in one location and then dropped off on either side as shown in Fig. 2 for phenytoin-induced clefting and markers on chromosome 19. However, in other cases the pattern was more irregular, with adjacent markers exhibiting inconsistent P values and differences in clefting frequency between parental allele groups. We believe this indicates either that double crossovers occurred in these chromosomal regions during development of the RI lines or that some of the markers may not be mapped accurately. Both possibilities have important implications for future studies aimed at more precisely localizing candidate genes within these regions.
Table 2.
Regions associated with clefting susceptibility
| Teratogen location* | Phenytoin
|
6-AN
|
Either P value | ||||
|---|---|---|---|---|---|---|---|
| A/B† | χ2 | P value | A/B† | χ2 | P value | ||
| 1 (9) | B | 26.2 | 0.078 | A | 58.6 | 0.1016 | 0.046 |
| 3 (54–64) | A | 35.0 | 0.023 | B | 52.8 | 0.1484 | 0.023 |
| 4 (36–45) | A | 35.5 | 0.019 | B | 82.3 | 0.0430 | 0.007 |
| 4 (55) | A | 8.8 | 0.176 | B | 82.3 | 0.0430 | 0.044 |
| 5 (28–34) | B | 79.8 | 0.004 | B | 43.7 | 0.1484 | 0.005 |
| 5 (66) | A | 17.1 | 0.125 | B | 94.9 | 0.0078 | 0.008 |
| 7 (43) | A | 53.4 | 0.031 | B | 71.4 | 0.0313 | 0.008 |
| 8 (15–31) | B | 21.2 | 0.047 | A | 78.0 | 0.0488 | 0.016 |
| 9 (27–39) | B | 5.0 | 0.316 | B | 96.5 | 0.0234 | 0.044 |
| 11 (16–33) | A | 5.2 | 0.297 | A | 105.7 | 0.0313 | 0.053 |
| 12 (3–15) | B | 62.6 | 0.008 | A | 72.9 | 0.0605 | 0.004 |
| 12 (52–60) | B | 62.6 | 0.008 | A | 5.6 | 0.7300 | 0.035 |
| 14 (9) | A | 3.1 | 0.445 | B | 73.5 | 0.0469 | 0.102 |
| 15 (57–59) | B | 26.1 | 0.062 | A | 96.1 | 0.0469 | 0.013 |
| 17 (1) | A | 4.0 | 0.359 | B | 124.1 | 0.0117 | 0.027 |
| 19 (6–22) | A | 24.7 | 0.031 | B | 39.9 | 0.3125 | 0.055 |
Chromosome (map distance in cM from centromere).
Clefting higher with A = A/J, or B = C57BL/6J allele.
Figure 2.
Evidence for phenytoin-induced clefting susceptibility for chromosome 19. P values are shown with negative log10 transformation.
DISCUSSION
Analyses of both human and mouse facial clefting suggest that multiple loci are involved. Our genome-wide analyses show that this also appears to be the case for susceptibility to clefting induced by phenytoin and 6-AN. While we view our findings as strongly supportive we believe that independent confirmation is necessary via studies using both additional RI lines from the AXB and BXA set and backcross and intercross experiments. Our proposal to follow up our findings obtained using RI lines with experimental crosses is consistent with that advocated recently for RI mapping studies of quantitative traits (60). Belknap et al. (60) show that in mapping studies of quantitative traits using the BXD set of 26 RI lines and 1,500 markers P values of 0.0001 are required to yield a 5% type I error rate on a genome-wide basis. This level of significance is probably overly conservative for the AXB data set used in our study, since we had far fewer RI lines and markers. Furthermore, their method of estimating genome wide type I error for a quantitative variable is not directly applicable to our work on clefting, since we are studying a discrete (binary) outcome. However, we fully endorse the two step mapping strategy that they advocate where findings at suggestive levels of statistical significance obtained using RI lines need to be confirmed with intercross or backcross data. This balances the risk of overlooking genes of moderate effect on the phenotype while guarding against too many false positives due to comparison of many linked and unlinked markers throughout the genome.
