Abstract
An Fcα receptor probe of human origin was used to identify novel members of the Ig gene superfamily in mice. Paired Ig-like receptors, named PIR-A and PIR-B, are predicted from sequence analysis of the cDNAs isolated from a mouse splenic library. Both type I transmembrane proteins possess similar ectodomains with six Ig-like loops, but have different transmembrane and cytoplasmic regions. The predicted PIR-A protein has a short cytoplasmic tail and a charged Arg residue in the transmembrane region that, by analogy with the FcαR relative, suggests the potential for association with an additional transmembrane protein to form a signal transducing unit. In contrast, the PIR-B protein has an uncharged transmembrane region and a long cytoplasmic tail containing four potential immunoreceptor tyrosine-based inhibitory motifs. These features are shared by the related killer inhibitory receptors. PIR-A proteins appear to be highly variable, in that predicted peptide sequences differ for seven randomly selected PIR-A clones, whereas PIR-B cDNA clones are invariant. Southern blot analysis with PIR-B and PIR-A-specific probes suggests only one PIR-B gene and multiple PIR-A genes. The PIR-A and PIR-B genes are expressed in B lymphocytes and myeloid lineage cells, wherein both are expressed simultaneously. The characteristics of the highly-conserved PIR-A and PIR-B genes and their coordinate cellular expression suggest a potential regulatory role in humoral, inflammatory, and allergic responses.
The Fc receptors (FcR) for antibodies of different isotypes are widely distributed on cells of the immune system and may couple humoral and cellular immunity by directing the interaction of antibodies with effector cells (1, 2). The immunological consequences of cell surface FcR binding of antigen/antibody complexes vary according to the Ig isotype specificity of the FcR and the cell type that expresses it. The different types of FcR share related ligand-binding domains, but differ in their transmembrane and intracellular domains, which determine intracellular signaling. Interestingly, the FcRs share with antigen receptors on T and B cells some of the same receptor subunits, protein motifs, and signal transduction pathways (1–4).
We and others have characterized an FcR for IgA antibodies that is expressed on phagocytic white blood cells in humans (5–7). A cDNA clone encoding the human FcαR α chain was isolated from a macrophage-derived library (8, 9), and the common FcR γ chain has since been found to associate with the FcαR α chain to form a signal-transducing receptor complex (10, 11). When the human FcαR α chain cDNA probe was used in the present studies to screen mouse genomic and splenic cDNA libraries, a novel gene family was discovered. This report describes some of the characteristics of this gene family, the members of which constitute a set of paired immunoglobulin-like receptor (PIR) genes, PIR-A and PIR-B.
MATERIALS AND METHODS
Preparation of Partial Genomic Clones as Probes.
High molecular weight BALB/c splenic DNA was digested to completion with EcoRI and fractionated by electrophoresis in a 1.0% agarose gel. DNA corresponding to 3–4 kb was extracted from the gel, ligated into the EcoRI site of a λ ZAP II vector (Stratagene), and packaged in vitro. The resultant phage library (≈7 × 105 plaque-forming unit/μg of DNA) was screened with a 32P-labeled human FcαR cDNA probe (≈0.7 kb BglII fragment) that encodes the 5′ untranslated region, the signal peptide, and the extracellular domains (9). Seven positive clones with inserts of two different sizes (≈3.3 and ≈3.6 kb) were identified among ≈7 × 104 clones, and the nucleotide sequence analysis of representative clones (M3.1 and M10.1) revealed that both inserts were closely related and contained several putative exons (not shown). Exon-containing DNA fragments (≈130-bp EcoRI/BamHI fragment and ≈660-bp PstI/EcoRI fragment) were excised from the 5′ and 3′ ends of the M3.1 genomic clone, and these were used as probes to screen mouse splenic cDNA libraries.
Isolation of PIR cDNA Clones.
The cDNA libraries were constructed in λ ZAP II using poly(A)+ RNA isolated from BALB/c (>25 weeks old) spleen or peritoneal lavage and oligo dT primers. The titers of the cDNA libraries were estimated as ≈106 plaque-forming units per 1.5 μg of poly(A)+ RNA. Duplicate lifts of phage plaques were screened with 5′ and 3′ mouse DNA probes, and clones hybridizing to both probes were subjected to secondary and tertiary screening. The cDNA inserts in positive clones were excised in vivo by helper phage-mediated circularization into pBluescript II SK(+) (Stratagene). Fourteen different cDNA clones were isolated: B1–B5 (≈2.7-kb insert); A1, A7, and A8 (≈3.4 kb); A2 (≈2.3 kb); A4 and A5 (≈2.1 kb); A3 and A9 (≈2.0 kb); and A6 (≈1.9 kb).
Sequence Analysis.
DNA sequencing was performed twice or more for each clone either on separate strands or from different primers on the same strand by the dideoxy chain termination method using sequenase 2.0 (United States Biochemical) and double-stranded DNA as a template. Nucleotide sequence homology searches and alignments were performed using Basic Local Alignment Search Tool (blast) programs (12) and the dnastar align program.
DNA and RNA Blot Analyses.
