Abstract
High-affinity binding was demonstrated between suppressor-T-cell-derived bioactive glycosylation-inhibiting factor (GIF) and helper T hybridomas and natural killer cell line cells. Inactive GIF present in cytosol of suppressor T cells and Escherichia coli-derived recombinant human GIF (rhGIF) failed to bind to these cells. However, affinity of rhGIF for the target cells was generated by replacement of Cys-57 in the sequence with Ala or of Asn-106 with Ser or binding of 5-thio-2-nitrobenzoic acid to Cys-60 in the molecule. Such mutations and the chemical modification of rhGIF synergistically increased the affinity of GIF molecules for the target cells. The results indicated that receptors on the target cells recognize conformational structures of bioactive GIF. Equilibrium dissociation constant (Kd) of the specific binding between bioactive rGIF derivatives and high-affinity receptors was 10–100 pM. Receptors for bioactive GIF derivatives were detected on Th1 and Th2 T helper clones and natural killer NK1.1+ cells in normal spleen but not on naive T or B cells. Neither the inactive rGIF nor bioactive rGIF derivatives bound to macrophage and monocyte lines or induced macrophages for tumor necrosis factor α production.
Keywords: conformational structure, macrophage migration inhibitory factor, suppressor T cells
In our previous experiments on isotype-specific regulation of the IgE antibody response, we described glycosylation-inhibiting factor (GIF), which inhibits N-glycosylation of IgE binding factors (IgE-BF) and renders the latter factors to selectively suppress the IgE synthesis (1). GIF is a 13-kDa cytokine (2), and the major cell source of GIF bioactivity is antigen-specific suppressor T (Ts) cells (3). After molecular cloning of this cytokine, however, we realized that essentially all murine and human cell line cells examined contained 0.6-kb mRNA that hybridized with murine GIF cDNA and human GIF cDNA, respectively (4). Many of these cell line cells secreted the 13-kDa peptide that reacted with polyclonal antibodies against rGIF; however, only the peptide secreted by Ts cells demonstrated GIF bioactivity (5). Escherichia coli-derived recombinant human (rh) GIF and GIF obtained by transfection of the hGIF cDNA in BMT10 cells were also inactive, but transfection of the same cDNA into a murine Ts hybridoma resulted in hGIF secretion, for which bioactivity was comparable to that produced by human Ts hybridoma cells (5). It was also found that both the murine Ts hybridoma and the stable transfectant of hGIF cDNA in the Ts hybridoma that secretes bioactive GIF contained a substantial quantity of inactive GIF in cytosol and that the amino acid sequence of the inactive GIF peptide was identical to that of the bioactive homologue (6). These findings suggested to us the possibility that bioactive GIF is generated by posttranslational modifications of the inactive GIF peptide in Ts cells and that heterogeneity of GIF in bioactivity is due to conformational transition of the same peptide (5, 6). This hypothesis was supported by our more recent experiments (7), which indicated that E. coli-derived inactive rhGIF could be converted to bioactive derivatives by chemical modification of a single cysteine residue at position 60 (Cys-60) with a sulfhydryl reagent, such as iodoacetate or 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB). The GIF bioactivity of the DTNB-derivative was comparable to that of the Ts-derived bioactive GIF. Separate experiments demonstrated that replacement of Cys-57 or Cys-60 in the inactive rhGIF with Ala by site-directed mutagenesis also resulted in the generation of bioactivity (T.T. and H. Watarai, T.M., and K.I., unpublished results).
We wondered if the heterogeneity of GIF peptide in bioactivity may be related to differences in the affinity of the molecules for target cells. In the present experiments, attempts were made to determine which type of cells have receptors for bioactive GIF. The results show high-affinity binding of bioactive GIF from Ts cells and bioactive derivatives of E. coli-derived rGIF to helper T (Th) hybridomas and Th clones, as well as natural killer (NK) cells, but not to unprimed T or B cells. In spite of previous publications that the nucleotide sequence of the cDNA encoding human macrophage migration inhibitory factor (MIF) is identical to that of human GIF except for one base (4, 8) and that E. coli-derived rMIF induced activation of macrophages for the formation of tumor necrosis factor α (TNF-α) (9, 10), high-affinity binding of either rhGIF or its bioactive derivatives to macrophages was not detectable.
