Abstract
We have investigated the protective role of the membrane-bound HLA-G1 and HLA-G2 isoforms against natural killer (NK) cell cytotoxicity. For this purpose, HLA-G1 and HLA-G2 cDNAs were transfected into the HLA class I-negative human K562 cell line, a known reference target for NK lysis. The HLA-G1 protein, encoded by a full-length mRNA, presents a structure similar to that of classical HLA class I antigens. The HLA-G2 protein, deduced from an alternatively spliced transcript, consists of the α1 domain linked to the α3 domain. In this study we demonstrate that (i) HLA-G2 is present at the cell surface as a truncated class I molecule associated with β2-microglobulin; (ii) NK cytolysis, observed in peripheral blood mononuclear cells and in polyclonal CD3− CD16+ CD56+ NK cells obtained from 20 donors, is inhibited by both HLA-G1 and HLA-G2; this HLA-G-mediated inhibition is reversed by blocking HLA-G with a specific mAb; this led us to the conjecture that HLA-G is the public ligand for NK inhibitory receptors (NKIR) present in all individuals; (iii) the α1 domain common to HLA-G1 and HLA-G2 could mediate this protection from NK lysis; and (iv) when transfected into the K562 cell line, both HLA-G1 and HLA-G2 abolish lysis by the T cell leukemia NK-like YT2C2 clone due to interaction between the HLA-G isoform on the target cell surface and a membrane receptor on YT2C2. Because NKIR1 and NKIR2, known to interact with HLA-G, were undetectable on YT2C2, we conclude that a yet-unknown specific receptor for HLA-G1 and HLA-G2 is present on these cells.
HLA-G is a nonclassical major histocompatibility complex (MHC) class I antigen selectively expressed on extra-villous trophoblast cells at the fetal–maternal interface (1–3). Although the fetus may be considered a semiallograft, fetal cells survive without immunological rejection. One possible explanation for the nonrejection of the fetus is the nonexpression of classical HLA class I and II molecules on the trophoblast, in which case maternal alloreactive T cells could neither recognize nor destroy fetal cells. However, the lack of classical HLA class I expression on trophoblast cells is known to not protect them from natural killer (NK) cytotoxicity. In fact, NK cells are specialized in killing cells that no longer express MHC class I molecules. It was thus proposed that the fetus is protected from maternal NK cell cytolysis by the expression of HLA-G on the trophoblast (4). According to this hypothesis, HLA-G mediates protection from NK cells, thereby playing an important role in maintaining maternal immune tolerance of the semiallogenic fetus (for review, see refs. 5 and 6). It is now well established that NK cells express receptors for MHC class I molecules, which are responsible for the inhibition of cytotoxicity, and that such HLA molecules act as ligands that are recognized by killer inhibitory receptors (KIRs) (7–9). Recently, Pazmany et al. (10) showed that expression of HLA-G protected an otherwise susceptible B lymphoma LCL 721.221 target cell line from lysis by NK clones and identified the receptors on NK cells that recognize HLA-G, namely NK inhibitory receptor 1 (NKIR1) and NKIR2 (11).
In contrast to classical HLA class I genes, the primary transcript of the HLA-G gene is alternatively spliced in cytotrophoblasts and other fetal and adult tissues (12), resulting in at least five different HLA-G mRNAs that potentially encode five HLA-G isoforms, namely, the HLA-G1, HLA-G2, HLA-G3, and HLA-G4 membrane-bound isoforms, as well as the soluble HLA-G5 form (13–15). Previous experiments have been carried out by other groups using target cells transfected with a full-length HLA-G locus genomic DNA clone that can potentially generate all alternatively spliced transcripts (16–17). Therefore, in these transformed eucaryotic target cells, expression of a given isoform at the cell surface cannot be predicted. To further characterize the immunological functions of HLA-G products, we studied the protective roles of the HLA-G1 and HLA-G2 isoforms with respect to NK lytic activity in 20 donors. Our results show that the HLA-G1 isoform that presents classical HLA class I structure, consisting of α1, α2, and α3 extracellular domains (the latter associated with a β2-microglobulin (β2m) domain) is capable of inhibiting the cytotoxicity of polyclonal NK cells. We have also demonstrated that the HLA-G2 isoform, which lacks the α2 domain, is capable of protecting target cells from NK lysis. We describe the presence of this isoform at the cell surface in the form of a truncated class I molecule associated with β2m. Our results suggest that the α1 extracellular domain common to HLA-G1 and HLA-G2 molecules mediates this protection from NK lysis. Furthermore, the fact that HLA-G inhibited the NK lytic activity of all the donors tested suggests a possible role for HLA-G as a public ligand for NKIRs. This hypothesis is supported by the fact that the HLA-G protein, as opposed to classical HLA class I molecules, exhibits limited polymorphism (18), thus stimulating all inhibitory NK receptors. Finally, HLA-G1 and HLA-G2 transfected into the K562 cell line abolished the cytotoxicity of the NK-like YT2C2 clone that expresses no KIR known to interact with HLA-G. This indicates the existence of a yet-unknown KIR(s) involved in HLA-G recognition.
MATERIALS AND METHODS
Cell Lines.