Because previous studies have shown that both maternal and fetal genomes influence risk of spontaneous (61) and teratogen-induced clefting (42, 62), we believe it is likely that maternal effects may also be relevant to our study. However, because RI lines are highly inbred, maternal and fetal genotypes are identical, and so it is not possible to disentangle these effects. In the future, we plan to address the maternal effect question using crossbreeding designs. We also plan to conduct genetic analyses focused on possible interactions of marker effects with litter size (i.e., some susceptibility genes may act primarily in small or large litters) and resorbtion of fetuses. However, the limited data available at present do not provide sufficient power to integrate these variables as a covariates. Therefore, we plan to first study litter size as an independent variable itself, and assess whether any of the same chromosomal regions implicated in clefting also appear to influence litter size or fetal resorbtion. We recognize the potential advantages of applying procedures such as stepwise logistic regression to simultaneously evaluate evidence of susceptibility for markers distributed across the genome (both linked and unlinked markers). However, entirely new statistical procedures would have to be developed to apply this method using our ranked outcome approach to estimating gene mapping P values. Furthermore, missing data for some RI lines for some marker loci also limit application of stepwise logistic regression. This latter problem may be addressed in future studies by typing additional makers ourselves using the many short tandem repeat loci now available.
An especially interesting aspect of our results relates to the fact that, as noted above, CL(P) and CPO are usually, though not always, considered to be distinct in etiology. A striking pattern questioning this assumption emerges from our study. In Table 2, 13 of the 16 chromosomal regions show the opposite parental alleles associated with CL(P) versus CPO, a pattern unlikely to occur by chance (McNemar’s χ2 = 6.3, 1 df, P < 0.014). This pattern occurs both where only one form of clefting exhibits statistically significant evidence at the P < 0.05 level for a susceptibility locus (9 of 12 cases) as well as where both forms are significant (3 of 3 cases). The implication of this observation, suggested for markers on chromosome 3 in a previous analysis (51), is that some genes may induce susceptibility to both CL(P) and CPO and that allelic variation may determine which form of clefting occurs. This conclusion is also supported by observations from a recent epidemiological study conducted using populations in France, Sweden, and the United States (63). Our finding of potential allelic heterogeneity has implications for gene mapping studies in humans in that it suggests that families with these alternative forms of clefting should be pooled for linkage tests but perhaps analyzed separately for tests of allelic association (linkage disequilibrium). On the other hand, some chromosomal regions appear to confer susceptibility to only one form of clefting indicating that locus heterogeneity also distinguishes the two subtypes of this disorder. This indicates that linkage tests in humans should also be performed for families with CL(P) separate from tests on families with CPO to maximize statistical power for detecting genes involved in only one type of clefting.
We identified homologous locations in the human genome (55, 64) corresponding to the clefting susceptibility regions we found in the mouse. Because it is well known that RI lines provide only approximate map localizations, we included 15 cM flanking the statistically significant regions in Table 2. We found several chromosomal regions implicated in clefting in humans are homologous to locations identified in our study. These include human chromosome 4q25-q31.3 (implicated in nonsyndromic clefting) homologous to mouse chromosomes 3, 5, and 8, and human chromosome 22q11 (containing the Velocardiofacial syndrome gene) homologous to mouse chromosome 5. (A complete table of all regions of homology is available on request.)
Based on our current understanding of the biology of clefting, knowledge of genes found in human syndromes displaying clefting, and mouse mutants with clefting phenotypes, we attempted to identify candidate loci located in the chromosomal regions (±15 cM) identified in this study. We searched the literature and information in the Mouse Genome Informatics databases (55). Many candidate genes found in our search are described below (a complete table is available on request). An especially striking finding is the occurrence of eight collagen genes within our candidate regions. These include two of the three forms of collagen associated with Stickler syndrome (65) with cleft palate (Col2a1 on chromosome 15 and Col11a1 on chromosome 3). Col2a2 mutations also cause this syndrome, but this collagen is not mapped in the mouse. Col3a1, located within our chromosome 1 candidate region, is expressed in the embryonic palate. In addition to Col11a1, the chromosome 3 region contains a cyclic nucleotide phosphodiesterase gene possibly relevant to the role of cAMP in the etiology of CPO (43) and several extracellular matrix/structural proteins. This region has previously been implicated in mice (51) but the homologous human region (1q21) does not appear to be linked to CL(P) in several large families (66). Proximal chromosome 4 contains the gene for tenascin C, an extracellular matrix protein, and several cell-signaling molecules; it, too, had previously been implicated. The more distal region on chromosome 4 includes more gap junction and cell signaling-related molecules.