Isolation of genomic DNA and total or poly(A)+ RNA, restriction enzyme digestions, agarose gel electrophoresis, DNA and RNA blotting procedures, preparation of random primed 32P-labeled DNA probes, and posthybridization washes were performed as described (13–15). The following probes were used: (i) an ≈1.5-kb EcoRI fragment of PIR-B1 cDNA as a common extracellular (EC) probe, (ii) an ≈1.6-kb SacI/XhoI fragment of PIR-A1 cDNA as a PIR-A-specific probe, and (iii) an ≈0.8-kb SacI/XhoI fragment of PIR-B1 cDNA as a PIR-B-specific probe.
Reverse Transcription–Polymerase Chain Reaction (RT-PCR).
Total RNA from cell lines and sorted cells was converted to first-strand cDNA with oligo(dT)18 primers and avian myeloblastosis virus reverse transcriptase (16) and amplified with a common forward primer (5′-CCTGTGGAGCTCACAGTCTCAG-3′) and the PIR-A-specific (5′-CCCAGAGTGTAGAACATTGAAGATG-3′) or PIR-B-specific (5′-GTGTTCAGTTGTTCCCTTGACATGA-3′) reverse primers. These primers correspond to the 3′ end of the sixth EC domain of PIR-A and PIR-B, and to the cytoplasmic regions of PIR-A or PIR-B, respectively, and yield fragments of 252 bp for PIR-A and 470 bp for PIR-B. Each amplification reaction underwent 30 cycles of: denaturation at 95°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1 min. A final extension was performed at 72°C for 7 min. Amplification of actin transcripts with the primers (5′-ATTGAACATGGCATTGTTACC-3′ and 5′-GGCATACAGGGACAGCACAGC-3′) was also performed as a control. To ensure linearity of the PCR amplifications, three different concentrations of the first-strand cDNA were used as templates for each pair of primers. Amplified products were electrophoresed in 2% agarose and stained with ethidium bromide.
Cell Preparation.
Splenic cells were sorted as B220+ B cells, Mac-1+ macrophages, and Gr-1+ granulocytes by a FACStarPlusinstrument (Becton Dickinson) (17).
Chromosomal Mapping.
Genomic DNA samples (10 μg each) from The Jackson Laboratory interspecific backcross DNA panel of 94 F2 segregants from the cross (C57BL/6JEi × SPRET/Ei)F1 × SPRET/Ei (18) were digested with BglI and subjected to Southern blot analysis using a 32P-labeled PIR-A1 cDNA probe. The resulting allele distribution was compared with those of 1,400 loci previously mapped in the cross, and the map position was determined by minimizing double recombinants.
RESULTS
Murine Relatives of the Human Fcα Receptor Gene.
In experiments aimed toward cloning a murine homologue of the human FcαR gene, we observed that the human FcαR probe cross-hybridizes with mouse genomic DNA to yield EcoRI-digested DNA fragments of ≈3.6 and ≈3.3 kb under relatively high stringency conditions (1× SSC, 60°C). When these DNA fragments were cloned from a BALB/c splenic DNA library, nucleotide sequence analysis of the inserts indicated these were closely related and appeared to contain multiple exons. Exon-containing DNA fragments excised from these genomic clones were used as probes to screen a BALB/c splenic cDNA library from which 13 cDNA clones with inserts of 2.0–3.4 kb were isolated. The incidence of the hybridizing cDNA clones was ≈1/10,000, suggesting moderate abundance of the transcripts. An additional cDNA clone (A6) was isolated from a mouse peritoneal cell library by cross-hybridization with the human FcαR probe.
Paired Ig-Like Receptors, PIR-A and PIR-B, Are Predicted by cDNA Sequence Analysis.
The 14 cDNA clones could be divided into two discrete groups on the basis of restriction enzyme digestion profiles and 3′ terminal nucleotide sequences. Prototypic full-length cDNA clones A1 and B1 were ≈3.4 and ≈2.7 kb, respectively. Nucleotide sequences of the two types of clones are available in GenBank (accession numbers U96682–U96693U96682U96683U96684U96685U96686U96687U96688U96689U96690U96691U96692U96693), and the amino acid sequences are shown in Fig. 1.
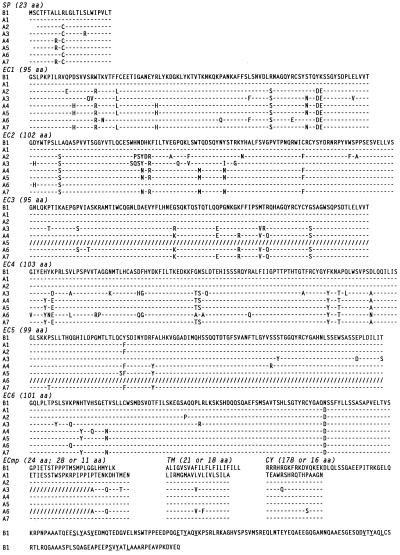
Figure 1.