The human erythroleukemia cell line K562 (American Type Culture Collection) and the nonadult T cell leukemia, NK-mediating YT2C2 clone (19) kindly provided by P. Paul (Hôpital Saint-Louis, Paris) were maintained in RPMI medium supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, and 1 μg/ml gentamicin, and fungizone (Sigma) and cultured in a 37°C, 5% CO2, humidified incubator. K562 transfectants were selected in media containing 1 mg/ml geneticin (G418 sulfate; Sigma). The human choriocarcinoma HLA-G-positive cell line JEG-3 (American Type Culture Collection) was cultured in DMEM (Sigma) supplemented with 10% heat-inactivated fetal calf serum with antibiotics and 2 mM l-glutamine. The cell lines used in this study were routinely tested for and found to be free from mycoplasma.
HLA-G Vector Constructions.
To generate HLA-G1 and HLA-G2 transfectants, the corresponding cDNAs were inserted into the pRc/RSV eukaryotic expression vector (Invitrogen) containing the Rous sarcoma virus (RSV) as the promoter and the neomycin gene as a selectable marker. The general strategy of vector construction is shown in the Fig. 1. Briefly, the 1.5-kb HLA-G1 full-length cDNA EcoRI fragment was blunted using a DNA polymerase I large fragment (Klenow) and ligated into HindIII-digested pRc/RSV vector blunted and dephosphorylated by calf intestinal phosphatase (GIBCO/BRL). The correct orientation of this construction, designated pRc/RSV-G1, was confirmed by restriction digest analysis and subsequent sequencing. To generate the HLA-G2 recombinant pRc/RSV plasmids, the full-length HLA-G1 cDNA EcoRI fragment was ligated into the EcoRI site of the pGEX-4T-3 vector (Pharmacia Biotech). The HLA-G2 cDNA fragment was obtained by PCR amplification, as described (14), using the HLA-G-specific primers G.53 (5′-CCCTGACCCTGACCGAGACCTGGG-3′) and G.1225 (14). It was then digested by NcoI/BsmI restriction enzymes and ligated into the pGEX-HLA-G1 vector, from which the corresponding NcoI/BsmI fragment containing the α2-domain coding region had been previously removed (pGEX-HLA-G2). The HLA-G2 expression plasmid, designated pRc/RSV-G2, was obtained after EcoRI digestion of the pGEX- HLA-G2 vector and the cDNA fragment blunted and ligated into HindIII-digested pRc/RSV that had already been blunted and dephosphorylated.
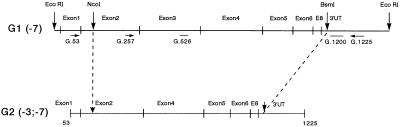
Figure 1.
Construction of the HLA-G2 cDNA fragment. Vertical arrowheads indicate restriction enzyme sites specific for the generation of HLA-G2 cDNA from HLA-G1 full-length cDNA. Horizontal arrowheads indicate PCR primers (G.53, G.257, and G.1225) and heavy bars without arrow indicate HLA-G-specific probes used for hybridization (G.526 and G.1200).
HLA-G Transfectant Cell Lines.
Vectors containing pRc/RSV-G1 and pRc/RSV-G2 as well as the pRc/RSV vector alone as a control were transfected into K562 cells by electroporation (Bio-Rad Gene Pulsar at 250V, 960 μF). After 2 days of culture, the cells were selected with 1 mg/ml geneticin. Transfectant cells were monitored by flow cytometry analysis to confirm the expression of HLA class I molecules (see below). To obtain the highest level of HLA-G expression in the K562 transfectants, the cells were isolated by cell-sorting (FACS Vantage; Becton Dickinson) after HLA class I-specific mAb staining.
Reverse Transcriptase–PCR (RT-PCR) Analysis.
Total mRNA from 107 cells was extracted using the RNA NOW reagent (Ozyme) according to manufacturer’s recommendations. The quality of RNA was checked by electrophoresis in 1.5% agarose denaturing gel. Complementary DNAs were prepared from 10 μg of total RNA using oligo(dT)12–18 primer and Moloney murine leukemia virus RT (GIBCO/BRL). RT-PCR amplifications were carried out using two HLA-G-specific primers, G.257 (exon 2) and G.1225 (3′-UT), as described (14). In all PCR amplifications we used the RT reaction mixture without Moloney murine leukemia virus RT (RT−) and the PCR mixture without a cDNA template (blank) as controls. Additional RT-PCR amplification of β-actin was performed to evaluate the amount of RNA in all samples. PCR products were analyzed by electrophoresis in 1% agarose gel and stained with ethidium bromide. The specificity of PCR products was confirmed by alkaline blotting of the fragments in 0.4 M NaOH onto nylon membranes (Hybond N+; Amersham). Hybridization was accomplished with G.1200 and G.526 (exon 3-specific) probes, as described (14). The filters were exposed to Kodak (Biomax) films with amplifying screens for 4–16 h at −80°C.
Immunoprecipitation and SDS/PAGE.