Over 20 oncogenes or tumor suppresser genes map within the candidate regions, as do 16 genes related to detoxification of xenobiotics such as glutathione S-transferases and members of the cytochrome p450 pathways. Ornithine decarboxylase is noteworthy, as it has been observed to undergo major changes in expression within the developing palate. N-acetyl transferase-1 is an especially strong candidate gene, since findings in RI lines have been confirmed by independent congenic strain analyses (50, 53). Several homeobox and other developmentally important genes lie within our candidate regions including Msx1, Msx2, and Pax9 whose absence (via knockout mutations) have been shown to cause CPO in the mouse. Five genes identified relate to retinoic acid including the beta and gamma receptors (on chromosomes 14 and 15, respectively). The α-receptor identified as possibly being involved in spontaneous clefting (45, 46) lies outside of our candidate region on chromosome 11. Growth factors and their receptors are noteworthy including epidermal growth factor and its receptor. Egf and Egfr are involved in the same pathways as TGFA and, as noted in the introduction, associations have been often found with CL(P) and TGFA. Epidermal growth factor receptor is also believed to play a role in palatal shelf growth (67). Other adrenergic receptors are also within candidate regions, and these are possibly related to cAMP levels. Several fibroblast growth factor receptors map within our candidate regions and these loci are involved with craniofacial syndromes with features including craniosynostosis (68). Other genes identified with previously known roles in clefting include γ-aminobutyric acid (GABA)-A receptors (36) and transforming growth factor β3 (37). Both platelet-derived growth factor and insulin-like growth factor binding proteins are expressed in the developing palate. Mutations in the aggrecan gene (which codes for the proteoglycan link in cartilage) cause cleft palate.
The candidate regions identified in this study also include several classical mutations (where the genes involved have not been cloned) that frequently include cleft lip and/or cleft palate as part of their phenotype. Examples where the association with clefting is especially strong include repeated epilation (CPO), pink-eyed dilution (CPO, GABA-A receptor related), Cleft palate-2 (hydrocortisone-induced CPO), Legless [CL(P) and CPO or both], Dancer [CL(P) and CPO or both] and Brachymorphic (hydrocortisone-induced CPO).
Because the dependent variable of interest in this study, clefting, is categorical in nature, previously developed quantitative trait locus (QTL) methods cannot be applied directly for statistical analyses. This motivated our development of the methods presented here. We noted above that the methods applied previously for analyses of these data for a limited number of marker loci did not directly test the gene mapping hypothesis and were generally anticonservative in nature. In fact, about half of the regions implicated as containing clefting susceptibility loci in these earlier analyses (47–53) were confirmed in the present study, although at less strongly supportive levels of significance than suggested previously. Methods used for analyses of quantitative behavioral traits using RI lines (69) similarly do not appear to directly test the gene mapping hypothesis. The analytical approach we developed here, based on ranking each result within the range of possible outcomes based on all possible combinations of RI lines (Fig. 1), can be applied to continuous dependent variables as well.
In conclusion, we have performed a genome wide search for loci contributing to susceptibility to teratogen-induced facial clefting in the mouse. We developed and applied a new analytical approach that is more conservative than methods applied when these data were analyzed previously using a limited number of marker and candidate loci. Our new analyses confirm that multiple loci contribute to liability to facial clefting. Some of these loci cause susceptibility to both CL(P) and CPO but with different parental strain alleles contributing to liability to the two different forms. Implications of these findings for future studies in humans include suggestions of candidate genes and chromosomal regions and justification for combined analyses of families affected by these different forms of facial clefting.
Acknowledgments
We thank J. Karolyi for collection of clefting phenotype data; M. Monsour for assistance in data management, conducting statistical analyses, and generating figures; C. Bock and D. Smith for computer programming and data management; B. Paigen for marker typing data and advice on analyses; L. Gillanders and A. Miller-Chisholm for database searches; and A. Kingman, D. Weeks, N. Schork, R. Hamer, T. Beaty, H. Slavkin, and K. Yamada for help with statistical analyses and suggestions for improving our manuscript. This is National Institute of Dental Research intramural research project Z01DE00633.