Sequence diversity among members of the activation-type (PIR-A) receptors. Variable amino acid sequences (one-letter code) deduced from the seven randomly picked, PIR activation-type cDNA clones (A1 to A7) are aligned with the invariable peptide sequence of the PIR inhibitory-type cDNA clone (B1). Amino acid identity is indicated by dashes (–); a change, by boldface letters; and a deletion introduced for optimal alignment, by slashes (/). A charged Arg residue in the transmembrane region of PIR-A-type clones and the ITIM-like motifs in the cytoplasmic tail of PIR-B clone are also in boldface letters. Sequences are divided into each putative region: SP, signal peptide; EC, extracellular domain; ECmp, membrane proximal extracellular; TM, transmembrane; and CY, cytoplasmic regions. The boundaries of EC1/EC2 to EC5/EC6 were established from the analysis of genomic clones. Numbers in parentheses indicate the amino acid (aa) length of each region.
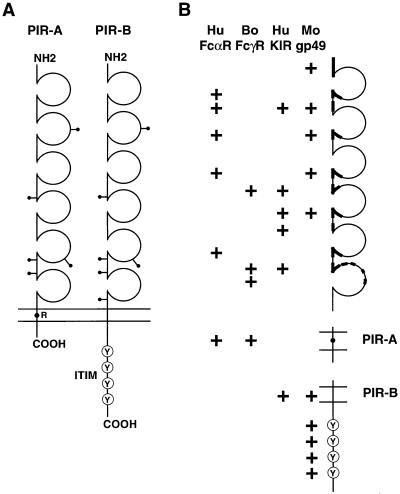
The PIR-A1 cDNA consists of a short (9 bp) 5′ untranslated region, a 2,040-bp open reading frame, a relatively long (≈1.4 kb) 3′ untranslated region containing four classical polyadenylylation signals and six RNA instability sequences (ATTTA; ref. 19) and terminates in a poly(A) tail. A Kyte–Doolittle plot analysis suggests that this cDNA encodes a type I transmembrane protein of 680 aa that begins with a 23-aa hydrophobic signal peptide containing a consensus cleavage site (20) at the junction with the predicted amino terminus of the mature protein. The mature protein would thus start at Gly1, resulting in a core peptide with an estimated Mr of 73,172. The 623-aa extracellular region includes five potential sites for N-linked glycosylation (Asn-Xaa-Ser/Thr, where Xaa is any amino acid except Pro) and six Ig-like domains, the second of which is a V set and the others being C2 sets (21–23). The extracellular region is followed by a hydrophobic stretch of 18 aa (Leu624 to Ala641) corresponding to a potential transmembrane region. This region contains a charged Arg residue, thus suggesting the potential for noncovalent association with an additional transmembrane protein. The predicted short cytoplasmic tail of 16 aa lacks a cluster of basic residues immediately following the hydrophobic transmembrane stretch and does not contain Tyr residues.
The PIR-B1 cDNA consists of a 39-bp 5′ untranslated region, a 2,523-bp open reading frame, and a 139-bp 3′ untranslated region containing a classical polyadenylylation signal (AATAAA), and terminates in a poly(A) tail. This cDNA encodes a type I transmembrane protein of 841 aa that, compared with the A1-type molecule, includes an identical signal peptide (23 aa) and an extracellular region (619 aa) with >90% homology, but very different transmembrane (21 aa) and cytoplasmic regions (178 aa). The Mr of the predicted core protein is 90,636. There are six potential sites for N-linked glycosylation in the extracellular domain. The putative transmembrane region (Ala620 to Leu640) is followed by several positively charged residues characteristic of the start of a cytoplasmic domain. The 178-aa cytoplasmic tail contains four copies (residues 688–693, 717–722, 769–774, and 799–804) of the sequence motif Ser/Glu/Val-Xaa-Tyr-Xaa-Xaa-Val/Leu, similar to the immunoregulatory tyrosine-based inhibitory motif (ITIM) recently identified in the cytoplasmic domains of the FcγRIIB (24–26) and CD22 molecules on B cells (27, 28) and the killer inhibitory receptors (KIR) on NK cells (29–32). This suggests that the PIR-B1 cDNA encodes a receptor molecule that may transduce an inhibitory signal via the interaction of its cytoplasmic ITIM with inositol or protein tyrosine phosphatases (33–36).
Diversity of the PIR-A cDNA Clones.
To determine the basis for the size heterogeneity observed for the PIR-A cDNA clones, we extended the nucleotide sequence analysis to other PIR-A members. One cause for heterogeneity was the alternative usage of polyadenylylation signals. The two largest cDNA clones (A1, A7) utilized the most 3′ polyadenylylation signal, the A2 and A5 clones used the second 5′ signal, and three other clones (A3, A4, A6) used the most 5′ signal. A second basis for cDNA size heterogeneity was alternate splicing of extracellular exons, such that the A5 and A6 clones lack an exon encoding the third and fifth Ig-like domains, respectively (see Fig. 1). In the A3 and A6 clones, the sequence corresponding to the amino side of the membrane-proximal extracellular region is missing, resulting in an amino acid change (Pro to Ala) 11 residues before the transmembrane region, where cleavage of the precursor and attachment of a glycosyl phosphatidyl inositol anchor may occur (37). In contrast with the PIR-A cDNAs, neither alternate polyadenylylation signals nor splice variants were observed among the five PIR-B cDNA clones.