The JEG-3 and K562 cell lines as well as the transfectants were surface-labeled with Biotin (Pharmacia), then washed and solubilized in lysis buffer containing 1% Triton X-100, 50 mM Tris⋅HCl (pH 7.4), 1 mM EDTA, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin (Sigma). Precleared cell lysates were divided in two samples and to each was added protein A-Sepharose beads incubated previously with either mouse anti-class I W6/32 mAb or irrelevant isotype-matched mAb (UPC-10, mouse IgG2a; Sigma). After incubation for 2 h at 4°C, the beads bearing the immune complexes were washed and treated with SDS sample buffer and the bound antigen eluted by boiling the beads. Precipitated proteins were analyzed by electrophoresis in 10% SDS/PAGE. The gels were blotted onto nitrocellulose filters, blocked in PBS/0.2% Tween 20/5% BSA (incubated overnight at 4°C) and washed in PBS/Tween 20. The filters were subsequently incubated for 40 min at room temperature with streptavidin-horseradish peroxidase conjugate reagent. After extensive washing of the membrane, the staining reaction was carried out, using ECL Western blotting detection reagent (Amersham), after which the membrane was exposed to Kodak film at room temperature.
mAbs and Flow Cytometry Analysis.
We used the following mAbs: W6/32, IgG2a anti-HLA class I α chains associated with β2m (Sigma); B1G6, IgG2a anti-β2m (Sigma); GL183, IgG1 anti-p58 (Immunotech, Marseille, France); EB6, IgG1 anti-p58 (Immunotech); HP-3B1, IgG2a anti-CD94 (Immunotech); UCHT-1, IgG1 anti-CD3 conjugated with quantum red (Sigma); 3G8, IgG1 anti-CD16 conjugated with fluorescein (Immunotech); and B159, IgG1 anti-CD56 conjugated with phycoerythrin (Immunotech).
For flow cytometry assays, cells were washed in PBS and stained with the corresponding mAb in PBS/1% BSA for 30 min at 4°C. After washing twice in PBS/1% BSA, cells were either directly analyzed in a flow cytometer (FACS Vantage; Becton Dickinson) if the mAb used was conjugated to fluorochrome or stained with an F(ab′)2 goat anti-mouse IgG antibody conjugated with phycoerythrin (Immunotech) before FACS analysis. Control aliquots were stained with an isotype-matched antibody to evaluate nonspecific binding to target cells.
NK Effectors.
Peripheral blood mononuclear cells (PBMC) from healthy adult volunteer donors (male and female, aged 30–60 years) were obtained by Ficoll/histopaque density gradient. NK cells were isolated from PBMC as follows. First, CD3+ cell-depletion was accomplished using anti-CD3-coated Dynabeads (Dynal, Oslo). The coated Dynabeads were mixed with the PBMC using a 4:1 ratio of beads to target CD3 cells. After incubation at 4°C for 30 min, the target cells rosetted with Dynabeads were depleted, using Dynal Magnetic Particle Concentrators. The CD3-depleted fraction was then stained with phycoerythrin-conjugated anti-CD56 mAb to select CD56+ cells by flow cytofluorometry cell sorting, using a FACS Vantage. To control the phenotype of sorted cell subsets (CD56+), a sample of cells was immediately stained with both fluorescein isothiocyanate-conjugated anti-CD16 mAb and quantum-red-conjugated anti-CD3 mAb, followed by three-color immunofluorescence analysis. The CD56+ population obtained was viable (>95%) and exhibited a phenotype that corresponded to that of NK cells (>90% CD3− CD16+ CD56+).
Cytotoxicity Assays.
The cytolytic activities of PBMC, NK cells, and YT2C2 cells against the HLA-G transfectants were assessed in 4-h 51Cr-release assays in which effector cells were mixed with 5 × 103 51Cr-labeled targets (100 μCi of 51Cr sodium chromate; 1 Ci = 37 GBq; Amersham) at different effector/target ratios in U-bottomed microtiter plates. After 4 h at 37°C in a humidified 5% CO2 incubator, 100 μl of the supernatant was collected for liquid scintillation counting (Wallac 1450 Microbeta; Pharmacia). The percentage of specific lysis was calculated as follows: percent specific lysis = [(cpm experimental well − cpm spontaneous release)/(cpm maximum release-cpm spontaneous release)] × 100. Spontaneous release was determined by incubation of labeled target cells with medium. Maximum release was determined by solubilizing target cells in 0.1 M HCl. In all experiments, the spontaneous release was <10% of maximum release. Results are presented as the means of triplicate samples. In experiments in which mAbs were used to block HLA-G–NK interaction, target cells were incubated with the corresponding mAb, then washed and incubated with an F(ab′)2 goat anti-mouse IgG antibody (Jackson ImmunoResearch) to prevent antibody-dependent cell cytotoxicity by interaction of NK cell Fc receptors with the first mAb used. mAb toxicities were checked in each assay and were always <3%.
RESULTS
Generation of Stable Transfectants Expressing HLA-G1 and HLA-G2 Molecules.
Analysis of the transfected HLA-G1 and HLA-G2 cDNA sequences demonstrated that the HLA-G1 cDNA did not contain the two GAC codons at amino acid positions 247 and 248 previously assigned (20), but only a single GAC codon at position 247, as recently described (10). The HLA-G2 cDNA sequence, obtained by RT-PCR amplification with G.53–G.1225 primers, presented a junction between exons 2 and 4, due to the lack of exon 3 in this alternatively spliced mRNA form (data not shown).