ABBREVIATIONS
- CL(P)
cleft lip with or without cleft palate
- CPO
cleft palate only
- 6-AN
6-aminonicotinamide
- RI
recombinant inbred
- TGFA
transforming growth factor-α
References
- 1.Ching C H S, Chung C S. Am J Hum Genet. 1974;26:162–176. [PMC free article] [PubMed] [Google Scholar]
- 2.WHO Scientific Group. WHO Tech Rep Ser. 1970;438:11–17. [PubMed] [Google Scholar]
- 3.Marazita M L, Spence M A, Melnick M. Am J Med Genet. 1984;19:9–19. doi: 10.1002/ajmg.1320190104. [DOI] [PubMed] [Google Scholar]
- 4.Chung C A S, Bixler D, Watanabe T, Koguchi H, Fogh-Anderson P. Am J Hum Genet. 1986;39:603–611. [PMC free article] [PubMed] [Google Scholar]
- 5.Chabora H J, Horowitz S L. Oral Surg. 1974;38:1818–186. doi: 10.1016/0030-4220(74)90053-x. [DOI] [PubMed] [Google Scholar]
- 6.Bonaiti C, Briard M L, Feingold J, Pavy B, Psaume J, Migne-Tufferaud G, Kaplan J. J Med Genet. 1982;19:8–15. doi: 10.1136/jmg.19.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nemana L J, Marazita M L, Melnick M. Am J Med Genet. 1992;42:5–9. doi: 10.1002/ajmg.1320420103. [DOI] [PubMed] [Google Scholar]
- 8.Marazita M L, Hu D-N, Spence M A, Liu Y-E, Melnick M. Am J Hum Genet. 1992;51:648–653. [PMC free article] [PubMed] [Google Scholar]
- 9.Ray A K, Field L L, Marazita M L. Am J Hum Genet. 1993;52:1006–1011. [PMC free article] [PubMed] [Google Scholar]
- 10.Hecht J T, Yang P, Michels V V, Buetow K H. Am J Hum Genet. 1991;49:674–681. [PMC free article] [PubMed] [Google Scholar]
- 11.Palomino H, Li S-C, Palomino H M, Barton S A, Chakraborty R. Am J Hum Genet. 1991;495:154. [Google Scholar]
- 12.Farrall M, Holder S. Am J Hum Genet. 1992;50:270–277. [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell L E, Risch N. Am J Hum Genet. 1992;51:323–332. [PMC free article] [PubMed] [Google Scholar]
- 14.Murray J C. Am J Hum Genet. 1995;57:227–232. [PMC free article] [PubMed] [Google Scholar]
- 15.Ardinger H H, Buetow K H, Bell G I, Bardach J, Van De Mark D R, Murray J C. Am J Hum Genet. 1989;45:348–353. [PMC free article] [PubMed] [Google Scholar]
- 16.Farrall M, Buetow K H, Murray J C. Am J Hum Genet. 1993;52:434–436. [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang S-J, Beaty T H, Panny S R, Street N A, Joseph J M, Gordon S, McIntosh L, Francomano C A. Am J Epidemiol. 1995;141:629–636. doi: 10.1093/oxfordjournals.aje.a117478. [DOI] [PubMed] [Google Scholar]
- 18.Shaw G M, Wasserman C R, Lammer E J, O’Malley C D, Murray J C, Basart A M, Tolarova M M. Am J Hum Genet. 1996;58:551–561. [PMC free article] [PubMed] [Google Scholar]
- 19.Shiang R, Lidral A C, Ardinger H H, Buetow K H, Romitti P, Munger R, Murray J C. Am J Hum Genet. 1993;53:1156–1157. [PMC free article] [PubMed] [Google Scholar]
- 20.Chenevix-Trench G, Jones K, Green A C, Duffy D L, Martin N G. Am J Hum Genet. 1992;51:1377–1385. [PMC free article] [PubMed] [Google Scholar]
- 21.Davies A F, Stephens R J, Olavesen M G, Heather L, Dixon M J, Magee A, Flinter F, Ragoussis J. Hum Mol Genet. 1995;4:121–128. doi: 10.1093/hmg/4.1.121. [DOI] [PubMed] [Google Scholar]
- 22.Jara L, Blanco R, Chiffelle I, Palomino H, Carreno H. Am J Hum Genet. 1995;56:339–341. [PMC free article] [PubMed] [Google Scholar]
- 23.Stein J, Mulliken J B, Stal S, Gasser D L, Malcolm S, Winter R, Blanton S H, Amos C, Seemanova E, Hecht J T. Am J Hum Genet. 1995;57:257–272. [PMC free article] [PubMed] [Google Scholar]