An interesting pattern of diversity was revealed by the comparison of nucleotide and deduced peptide sequences among the members of the two types of PIR cDNA clones. The nucleotide sequences of the open reading frame were invariant for the five PIR-B cDNA clones, whereas each of the seven PIR-A cDNA clones was found to have a different sequence. Our comparison of these differences, based on the invariant PIR-B sequence because complete germline sequences for this gene family are presently unavailable, indicates that the nucleotide sequence variations are distributed throughout the open reading frame, although they are more abundant in the region of the first four extracellular domains than in other regions. The frequency of overall nucleotide variations ranges from 0.2% to 4.7%. Some of the nucleotide variants are silent while others result in amino acid differences, which are dispersed throughout the extracellular region, but are more frequent in the first four domains (Fig. 1). These modifications are present both within and outside the loops of the Ig-like domains, and nearly half are shared by one or more of the seven cDNA clones. Although most changes occurring at the same position in multiple clones involve a single amino acid, changes involving several amino acids at the same position are found more frequently within the loop of the second to fourth Ig domains. The overall frequency of amino acid changes is 0.3–8.2%. The fact that, unlike the PIR-B cDNAs, identical cDNA clones were not observed among the seven randomly selected isolates implies a remarkable degree of diversity for the PIR-A molecules.
Multiple PIR-A Genes and a Single PIR-B Gene.
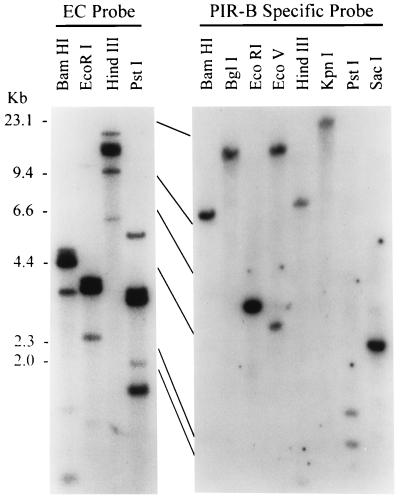
As a first step in determining the extent of the genomic diversity of the PIR gene family, genomic DNA from a BALB/c mouse was digested with different restriction enzymes and subjected to Southern blot analysis. The three probes employed were 32P-labeled cDNA fragments corresponding to the common extracellular region or to each type-specific transmembrane and cytoplasmic region. Four to six discrete bands hybridizing to the extracellular probe were observed for each enzyme digestion, although the intensity of each band was highly variable (Fig. 2). Hybridization obtained with the PIR-A-specific probe encoding the membrane-proximal extracellular, transmembrane, cytoplasmic, and 3′ untranslated regions yielded a similar pattern. In contrast, when the same DNA blot was hybridized with the PIR-B-specific probe, only one or two hybridizing bands were observed for each digestion. The DNA blot analysis suggests the presence of multiple PIR-A genes but only one PIR-B gene in the mouse genome.
Figure 2.
Southern blot analysis of PIR gene family. DNA from BALB/c testis was analyzed with PIR probes corresponding to the common extracellular (EC) region (Left) or to the PIR-B-specific region (Right).
Chromosomal Location of the PIR Genes.
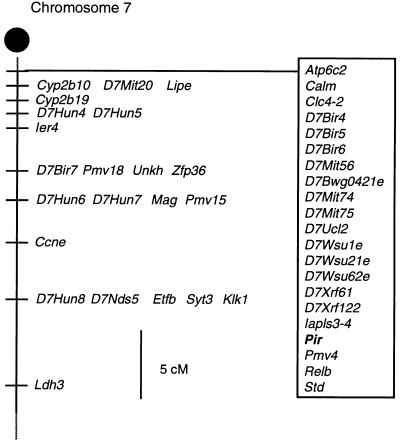
The PIR gene family was mapped to the proximal end of mouse chromosome 7 (Fig. 3) in a region syntenic with the human chromosome 19q13 region where the FcαR gene is mapped (38). Interestingly, the human KIR genes are also located on the same chromosome (39–41).
Figure 3.
Chromosomal localization of the PIR gene family. Partial chromosome 7 linkage map showing the location of Pir in relation to linked genes. cM, centiMorgans.
Cell Lineage Restriction of PIR Gene Expression.
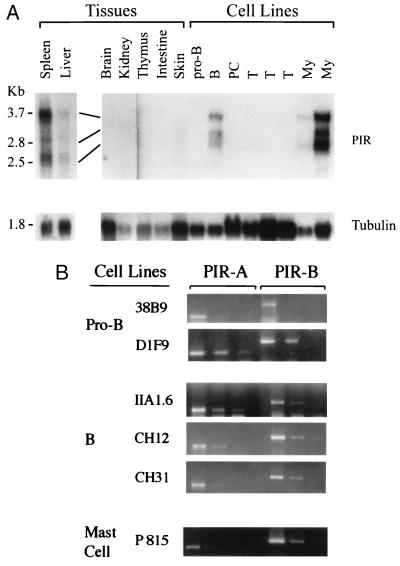
Three major PIR transcripts of ≈3.5, ≈2.7, and ≈2.5 kb were detected in bone marrow and spleen, but not in other tissues including the thymus, brain, kidneys, intestine, skin, heart, and skeletal muscle, when RNA was analyzed by Northern blots with a 32P-labeled common extracellular probe (Fig. 4A). Weak signals were observed for adult liver and lung samples, possibly reflecting transcripts in cells derived from the circulation. The same PIR transcripts were expressed by B cell lines (WEHI231, WEHI279) and myeloid cell lines (WEHI3, DAGM), and not by pro-B (HAFTL-1), pre-B (18–81), plasmacytoma (P3X, Ag8), T (CAK4.4, YAC1, SCID thymoma), and fibroblast (LTK) cell lines in levels detectable by Northern blot analysis. These findings suggest that expression of the PIR gene family is restricted to cells of B lineage and myeloid lineages.