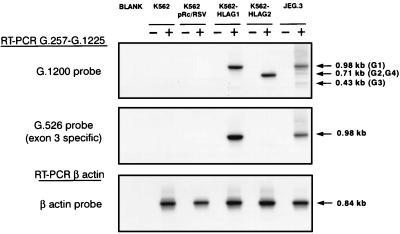
Characterization of the transfectants was carried out by Southern blot analysis of the RT-PCR products obtained using the G.257-G.1225 primers with the K562 parental cell line, transfectants, and JEG-3 cells (Fig. 2). Hybridization of the corresponding RT-PCR products obtained with the G.1200 probe revealed a band at 0.98 kb for K562-HLA-G1, assigned to the HLA-G1 mRNA and a band at 0.71 kb, assigned to the alternatively spliced form, which lacks exon 3, according to our previous results (21). As expected in JEG-3, three bands were observed at 0.98 kb (HLA-G1), 0.7 kb (HLA-G2 and HLA-G4), and 0.43 kb (HLA-G3) (22). In contrast, no HLA-G bands were detected in the K562 parental cell line or in the K562 pRc/RSV control. Hybridization of the same RT-PCR with the exon 3-specific G.526 probe revealed a single band in the HLA-G1-transfected cells, whereas no hybridization was observed in the HLA-G2 transfectant, due to the lack of exon 3 in the HLA-G2 form (Fig. 2). RT-PCR amplification of the constitutively expressed β-actin gene demonstrated that the absence of HLA-G bands in the K562 controls was not due to the absence of RNA (Fig. 2).
Figure 2.
Detection of HLA-G transcripts in the K562 parental cell line, transfectants, and the JEG-3 cell line. Reversed RNAs were amplified using G.257 and G.1225 HLA-G-specific primers and Southern blot was hybridized with G.1200 or G.526 32P-labeled probe. Positive (+) and negative (−) lanes correspond to the RT+ and RT− templates, and the blank is a control performed using PCR mixture without the cDNA template, as described. To control the amount of RNA in all samples, RT-PCR amplification results obtained with β-actin-specific primers and Southern blots were hybridized with β-actin 32P-labeled probe.
Immunoprecipitation and SDS/PAGE Analysis of Labeled Cell-Surface Proteins.
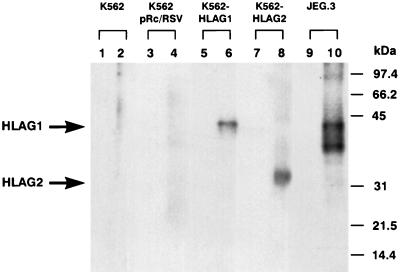
Immunoprecipitation of the HLA proteins with the W6/32 mAb was carried out to establish whether the transfections led to the specific expression of HLA-G proteins. As presented in Fig. 3, the corresponding Western blot revealed that the W6/32 mAb immunoprecipitated a 39-kDa molecule in K562 cells that expressed HLA-G1 protein (lane 6) and in JEG-3 (lane 10). For the HLA-G2 transfectant, W6/32 immunoprecipitation showed one band of expected size for the G2 protein. No classical HLA class I, HLA-G1, or HLA-G2 molecules were found in either the K562 parental cell line or in the K562 pRc-RSV control cell line.
Figure 3.
Immunoprecipitation, SDS/PAGE, and Western blot analysis of biotin-labeled cell surface proteins. Biotinylated membrane lysates from K562 (lanes 1 and 2), K562-pRc/RSV (lanes 3 and 4), K562-HLA-G1 (lanes 5 and 6), K562-HLA-G2 (lanes 7 and 8), and JEG-3 (lanes 9 and 10) were immunoprecipitated with control mAb UPC-10 (lanes 1, 3, 5, 7, and 9) or with mAb W6/32 (lanes 2, 4, 6, 8, and 10) and analyzed by 10% SDS/PAGE under reducing conditions. Size markers in kDa are indicated at the right.
Flow Cytometry Analysis of HLA-G Transfectants.
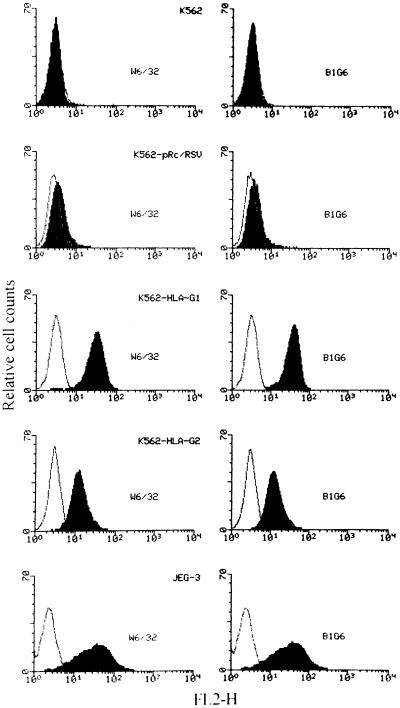
To determine the levels of HLA-G cell surface expression, flow cytometry analysis profiles of the K562 parental cell line, the pRc/RSV, HLA-G1, and HLA-G2 transfectants, as well as of the HLA-G- positive choriocarcinoma JEG-3 cell line are shown in Fig. 4. The K562 parental cell line did not express detectable levels of HLA class I surface molecules, as assessed by the W6/32 pan-HLA class I and the B1G6 anti-β2m mAbs. Also, no HLA class I molecule was detected in the K562 cell line that had been transfected with the empty pRc/RSV vector. The HLA-G1 transfectant was stained by the W6/32 and B1G6 mAbs to approximately the same extent as obtained with JEG-3 cells. The HLA-G2 transfectant was also stained by W6/32, as well as by the B1G6 mAb. This result demonstrates for the first time that the membrane-bound HLA-G2 isoform is present at the cell surface in association with β2m in the same way that it is in classical HLA class I molecules.