- 24.Beiraghi S, Foroud T, Dlouhy S, Bixler D, Delohter-Blanchet D, Conneally P M, Hodes M E. Clin Genet. 1994;46:255–256. doi: 10.1111/j.1399-0004.1994.tb04236.x. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell L E, Healey S C, Chenevix-Trench G. Am J Hum Genet. 1995;57:1130–1136. [PMC free article] [PubMed] [Google Scholar]
- 26.Sander A, Murray J C, Scherpbier-Heddema T, Buetow K H, Weissenbach J, Zingg M, Ludwig K, Schmelzle R Am. Am J Hum Genet. 1995;56:310–318. [PMC free article] [PubMed] [Google Scholar]
- 27.Gorski S M, Adams K J, Birch P H, Friedman J M, Goodfellow P J. Am J Hum Genet. 1992;50:1129–1136. [PMC free article] [PubMed] [Google Scholar]
- 28.Francomano C A, Liberfarb R M, Hirose T, Maumenee I H, Streeten E A, Meyers D A, Pyeritz R E. Genomics. 1987;1:293–296. doi: 10.1016/0888-7543(87)90027-9. [DOI] [PubMed] [Google Scholar]
- 29.Driscoll D A, Spinner N B, Budarf M L, McDonald-McGinn D M, Zackai, et al. Am J Med Genet. 1992;44:261–268. doi: 10.1002/ajmg.1320440237. [DOI] [PubMed] [Google Scholar]
- 30.Gorlin R J, Cohen M M, Levin L S. Syndromes of the Head and Neck. New York: Oxford Univ. Press; 1990. [Google Scholar]
- 31.Satokata I, Maas R. Nat Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- 32.Chisaka O, Capecchi M R. Nature (London) 1991;350:473–479. doi: 10.1038/350473a0. [DOI] [PubMed] [Google Scholar]
- 33.Condie B G, Capecchi M R. Nature (London) 1994;370:304–307. doi: 10.1038/370304a0. [DOI] [PubMed] [Google Scholar]
- 34.Damm K, Heyman R A, Umesono K, Evans R M. Proc Natl Acad Sci USA. 1993;90:2989–2993. doi: 10.1073/pnas.90.7.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Culiat C T, Stubbs L J, Woychik R P, Russell L B, Johnson D K, Rinchik E M. Nat Genet. 1995;11:344–346. doi: 10.1038/ng1195-344. [DOI] [PubMed] [Google Scholar]
- 36.Proetzel G, Pawlowski S A, Wiles M V, Yin M, Boivin G P, Howles P N, Ding J, Ferguson M W J, Doetschman T. Nat Genet. 1995;11:409–414. doi: 10.1038/ng1295-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaartinen V, Voncken J W, Shuler C, Warburton D, Bu D, Heisterkamp N, Groffen J. Nat Genet. 1995;11:415–421. doi: 10.1038/ng1295-415. [DOI] [PubMed] [Google Scholar]
- 38.Kimata K, Line S, Strong D, Gao L Y, Kozak C A, Yamada Y. Nat Genet. 1994;7:154–157. doi: 10.1038/ng0694-154. [DOI] [PubMed] [Google Scholar]
- 39.Matzuk M M, Kuman T R, Vassalli A, Bickenbach J R, Roop D R, Jaenisch R, Bradley A. Nature (London) 1995;384:354–356. doi: 10.1038/374354a0. [DOI] [PubMed] [Google Scholar]
- 40.Lyon M F. J Embryol Exp Morphol. 1958;6:105–116. [PubMed] [Google Scholar]
- 41.Deol M S, Lane P W. J Embryol Exp Morphol. 1966;16:543–558. [PubMed] [Google Scholar]
- 42.Bonner J J, Slavkin H C. Immunogenetics. 1975;2:213–218. [Google Scholar]
- 43.Erickson R P, Butley M S, Sing C F. J Immunogenet. 1979;6:253–262. doi: 10.1111/j.1744-313x.1979.tb00682.x. [DOI] [PubMed] [Google Scholar]
- 44.Gasser D, Mele L, Lees D, Goldman A. Proc Natl Acad Sci USA. 1981;78:3147–3150. doi: 10.1073/pnas.78.5.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Juriloff D M, Mah D G. Mamm Genome. 1995;6:63–69. doi: 10.1007/BF00303246. [DOI] [PubMed] [Google Scholar]
- 46.Juriloff D M. J Craniofacial Genet Dev Biol. 1995;15:1–12. [PubMed] [Google Scholar]