Figure 4.
Tissue specificity and cell lineage restriction of PIR gene expression. (A) RNA blot analysis. One microgram of poly(A)+ RNA (spleen, liver) or 10 μg of total RNA from the indicated murine tissues and cell lines was analyzed with 32P-labeled PIR (Upper) and tubulin (Lower) probes. Similar transcripts were also observed in bone marrow. (B) RT-PCR analysis. Three different concentrations of first-strand cDNA products were subjected to PCR amplification of PIR-A, PIR-B, and actin transcripts (not shown). Amplified products electrophoresed in 2% agarose were stained with ethidium bromide. Similar coexpression patterns were observed for macrophage cell lines.
To determine whether the PIR-A and PIR-B genes are simultaneously or differentially expressed by B cells and myeloid cells, we conducted RT-PCR analysis using primer pairs specific for each receptor type. The PCR products of both PIR-B (470 bp) and PIR-A (252 bp) were identified in pro-B, B, myeloid, and mast cell lines (Fig. 4B). A similar expression pattern was observed for normal B220+ B cells, Mac-1+ macrophages, and Gr-1+ granulocytes (not shown). These findings indicate that both the activating- and inhibitory-type PIR genes are coordinately expressed by B, myeloid, and mast cell lines.
Evolutionary Conservation of the PIR Gene Family.
To explore the potential conservation of this gene family, we performed a zoo blot analysis employing EcoRI-digested DNA from human, monkey, cow, dog, mouse, rat, rabbit, chicken, and yeast with a 32P-labeled cDNA fragment corresponding to the extracellular region. All samples, except for the yeast DNA, were found to cross-hybridize with the murine probe (not shown). When the same mouse probe was used for Southern blot analysis of human placental DNA digested with other common restriction enzymes (BamHI, HindIII, PstI), 11 discrete fragments were found to cross-hybridize with the mouse probe, only one of which could hybridize with the human FcαR probe (data not shown), suggesting that the human PIR counterparts also comprise a multigene family.
DISCUSSION
Members of the PIR gene family described here encode two types of transmembrane proteins, models of which are illustrated in Fig. 5. PIR-A and PIR-B share similar extracellular domains while having distinctive transmembrane and cytoplasmic regions. Nucleotide sequence analysis of PIR cDNAs suggests that the PIR proteins are cell surface receptors with related ligand-binding specificity but that have different signaling capabilities. PIR-A may have cellular activating potential, whereas PIR-B is likely to have inhibitory potential.
Figure 5.
Model of PIR-A and PIR-B and their relationship with other Ig gene superfamily members. (A) PIR-A and PIR-B cDNA clones encode type I transmembrane proteins with similar extracellular domains, but different transmembrane and cytoplasmic regions. The extracellular region has six Ig-like domains, the second of which is V-like and the others C2-like. There are five or six potential sites for N-linked glycosylation (—•). The predicted PIR-A protein contains a relatively short cytoplasmic tail and a charged arginine (R) residue in the transmembrane region, whereas the PIR-B protein has a typical nonpolar transmembrane region and a long cytoplasmic tail with four potential tandem immunoreceptor tyrosine-based inhibitory motifs (Y). (B) Schematic depiction of the regions of PIR-A and PIR-B that share significant homology with other members of the Ig gene superfamily, the human FcαR (8), bovine FcγR (42), human KIRs (39–41), and mouse gp49 (43).
PIR Family Relationship with Other Ig Gene Superfamily Members.
A database search of available sequences indicated that PIR-A and PIR-B represent a new gene family sharing limited homology with other Ig gene superfamily members (see Fig. 5). As would be expected, one PIR relative is the gene for the human FcαR α chain that has two Ig-like domains in its extracellular region. The homology with this FcαR gene is restricted to portions of the Ig loops, interloop sequences, and the transmembrane region, where the greatest degree of homology (≈50%) is seen with PIR-A. The positively charged Arg residue common to the transmembrane regions of both proteins is noteworthy because of its importance for FcαR α chain association with the common FcR γ chain, which contains an immunoreceptor tyrosine-based activation motif (ITAM), to form a signal-transducing unit (10, 11). The bovine FcγR gene (42), another two-Ig-domain relative of PIR-A, shares limited homology in the extracellular region and a higher degree of homology in its transmembrane region (≈60%) that includes the conserved Arg residue.
Human KIRs share homology with PIR-B in the extracellular and transmembrane regions. The highest level of homology was observed for the murine gp49B1, an inhibitory-type receptor expressed by mast cells and NK cells that contains consensus ITIMs in the cytoplasmic region (43–46). Receptors with ITIMs, such as the KIR molecules on NK cells and the FCγRIIB molecules on B cells, deliver an inhibitor signal when they are coligated with ITAM-bearing activation receptor complexes like the B cell antigen receptor. The coligation of the activating and inhibitory types of receptors facilitates the ITIM interaction with inositol or protein tyrosine phosphatases, such as SHIP and SHP-1, thereby blocking elevation of the intracellular calcium pool and effective cell signaling (33–36).