Figure 4.
Expression of HLA molecules on the transfectants of the K562 cell line detected by cytofluorometry. Wild-type K562 cells (K562), K562 cells transfected with either the vector alone (K562 pRc/RSV), HLA-G1 (K562-HLA-G1) or with HLA-G2 (K562-HLA-G2), and JEG-3 cells were labeled by indirect immunofluorescence with the following primary mAbs (bold profiles): W6/32 (monomorphic anti-class I) (Left) and B1.G6 (anti-human β2m) (Right). Controls were the same cells stained with an isotype-matched control antibody (light profiles). After washing, cells were stained with phycoerythrin-conjugated F(ab′)2 goat anti-mouse IgG. One of four representative experiments is shown.
Resistance of HLA-G1 and HLA-G2 Transfectants to NK Cytolysis.
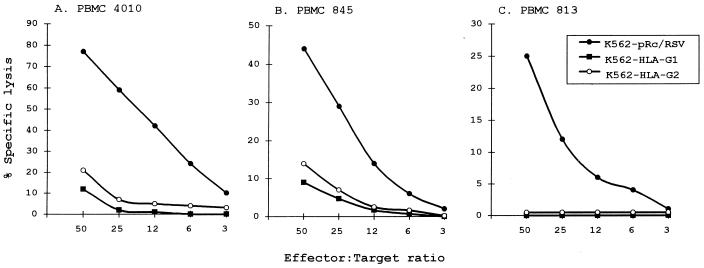
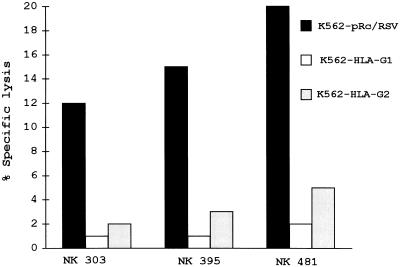
The HLA-G1 and HLA-G2 transfectants were tested along with their vector-carrying parental line for sensitivity to NK killing. Twenty experiments were carried out, each recorded with mononuclear cells from a different donor. Fig. 5 shows that NK lytic activity present in PBMC isolated from three healthy adult donors was inhibited by both K562 HLA-G1 and HLA-G2 transfectants, whereas the K562 pRc/RSV control cell line remained sensitive to NK lytic activity. Although absolute levels of lysis varied from donor to donor, as presented in Fig. 5 for donors 4010, 845, and 813, who exhibited high, medium, and low levels of lysis, respectively, the sensitivity of both HLA-G1- and HLA-G2-expressing cells was reduced by at least 70%. These results were confirmed using polyclonal CD3− CD16+ CD56+ NK cells obtained from the PBMC of three other donors as effectors (Fig. 6). No difference was found between male and female donors.
Figure 5.
Effect of HLA-G1 and HLA-G2 expression on sensitivity to NK lysis of PBMC. K562 transfected with either the vector alone, with HLA-G1, or with HLA-G2 were used as targets. Freshly isolated PBMC from (A) donor 4010 (HLA-A3, -B44, -B56, and -Cw1), (B) donor 845 (HLA-A1, -A28, -B8, -B51, and -Cw7), and (C) donor 813 (HLA-A2, -B27, -B51, and -Cw1) were used as effectors. The results are expressed as the percentage lysis recorded in a 4-h 51Cr-release assay. The standard deviation of the mean of the triplicates was below 5%, and the spontaneous release never exceeded 10% of the maximum release.
Figure 6.
Effect of HLA-G1 and HLA-G2 expression on sensitivity to lysis of polyclonal NK cells. K562 transfected with either the vector alone, with HLA-G1, or with HLA-G2, were used as targets for NK cells (>90% CD3− CD16+ CD16+) isolated from donor 303 (HLA-A29, -B44, -B62, and -Cw3, -Cw5) at 1:1 effector/target (E/T) ratio, donor 395 (HLA-A2, -B13, -B44, and -Cw1) at 1:1 E/T ratio, and donor 481 (HLA-A3, -A26, -B27, -B35, and -Cw4, -Cw2) at 5:1 E/T ratio. The results are expressed as the percentage lysis recorded in a 4-h 51Cr-release assay. The standard deviation of the mean of the triplicates was below 5%, and the spontaneous release never exceeded 10% of the maximum release.