- 47.Liu S J, Erickson R P. Genetics. 1986;113:745–754. doi: 10.1093/genetics/113.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu S J, Erickson R P. Genetics. 1986;113:735–744. doi: 10.1093/genetics/113.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lovell D P, Erickson R P. Genetics. 1986;113:755–764. doi: 10.1093/genetics/113.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karolyi J, Erickson R P, Liu S, Killewald L. Genetics. 1990;126:201–205. doi: 10.1093/genetics/126.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karolyi J, Erickson R P. J Craniofacial Genet Dev Biol. 1993;13:1–5. [PubMed] [Google Scholar]
- 52.Karolyi I J, Liu S, Erickson R P. Genet Res. 1987;49:43–49. doi: 10.1017/s0016672300026719. [DOI] [PubMed] [Google Scholar]
- 53.Karolyi J, Erickson R P, Liu S. Teratology. 1988;37:283–287. doi: 10.1002/tera.1420370314. [DOI] [PubMed] [Google Scholar]
- 54.Nesbitt M N, Skamene E. J Leukocyte Biol. 1984;36:357–364. doi: 10.1002/jlb.36.3.357. [DOI] [PubMed] [Google Scholar]
- 55.Mouse Genome Database. Mouse Genome Informatics. Bar Harbor, ME: The Jackson Laboratory; 1996. http://www.informatics.jax.org/ , World Wide Web (URL: http://www.informatics.jax.org/), Version 3.1. ), Version 3.1. [Google Scholar]
- 56.Marshall J D, Mu J-L, Cheah Y-C, Nesbitt M N, Frankel W N, Paigen B. Mamm Genome. 1992;3:669–680. doi: 10.1007/BF00444361. [DOI] [PubMed] [Google Scholar]
- 57.Manly K, Cudmore R. map manager: A Program for Genetic Mapping. Buffalo: State Univ. of New York; 1994. http://mcbio.med.buffalo.edu/mapmgr.html , World Wide Web (URL: http://mcbio.med.buffalo.edu/mapmgr.html) Version 2.6. ) Version 2.6. [Google Scholar]
- 58.SAS Institute. SAS/STAT Software Changes and Enhancement Through Release 6.11. Cary, NC: SAS Institute; 1996. [Google Scholar]
- 59.Fisher R A. Statistical Methods for Research Workers. 12th Ed. Edinburgh: Oliver & Boyd; 1954. [Google Scholar]
- 60.Belknap J K, Mitchell S R, O’Toole L A, Helms M L, Crabbe J C. Behav Genet. 1996;26:149–160. doi: 10.1007/BF02359892. [DOI] [PubMed] [Google Scholar]
- 61.Davidson J G, Fraser F C, Schlager G. Teratology. 1969;2:371–376. doi: 10.1002/tera.1420020411. [DOI] [PubMed] [Google Scholar]
- 62.Melnick M, Jaskoll T, Slavkin H C. Immunogenetics. 1981;13:443–450. doi: 10.1007/BF00346025. [DOI] [PubMed] [Google Scholar]
- 63.Källén B, Harris J, Robert E. J Craniofacial Genet Dev Biol. 1996;16:242–248. [PubMed] [Google Scholar]
- 64.DeBry R W, Seldin M F. Genomics. 1996;33:337–351. doi: 10.1006/geno.1996.0209. [DOI] [PubMed] [Google Scholar]
- 65.Vikkula M, Mariman E C M, Lui V C H, Zhidkova N I, Tiller G E, Goldring M B, van Beersum S E C, de Waal Malefijt M C, van den Hoogen F H J, Ropers H-H, Mayne R, Cheah K S E, Olsen B R, Warman M L, Brunner H G. Cell. 1995;80:431–437. doi: 10.1016/0092-8674(95)90493-x. [DOI] [PubMed] [Google Scholar]
- 66.Pierpont J W, Storm A L, Erickson R P, Kohn B R, Pettijohn L, DePaepe A. J Craniofacial Genet Dev Biol. 1995;15:66–71. [PubMed] [Google Scholar]
- 67.Nexo E, Hollenberg M D, Figueroa A, Pratt R M. Proc Natl Acad Sci USA. 1980;77:2782–2785. doi: 10.1073/pnas.77.5.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gorry M C, Preston R A, White G J, Zhang Y, Singhal V K, Losken H W, Parker M G, Nwokoro N A, Post J C, Ehrlich G D. Hum Mol Genet. 1995;4:1387–1390. doi: 10.1093/hmg/4.8.1387. [DOI] [PubMed] [Google Scholar]
- 69.Crabbe J C, Belknap J K, Buck K J. Science. 1994;264:1715–1723. doi: 10.1126/science.8209252. [DOI] [PubMed] [Google Scholar]