Cellular Distribution of PIR-A and PIR-B.
The expression of PIR-A and PIR-B transcripts is restricted to cells of B and myeloid lineages, including granulocytes, macrophages, and mast cells. This expression pattern has been confirmed by the reactivity profile of a monoclonal anti-PIR antibody directed against an extracellular epitope shared by PIR-A and PIR-B (unpublished observations). PIR cellular distribution thus differs from its presently known relatives, in that KIR expression is restricted to NK cells and a subpopulation of T cells (47) and FcαR expression is restricted to myeloid cells with phagocytic capability (5–7).
Another informative feature of PIR-A and PIR-B expression is the mutual expression of both types of transcripts in every clonal cell line of B cell or myeloid cell lineage that has been examined. PIR-A and PIR-B thus represent a matched receptor pair, the characteristics of which suggest that they exert a cellular regulatory role via opposing activation and inhibition capabilities.
PIR-A Diversity.
The KIRs display considerable variability, both in their ligand specificities and clonal expression patterns (47). In this context, one of the most striking features of the PIR gene family is the remarkable diversity of the PIR-A family members vis-à-vis the invariance of the PIR-B gene. PIR-A diversity exists at multiple levels, the most structurally consequential of which result from RNA processing events. Alternate splicing can result in the deletion of entire Ig domains, and the truncated protein products may have altered interactions with intracellular chaperones or with other cell surface proteins. Putatively glycosyl phosphatidyl inositol-linked PIR proteins may function to fine-tune the signal delivered by the transmembrane form of PIR-A. The use of three alternate polyadenylylation sites has no structural consequences but will likely affect mRNA stability because the three forms contain varying copy numbers of the ATTTA instability motif (19). The frequency, cellular distribution, and expression levels of these alternatively spliced and polyadenylylated transcripts may provide important functional clues.
The most intriguing aspect of the PIR-A family diversity lies in the relatively subtle changes in amino acid sequences of the different receptor isoforms. The full extent of this diversity is still unknown, but statistical estimates based on our failure to detect repeat sequences in seven randomly selected cDNAs suggest a PIR-A gene copy number in the range of 33–500 (50–95% confidence). Southern blot analysis indicates the existence of multiple PIR-A genes, and the nucleotide sequences of partial genomic clones supports this conclusion. If each band on the DNA blot (see Fig. 2) corresponds to a single gene, the number of PIR genes is quite limited. However, the relatively high intensity of some hybridization bands could reflect multiple PIR-A genes with conserved restriction enzyme sites, thereby leading to an underestimate of actual PIR-A gene frequency. Precedence for such restriction site conservation is seen in other Ig gene superfamilies such as the variable region gene segments of Ig and the T cell receptor. Ongoing studies will provide a more accurate estimate of the germline complexity of the PIR-A gene family, address the possibility that diversity is generated by somatic modification of a relatively limited number of genes, and determine the extent of intraclonal diversity of PIR-A expression in B cells and myeloid cells.
Potential Ligands for PIR-A and PIR-B.
Although the nature of the PIR ligands is presently unknown, the considerable PIR-A diversity may imply a corresponding diversity in ligand structures. However, general conservation of ligand structure is suggested by the monomorphic nature of PIR-B. The PIR-B molecule shares >90% identity with the PIR-A molecules in the extracellular region and thus may recognize the same or related ligands. Major histocompatibility complex (MHC) molecules could exhibit the requisite structural conservation and population diversity, since the different polymorphic forms of the MHC class I molecules are known to serve as ligands for the KIR molecules on NK cells. Although an obvious rationale for PIR–MHC interaction cannot readily be deduced from the cellular expression pattern of the PIR proteins, this possibility merits exploration. The immense diversity of carbohydrate structures also makes these attractive PIR-ligand candidates, either as glycoproteins on self-components or complex carbohydrates on microbial pathogens. One could envision that pathological alterations in the ligand would result in loss of recognition by PIR-B in the face of continued recognition by the more versatile PIR-A. The resultant imbalance could provoke a response by the B cells or phagocytic myeloid cells, but suffice to say the nature of the ligand changes and cellular responses remain intriguing puzzles for the present.
Conservation of the PIR Genes.
It is likely that the coordinate expression of activating and inhibitory receptors on NK cells represents a highly conserved means for regulating the activity of these important host defense elements since NK-like cells have been characterized in birds (48), snails (49), and earthworms (50), as well as in mammals. Presently, however, the KIR molecules and their activation receptor partners have been identified only in mammals. The observation that mouse PIR cDNA probes cross-hybridize with DNA from both mammalian and avian species raises the possibility that the PIR genes arose early in vertebrate evolution. Tracing the evolutionary origin of the PIR gene family will indicate whether these genes arose after the Ig and the T cell receptor genes or before, when primitive macrophage-like cells constituted the main line of defense against non-self-intrusion.