MATERIALS AND METHODS
Cell Line Cells.
Ovalbumin (OVA)-specific murine Th2 hybridoma 12H5 cells (11) and the pigeon cytochrome c-specific murine Th1 clone AD10 (12) have been described. OVA-specific Th2 clone A1C6 was established from OVA-primed BALB/c lymph node cells by the method of Gajewski et al. (13). Human NK-like cell line YT cells (14) were supplied by Junji Yodoi, Kyoto University, Japan. Murine macrophage cell lines RAW264.7 and P388D1 cells and human monocyte cell line U937 cells were obtained from the American Type Culture Collection. The 2FH2 cell line is a stable transfectant of human GIF cDNA in murine Ts hybridoma 231F1 cells (5). The 2FH2 cells were cultured in high glucose Dulbecco’s modified Eagle’s medium supplemented with 10% Nu-Serum (Collaborative Biochemical Products, Bedford, MA) (5). The other cells were cultured in RPMI 1640 medium supplemented with 2 mM l-glutamine, 50 μM 2-mercaptoethanol, 0.1 mM nonessential amino acids (GIBCO/BRL), 1 mM sodium pyruvate, 10% fetal calf serum (Harlan Bioproducts for Science, Indianapolis, IN), 100 units/ml penicillin, and 100 μg/ml streptomycin.
Animals.
DO11.10 T cell receptor αβ transgenic mice on BALB/c genetic background (15) were supplied by Kenneth M. Murphy (Washington University). B10.A/SgSnJ mice were purchased from The Jackson Laboratories.
Antibodies.
All of the biotinylated monoclonal antibodies specific for mouse CD8, I-Ad, MAC-1, CD62L, or NK1.1, monoclonal anti-mouse CD16/32, biotinylated polyclonal goat anti-mouse immunoglobulin, and biotinylated rabbit anti-mouse TNF-α were purchased from PharMingen. Monoclonal anti-human GIF 388F1 (16) was purified from culture supernatant of hybridoma cells, with protein A-Sepharose (Pharmacia).
Purification of Naive T Cells, Ia+ Cells, and NK1.1+ Cells.
Naive T cells were obtained from erythrocyte-free splenocytes of the T cell receptor αβ transgenic mice described above. Cells nonadherent to nylon wool were treated with a mixture of biotinylated anti-CD8, anti-I-Ad, anti-mouse immunoglobulin, and anti-MAC-1 antibodies. The antibody-labeled cells were tagged with streptavidin-microbeads (Miltenyi Biotech, Auburn, CA), and CD4+ cells were purified by negative selection using a magnetic cell sorting system (MiniMACS, Miltenyi Biotech). They were further treated with biotinylated anti-CD62L, and the cells were incubated with streptavidin-coated microbeads for positive selection by magnetic sorting. More than 90% of the cells in the final cell preparation were CD4+ CD62Lhigh, which is consistent with the naive T cell phenotype. To purify Ia+ cells, splenocytes of the transgenic mice were treated with the biotinylated anti-I-Ad antibody and mixed with streptavidin-microbeads for positive selection. The NK1.1+ cells were obtained by treatment of splenocytes of normal B10.A mice with biotinylated anti-NK1.1 antibody, followed by incubation with streptavidin-microbeads for positive selection.
rGIF and Its Mutated Proteins.