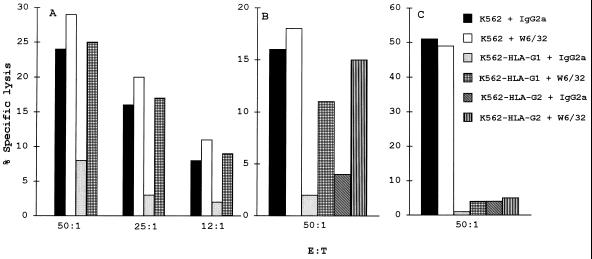
To show that the inhibition of NK lysis was due to the presence of HLA-G1 and HLA-G2 molecules on the cell surface, cytotoxicity assays were performed in the presence of target cells preincubated with either the W6/32 mAb, which recognizes HLA-G1 and HLA-G2 molecules, or an isotype-matched control mAb. The addition of W6/32 reversed HLA-G1- and HLA-G2-mediated inhibition of NK cell activity, thus restoring lysis of the transfected target cells (Fig. 7 A and B). Because the mechanism of antibody-dependent cell cytotoxicity might likely be involved in this reaction by interaction with NK cell Fc receptors, the Fc fragment of W6/32 was blocked by F(ab′)2 goat anti-mouse IgG.
Figure 7.
Effect of mAb treatment on the resistance of HLA-G1- and HLA-G2-transfected target cells from NK lysis. Cytotoxicity were set up with the indicated targets and (A) PBMC from donor 382 (HLA-A11, -A29, -B44, and -Cw5), (B) PBMC from donor 845 (HLA-A1, -A28, -B8, -B51, and -Cw7), and (C) the YT2C2 clone as effector cells. The W6/32 and the IgG2a isotype-matched control antibodies were added to the assay at 10 μg/ml. mAb toxicities were checked in each assay and were always <3%.
Total Inhibition of the NK-Like YT2C2 Clone-Mediated Lysis by HLA-G1 and HLA-G2 Transfectants.
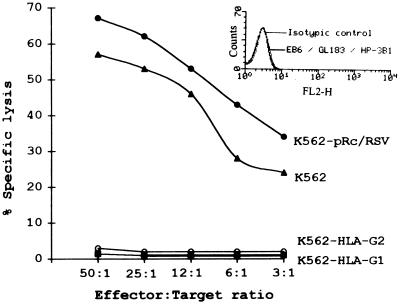
The nonadult T-cell leukemia NK-like YT2C2 clone was used as an effector against HLA-G transfectants. Fig. 8 shows that both HLA-G1 and HLA-G2 transfectants abolished YT2C2 lysis, whereas the control cell lines did not. However, in contrast to what was observed with PBMC, the inhibition of YT2C2 lysis was not reversed by the use of transfected target cells treated with the W6/32 mAb (Fig. 7C), suggesting that the epitope recognized by the mAb on HLA-G is not involved in HLA-G recognition by the receptor that is present on YT2C2 cells. Indirect immunofluorescence analysis was carried out to check the phenotype of the YT2C2 cells. We used the EB6 and GL183 mAbs, which (respectively) recognize NKIR 1 and NKIR 2, expressed on subsets of NK cells, as well as the HP-3B1 mAb that recognizes the CD94 receptor, expressed on most NK cells. These receptors are involved in the recognition by NK cells of the HLA-A, -B, or -C allotypes (13, 23). Fig. 8 shows that YT2C2 cells present the following negative phenotype: EB6−, GL183−, and HP3B1−. These mAbs were previously checked by indirect immunofluorescence and found to positively stain 3–10% of the PBMC from healthy adult donors (data not shown).
Figure 8.
Effect of HLA-G1 and HLA-G2 expression on sensitivity to cytotoxicity of the YT2C2 clone. K562 transfected with either the vector alone, HLA-G1, or HLA-G2 were used as targets. The results are expressed as the percentage lysis recorded in a 4-h 51Cr-release assay. The standard deviation of the mean of the triplicates was below 5%, and the spontaneous release never exceeded 10% of the maximum release. This experiment was repeated at least five times, giving the same pattern of protection. The expression of NKIRs on the YT2C2 clone was assessed by cytofluorometry. YT2C2 cells were labeled by indirect immunofluorescence with the following primary mAbs: EB6 (IgG1, anti-p58.1 or NKIR1), GL183 (IgG1, anti-p58.2 or NKIR2), and HP-3B1 (IgG2a, anti-CD94). Controls were the same cells stained with an isotype-matched control antibody. After washing, cells were stained with phycoerythrin-conjugated F(ab′)2 goat anti-mouse IgG. One of four representative experiments is shown.