Acknowledgments
We thank Dr. Charles Maliszewski (Immunex) for providing the FcαR probe; Ms. Lucy Rowe (The Jackson Laboratory) for chromosomal mapping; Dr. Matthew Mayo for statistical analysis; Dr. G. Larry Gartland for flow cytometric analysis; Jin-Yi Wang, Jennifer S. Glass, Dong-Won Kang, Ming Chen, and Haitao Li for excellent technical assistance; Ann Brookshire and Marsha Flurry for help in manuscript preparation; and Drs. Irving Weissman, Frederick Alt, Eric Vivier, Louis Justement, and Vincent Hurez for critical comments on the manuscript. This work was supported in part by National Institute of Allergy and Infectious Diseases Grants AI34568 and AI39816. M.D.C. is an investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- RT-PCR
reverse transcription–polymerase chain reaction
- KIR
killer inhibitory receptors
- PIR
paired Ig-like receptors
- FcR
Fc receptors
- ITIM
immunoreceptor tyrosine-based inhibitory motif
- ITAM
immunoreceptor tyrosine-based activation motif
- EC
extracellular
Note Added in Proof
Hayami et al. (51) have very recently reported a cDNA clone derived from B10A mouse macrophages that has 98% nucleotide sequence identity with the BALB/c PIR-B cDNA.
Footnotes
References
- 1.Ravetch J V. Cell. 1994;78:553–560. doi: 10.1016/0092-8674(94)90521-5. [DOI] [PubMed] [Google Scholar]
- 2.Jouvin M-H, Numerof R P, Kinet J-P. Semin Immunol. 1995;7:29–35. doi: 10.1016/1044-5323(95)90005-5. [DOI] [PubMed] [Google Scholar]
- 3.Weiss A, Littman D R. Cell. 1994;76:263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 4.Cambier J C. J Immunol. 1995;155:3281–3285. [PubMed] [Google Scholar]
- 5.Albrechtsen M, Yeaman G R, Kerr M A. Immunology. 1988;64:201–206. [PMC free article] [PubMed] [Google Scholar]
- 6.Monteiro R C, Kubagawa H, Cooper M D. J Exp Med. 1990;171:597–613. doi: 10.1084/jem.171.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen L. Immunol Res. 1992;11:273–282. doi: 10.1007/BF02919133. [DOI] [PubMed] [Google Scholar]
- 8.Maliszewski C R, March C J, Schoenborn M A, Gimpel S, Shen L. J Exp Med. 1990;172:1665–1672. doi: 10.1084/jem.172.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maliszewski C R, VandenBos T, Shen L, Schoenborn, Kubagawa H, Beckmann P, Monteiro R C. J Leukocyte Biol. 1993;53:223–232. doi: 10.1002/jlb.53.3.223. [DOI] [PubMed] [Google Scholar]
- 10.Pfefferkorn L C, Yeaman G R. J Immunol. 1994;153:3228–3236. [PubMed] [Google Scholar]
- 11.Morton E C, Van den Herik-Oudijk I E, Vossebeld P, Snijders A, Verhoeven A J, Capel P J A, Van de Winkel J G J. J Biol Chem. 1995;270:29781–29787. doi: 10.1074/jbc.270.50.29781. [DOI] [PubMed] [Google Scholar]
- 12.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 13.Kubagawa H, Burrows P D, Grossi C E, Mestecky J, Cooper M D. Proc Natl Acad Sci USA. 1988;85:875–879. doi: 10.1073/pnas.85.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 15.Aruffo A. In: Cell Adhesion: A Practical Approach. Stevenson B R, Gallin W J, Paul D L, editors. Oxford: IRL–Oxford Univ. Press; 1992. pp. 55–74. [Google Scholar]
- 16.Bertrand F E, Billips L G, Gartland G L, Kubagawa H, Schroeder H W. J Immunol. 1996;156:4240–4244. [PubMed] [Google Scholar]
- 17.Billips L G, Nunez C A, Bertrand F E, Stankovic A K, Gartland G L, Burrows P D, Cooper M D. J Exp Med. 1995;182:973–982. doi: 10.1084/jem.182.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowe L, Nadeau J H, Turner R, Frankel W N, Letts V A, Eppig J T, Ko M S H, Thurston S J, Birkenmeier E H. Mamm Genome. 1994;5:253–274. doi: 10.1007/BF00389540. [DOI] [PubMed] [Google Scholar]
- 19.Shaw G, Kamen R. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 20.Von Heijne G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams A F, Barclay A N. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 22.Bork P, Holm L, Sander C. J Mol Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 23.Vaughn D E, Bjorkman P J. Neuron. 1996;16:261–273. doi: 10.1016/s0896-6273(00)80045-8. [DOI] [PubMed] [Google Scholar]
- 24.Amigorena S, Bonnerot C, Drake J R, Choquet D, Hunziker W, Guillet J G, Webster P, Sautes C, Mellman I, Fridman W H. Science. 1992;256:1808–1812. doi: 10.1126/science.1535455. [DOI] [PubMed] [Google Scholar]