The rhGIF (wild-type rGIF) was expressed in E. coli and purified from the soluble fraction of the cells by the method previously described (6, 7). A mutated hGIF protein with a Cys → Ala substitution at position 57, i.e., C57A, and the protein with an Asn → Ser substitution at position 106, i.e., N106S, were prepared by site-directed mutagenesis. Another mutated GIF employed in the present experiment was the protein C57A/N106S, in which both the Cys-57 → Ala and Asn-106 → Ser replacements were made in the wild-type rGIF. Detailed method for the site-specific point mutation of human GIF cDNA will be described elsewhere (T.T., H. Watarai, T.M., and K.I., unpublished results). The mutant cDNAs were expressed in E. coli and recombinant proteins were recovered by the same method as that employed for partial purification of the wild-type rGIF (6). Each of the partially purified mutated GIF was dialyzed against 20 mM sodium acetate (pH 5.5) and fractionated on a CM-5PW column (Tosoh, Tokyo) with a NaCl gradient by HPLC to recover the major protein peak. The preparations contained less than 7 pg of LPS per μg of GIF, as determined by the Limulus amoebocyte assay (Sigma). GIF in culture supernatants and cytosol of the 2FH2 cells were purified by affinity chromatography on anti-GIF (388F1)-coupled Affi-Gel 10 (Bio-Rad) (5). All purified GIF preparations gave a single 13-kDa band in SDS/PAGE analysis.
Chemical Modifications of GIF Proteins.
Purified C57A was treated with iodoacetate under the conditions used for carboxymethylation of wild-type rhGIF (7). For the treatment with DTNB, wild-type rhGIF or C57A at 1 mg/ml in 50 mM Tris⋅HCl (pH 8.5) was incubated overnight at room temperature with 3.3 mM DTNB (Pierce) in the presence of 4% acetonitrile (7). Each of the carboxymethylated C57A (C57A-CM) and DTNB derivatives of rhGIF and C57A gave a single peak upon fractionation on a CM-5PW column with NaCl gradient.
Binding Assay.
Five micrograms of an appropriate GIF sample was radiolabeled with 0.25 mCi of 125I-labeled Bolton–Hunter reagent (NEN; 1 Ci = 37 GBq). Procedures for the binding assay were similar to those employed for the binding of interleukin 2 to interleukin 2 receptors (17, 18). Cells were suspended in RPMI 1640 medium containing 10% fetal calf serum and 2 mM l-glutamine at the concentration of 1 × 107 cells per ml. Serial dilutions of 125I-labeled GIF in the same medium were mixed with an equal volume of the cell suspension. To determine nonspecific binding, a 300-fold excess of unlabeled GIF was added to radiolabeled GIF, and the same cell suspension was added to the mixtures. After incubation for 20 min at 37°C, 200 μl of the mixtures was layered on 20% olive oil/80% di-n-butyl phthalate (Sigma) in microtubes and centrifuged at 3,500 × g for 90 sec. After subtraction of nonspecific binding, the number of binding sites per cell and the equilibrium dissociation constant (Kd) were calculated by Scatchard analysis using the ligand computer program (19).
To compare the affinity of mutated GIF proteins to target cells, we determined the ability of various mutated proteins to inhibit the binding of radiolabeled C57A/N106S to either 12H5 or YT cells. Serial dilutions of a GIF sample to be tested were mixed with an equal volume of 4 nM 125I-labeled C57A/N106S, and each mixture was added to an equal volume of a cell suspension (1 × 107 cells per ml). After incubation for 20 min at 37°C, cell-bound radioactivity was determined by the procedures described above. Nonspecifically bound radioactivity was subtracted from cell-bound cpm, and inhibition of the binding of radiolabeled C57A/N106S by an appropriate GIF derivative was determined by the ratio: (specifically bound cpm in the presence of a sample)/(specifically bound cpm in the absence of the sample).
Determination of GIF Bioactivity.