DISCUSSION
In this study, we have investigated the role of HLA-G antigens in the protection of cells that express these proteins on their surfaces against NK cell-mediated cytotoxicity. In contrast with previous authors who transfected genomic HLA-G into target cells to study the effects on the lytic activity of NK clones (7, 8, 12), we separately evaluated membrane-bound HLA-G1 and HLA-G2 isoforms under physiological conditions, using polyclonal NK cells obtained from healthy adult donors. We constructed two eukaryotic pRc/RSV expression vectors, containing either HLA-G1 or HLA-G2 cDNA. Transfection of K562 cells with these constructs enabled us to generate two stable cell lines that express HLA-G1 or HLA-G2 molecules on the cell surface. The membrane-bound protein level was assessed by cytofluorometry, using the pan anti-HLA class I W6/32 and the anti-β2m B1G6 mAbs that stained only HLA-G1 and HLA-G2 antigens present on the cell surfaces of their respective transfected cell lines (Fig. 4). These antigens were also detected by immunoprecipitation with W6/32 (Fig. 3) and the specificity of the HLA-G isoform was confirmed by RT-PCR amplification, using HLA-G-specific primers (Fig. 2). To analyze how the HLA-G1 and HLA-G2 molecules act on NK cell-mediated cytotoxicity, we used either PBMC or polyclonal NK cells (CD3− CD16+ CD56+) isolated from 20 donors as effector cells (Figs. 5 and 6). Our results clearly demonstrated that both membrane-bound HLA-G1 and HLA-G2 isoforms strongly inhibit NK cell-mediated lysis. This inhibition was reversed when the transfectant cells that express HLA-G1 or HLA-G2 were incubated with the pan class I W6/32 mAb, indicating that inhibition of NK lysis was due to the presence of HLA-G on the K562 transfected target cells (Fig. 7). The fact that NK lytic activity in all 20 donors tested was inhibited by HLA-G-postive target cells suggests that HLA-G could be the public ligand for a NK inhibitory receptor(s) present in all individuals.
The original approach we have developed has enabled us to demonstrate the role of each extracellular domain in protection from NK lysis. In fact, we observed that the HLA-G2 isoform, which lacks the α2 domain, acts as a protective molecule, as does HLA-G1. Furthermore, our results demonstrate for the first time that HLA-G2 is present at the cell surface as a truncated class I molecule associated with β2m. However, the membrane-bound HLA-G2 isoform could also be present as a G2/G2 homodimer similar to HLA class II molecules. We are currently considering the hypothesis that this homodimeric form may also inhibit NK cytolysis by interacting with NKIR(s). In view of our results, we conclude that the α1 domain common to both isoforms could play an important role in NK protection. This is supported by the fact that the amino acid sequences on HLA class I molecules necessary for KIR interaction are located in the α1 domain (24, 25). It is noteworthy that although polymorphism in the HLA-G locus was observed, especially in exon 3 that codes for the α2 extracellular domain, most of the single base substitutions are silent, thereby reducing HLA-G protein polymorphism (18).
Interestingly, we also showed that T-cell leukemia YT2C2 cell-mediated lysis was totally abolished when HLA-G1 and HLA-G2 were present on K562 transfectants, although none of the known KIRs capable of binding HLA-G (i.e., p58.1 and p58.2) is present on YT2C2 cells (Fig. 8). These results suggest that a new KIR(s), distinct from those already described, is involved in HLA-G1 and HLA-G2 recognition. An alternative explanation proposes that YT2C2 exhibits classical NKIRs that are modified in this T cell leukemia clone, thus not recognized by the anti-NKIR mAbs used. However, addition of the pan class I W6/32 mAb to transfectants cells did not restore the YT2C2 cytotoxicity, suggesting that the epitope recognized by W6/32 is not involved in the interaction between HLA-G and its receptor counterpart in the YT2C2 clone. It is of interest to note that, as we previously showed, NK cells as well the YT2C2 clone do not present HLA-G transcripts and treatment with several cytokines did not modify this transcription pattern (35).
On the other hand, our study allowed us to determine the immune function of each isoform. We previously showed that all HLA-G alternative transcripts are expressed at the mRNA level in trophoblast, with a predominance of the HLA-G1 transcript (200-fold) over the HLA-G2 transcript (16). Finally, if HLA-G2 is expressed on the trophoblast cell surface along with HLA-G1, it could contribute to protecting the fetus from maternal NK lytic activity. Thus, the fetus protects itself from rejection by alloreactive maternal T cells by shielding itself with a layer of cells that lack HLA-A and HLA-B expression. To avoid destruction of this layer by NK cells, trophoblast cells express weakly polymorphic HLA-G molecules, thereby interacting with all inhibitory NK receptors, shifting the NK balance toward inhibition. In this regard, HLA-G acts as the public ligand for NKIR(s), thus protecting fetal trophoblasts from maternal NK lytic activity and conferring immunological tolerance to fetal tissues.
Our findings strongly support the assignment of an important role for HLA-G in materno–fetal tolerance, and its extension to tissue-graft tolerance and the escape of tumors from immunosurveillance. Finally, the major histocompatibility complex could consist of two classes of antigens: classical ones involved in self and non-self-recognition, and the nonclassical ones, such HLA-G, responsible for immune tolerance.
Acknowledgments
We thank Jean-Gérard Guillet for helpful discussions and Noah Hardy for rereading and correcting the manuscript. We are grateful to Catherine Menier for flow cytometry sorting and analysis. The HLA-G1 cDNA was kindly provided by Dr. Shirley A. Ellis (Institute for Animal Health, Compton, Nr Newbury, Berks, U.K.). This study was supported by grants from the Association pour la Recherche sur le Cancer and the Ligue Nationale contre le Cancer.