- 25.Choquet D, Partiseti M, Amigorena S, Bonnerot C, Fridman W H. J Cell Biol. 1993;121:355–363. doi: 10.1083/jcb.121.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muta T, Kurosaki T, Misulovin Z, Sanchez M, Nussenzweig M C, Ravetch J V. Nature (London) 1994;368:70–73. doi: 10.1038/368070a0. [DOI] [PubMed] [Google Scholar]
- 27.Thomas M L. J Exp Med. 1995;181:1953–1956. doi: 10.1084/jem.181.6.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doody G M, Justement L B, Delibrias C C, Matthews R J, Lin J, Thomas M L, Fearon D T. Science. 1995;269:242–244. doi: 10.1126/science.7618087. [DOI] [PubMed] [Google Scholar]
- 29.Campbell K S, Dessing M, Lopez-Botet M, Cella M, Colonna M. J Exp Med. 1996;184:93–100. doi: 10.1084/jem.184.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olcese L, Lang P, Vély F, Cambiaggi A, Marguet D, Bléry M, Hippen K L, Biassoni R, Moretta A, Moretta L, Cambier J C, Vivier E. J Immunol. 1996;156:4531–4534. [PubMed] [Google Scholar]
- 31.Burshtyn D N, Scharenberg A M, Wagtmann N, Rajagopalan S, Berrada K, Yi T, Kinet J P, Long E O. Immunity. 1996;4:77–85. doi: 10.1016/s1074-7613(00)80300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fry A M, Lanier L L, Weiss A. J Exp Med. 1996;184:295–300. doi: 10.1084/jem.184.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono M, Bolland S, Tempst P, Ravetch J V. Nature (London) 1996;383:263–266. doi: 10.1038/383263a0. [DOI] [PubMed] [Google Scholar]
- 34.Scharenberg A M, Kinet J P. Cell. 1996;87:961–964. doi: 10.1016/s0092-8674(00)81790-0. [DOI] [PubMed] [Google Scholar]
- 35.Renard V, Cambiaggi A, Vély F, Bléry M, Olcese L, Olivero S, Bouchet M, Vivier E. Immunol Rev. 1997;155:205–221. doi: 10.1111/j.1600-065x.1997.tb00953.x. [DOI] [PubMed] [Google Scholar]
- 36.Tonks N K, Neel B G. Cell. 1996;87:365–368. doi: 10.1016/s0092-8674(00)81357-4. [DOI] [PubMed] [Google Scholar]
- 37.Barclay A N, Birkeland M L, Brown M H, Beyers A D, Davis S J, Somoza C, Williams A F. The Leukocyte Antigen Facts Book. New York: Academic; 1993. pp. 13–37. [Google Scholar]
- 38.Kremer E J, Kalatzis V, Baker E, Callen D F, Sutherland G R, Maliszewski C R. Hum Genet. 1992;89:107–108. doi: 10.1007/BF00207054. [DOI] [PubMed] [Google Scholar]
- 39.Colonna M, Samaridis J. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 40.Wagtmann N, Biassoni R, Cantoni C, Verdiani S, Malnati M S, Vitale M, Bottino C, Moretta L, Moretta A, Long E O. Immunity. 1995;2:439–449. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 41.D’Andrea A, Chang C, Franz-Bacon K, McClanahan T, Phillips J H, Lanier L L. J Immunol. 1995;155:2306–2310. [PubMed] [Google Scholar]
- 42.Zhang G, Young J R, Tregaskes C A, Scopp P, Howard C J. J Immunol. 1995;155:1534–1541. [PubMed] [Google Scholar]
- 43.Castells M, Wu X, Arm J P, Austen K F, Katz H R. J Biol Chem. 1994;266:8393–8401. [PubMed] [Google Scholar]
- 44.Katz H R, Vivier E, Castells M C, McCormick M J, Chambers J M, Austen K F. Proc Natl Acad Sci USA. 1996;93:10809–10814. doi: 10.1073/pnas.93.20.10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rojo S, Burshtyn D N, Long E O, Wagtmann N. J Immunol. 1997;158:9–12. [PubMed] [Google Scholar]
- 46.Wang L L, Mehta I K, LeBlanc P A, Yokoyama W M. J Immunol. 1997;158:13–17. [PubMed] [Google Scholar]
- 47.Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari M C, Moretta L. Annu Rev Immunol. 1996;14:699–648. doi: 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- 48.Göbel T W F, Chen C H, Shrimpf J, Grossi C E, Bernot A, Bucy R P, Affray C, Cooper M D. Eur J Immunol. 1994;24:1685–1691. doi: 10.1002/eji.1830240734. [DOI] [PubMed] [Google Scholar]
- 49.Franceschi C, Cossarizza A, Monti D, Ottaviani E. Eur J Immunol. 1991;21:489–493. doi: 10.1002/eji.1830210235. [DOI] [PubMed] [Google Scholar]
- 50.Cossarizza A, Cooper E L, Suzuki M M, Salvioli S, Capri M, Gri G, Quaglino D, Franceschi C. Exp Cell Res. 1996;224:174–182. doi: 10.1006/excr.1996.0125. [DOI] [PubMed] [Google Scholar]
- 51.Hayami K, Fukuta D, Nishikawa Y, Yamashita Y, Inui M, Ohyama Y, Hikida M, Ohmori H, Takai T. J Biol Chem. 1997;272:7320–7327. doi: 10.1074/jbc.272.11.7320. [DOI] [PubMed] [Google Scholar]