ABBREVIATIONS
- β2m
β2-microglobulin
- KIR(s)
killer inhibitory receptor(s)
- NK
natural killer
- NKIR(s)
NK inhibitory receptor(s)
- PBMC
peripheral blood mononuclear cells
- RSV
Rous sarcoma virus
- RT-PCR
reverse transcriptase–PCR
References
- 1.Ellis S A, Sargent I L, Redman C W G, McMichael A J. Immunology. 1986;59:595–601. [PMC free article] [PubMed] [Google Scholar]
- 2.Kovats S, Main E K, Librach C, Stubblebine M, Fisher S J, DeMars R. Science. 1990;248:220–223. doi: 10.1126/science.2326636. [DOI] [PubMed] [Google Scholar]
- 3.Mcmaster M T, Librach C L, Zhou Y, Lim K H, Janatpour M J, DeMars R, Kovats S, Damsky C, Fisher S J. J Immunol. 1995;154:3771–3778. [PubMed] [Google Scholar]
- 4.King A, Birkby C, Loke S L. Cell Immunol. 1989;118:337–344. doi: 10.1016/0008-8749(89)90382-1. [DOI] [PubMed] [Google Scholar]
- 5.Carosella E D, Dausset J, Kirszenbaum M. Immunol Today. 1996;17:407–409. doi: 10.1016/0167-5699(96)30055-8. [DOI] [PubMed] [Google Scholar]
- 6.Carosella E D, Kirszenbaum M, Dausset J. C R Acad Sci. 1995;318:827–830. [PubMed] [Google Scholar]
- 7.Colonna M, Borsellino G, Falco M, Ferrara G B, Strominger J L. Proc Natl Acad Sci USA. 1993;90:12000–12004. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phillips J H, Chang C W, Mattson J, Gumperz J E, Parham P, Lanier L L. Immunity. 1996;5:163–172. doi: 10.1016/s1074-7613(00)80492-6. [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Botet M, Moretta L, Strominger J. Immunol Today. 1996;17:212–214. doi: 10.1016/0167-5699(96)30009-1. [DOI] [PubMed] [Google Scholar]
- 10.Pazmany L, Mandelboim O, Vales-Gomez M, Davis D M, Reyburn H T, Strominger J L. Science. 1996;274:792–795. doi: 10.1126/science.274.5288.792. [DOI] [PubMed] [Google Scholar]
- 11.Colonna M, Samaridis J. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 12.Moreau P, Carosella E D, Gluckman E, Gourand L, Prost S, Dausset J, Kirszenbaum M. C R Acad Sci. 1995;318:837–842. [PubMed] [Google Scholar]
- 13.Ishitani A, Geraghty D E. Proc Natl Acad Sci USA. 1992;89:3947–3951. doi: 10.1073/pnas.89.9.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirszenbaum M, Moreau P, Gluckman E, Dausset J, Carosella E D. Proc Natl Acad Sci USA. 1994;91:4209–4213. doi: 10.1073/pnas.91.10.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreau P, Carosella E D, Teyssier M, Prost S, Gluckman E, Dausset J, Kirszenbaum M. Hum Immunol. 1995;43:231–236. doi: 10.1016/0198-8859(95)00009-s. [DOI] [PubMed] [Google Scholar]
- 16.Chumbley G, King A, Robertson K, Holmes N, Loke Y W. Cell Immunol. 1994;155:312–322. doi: 10.1006/cimm.1994.1125. [DOI] [PubMed] [Google Scholar]
- 17.Deniz G, Christmas S E, Brew R, Johnson P M. J Immunol. 1994;152:4255–4261. [PubMed] [Google Scholar]
- 18.Kirszenbaum, M., Djoulah, S., de Oliveira, E. B., Prost, S., Legall, L., Dausset, J., Hors, J. & Carosella, E. D. (1997) Hum. Immunol., in press. [DOI] [PubMed]
- 19.Yoneda N, Tatsumi E, Kawano S, Teshiwagara K, Oka T, Fukuda M, Yamaguchi N. Leukemia. 1992;6:136–141. [PubMed] [Google Scholar]
- 20.Ellis S A, Palmer M S, McMichael A J. J Immunol. 1990;144:731–735. [PubMed] [Google Scholar]
- 21.Kirszenbaum M, Moreau P, Teyssier M, Lafon C, Gluckman E, Dausset J, Carosella E D. Hum Immunol. 1995;43:237–241. doi: 10.1016/0198-8859(95)00008-r. [DOI] [PubMed] [Google Scholar]
- 22.Moreau P, Paul P, Gourand L, Prost S, Dausset J, Carosella E D, Kirszenbaum M. Hum Immunol. 1997;52:41–46. doi: 10.1016/S0198-8859(96)00242-X. [DOI] [PubMed] [Google Scholar]
- 23.Moretta A, Vitale M, Sivori S, Bottino C, Morelli L, Augugliaro R, Barbaresi M, Pende D, Ciccone E, Lopezbotet M, Moretta L. J Exp Med. 1994;180:545–555. doi: 10.1084/jem.180.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colonna M, Spies T, Strominger J L, Ciccone E, Moretta A, Moretta L, Pende D, Viale O. Proc Natl Acad Sci USA. 1992;89:7983–7985. doi: 10.1073/pnas.89.17.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandelboim O, Reyburn H T, Vales-Gomez M, Pazmany L, Colonna M, Borsellino G, Strominger J. J Exp Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teyssier M, Bensussan A, Kirszenbaum M, Moreau P, Gluckman E, Dausset J, Carosella E D. Nat Immun. 1996;14:262–270. [PubMed] [Google Scholar]