Bioactivity of GIF proteins was detected by their ability to cause mouse Th hybridoma 12H5 cells to switch from forming glycosylated IgE-BF to forming unglycosylated IgE-BF. Detailed procedures for the assay have been described (20). IgE-BF in culture filtrates was fractionated on lentil lectin-Sepharose. When the 12H5 cells were cultured in the absence of GIF, essentially all IgE-BF formed by the cells bound to the lectin and was recovered by elution with 0.2 M methyl α-d-mannoside. In the presence of a sufficient concentration of GIF, most of the IgE-BF formed by the cells failed to be retained on the column. To titrate bioactivity of a sample, aliquots of the 12H5 cells were cultured with serial 1:2 dilutions of the sample, and the bioactivity was expressed by a minimum concentration of the 13-kDa peptide required for switching the cells for the formation of unglycosylated IgE-BF.
TNF-α Measurement.
Formation of TNF-α by RAW264.7 cells was determined by a previously described method (9, 10). Briefly, 2 ml of the cell suspension in RPMI 1640 medium containing 10% fetal calf serum and 2 mM l-glutamine (1 × 106 cells per ml) were plated in 3.5-cm tissue culture dishes (Falcon). After incubation for 3 hr at 37°C in a humidified atmosphere with 5% CO2/95% air, nonadherent cells were removed and adherent cells were cultured in RPMI/1% fetal calf serum for 12 hr in the presence or absence of a test sample. As a positive control, the same adherent cells were stimulated with various concentrations of lipopolysaccharide (LPS) from E. coli O111:B4 or from Salmonella typhosa (Sigma). The concentration of TNF-α in culture supernatants was determined by sandwich ELISA (Endogen, Woburn, MA) by following manufacturer’s protocol. Membrane form of TNF-α was quantitated by immunofluorescence (21). After stimulation with GIF, the cells were incubated with monoclonal anti-CD16/32 at 50 μg/ml to block Fc receptors and then treated with biotinylated rabbit anti-mouse TNF-α antibody at 50 μg/ml. The cells were washed, treated with phycoerythrin-conjugated streptavidin (PharMingen), and analyzed by a flow cytometer FACScan (Becton Dickinson).
RESULTS
High-Affinity Binding of Bioactive GIF to Cell Line Cells.
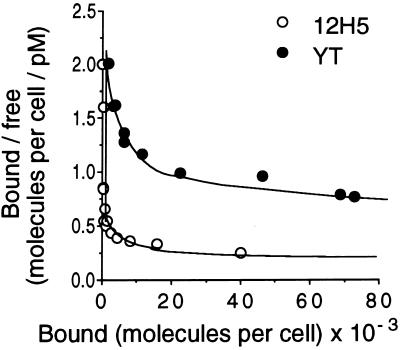
Attempts were made to determine the affinity of bioactive GIF to the 12H5 cells, which have been employed for the detection of GIF bioactivity. Human GIF was purified from the culture supernatant of 2FH2 cells. The minimum concentration of GIF in the preparation for the detection of bioactivity was 20 ng/ml. The protein was labeled with 125I, and binding of the protein to the 12H5 cells was determined. Fig. 1 shows the Scatchard analysis of the binding data, which indicates that 12H5 cells have two kinds of binding sites. In the experiment shown in Fig. 1, the number of high-affinity binding sites (Kd = 950 pM) was 600 per cell, whereas the number of low-affinity binding sites (Kd = 1,400 nM) was 500 × 103 per cell.
Figure 1.
Scatchard plot of the binding of 125I-labeled bioactive GIF from 2FH2 cells to 12H5 cells and YT cells. The abscissa represents the number of GIF molecules bound to a cell. The ordinate represents the number of GIF molecules bound to a cell divided by the concentration of unbound GIF. Similar experiments of the same design gave comparable results.
Similar experiments were carried out with the OVA-specific Th2 clone A1C6 and a human NK-like cell line YT cells with comparable results. Dissociation constant (Kd) between 2FH2-derived GIF and high-affinity receptors and low-affinity receptors on A1C6 cells were 12 pM and 2,500 nM, respectively. It was also found that the bioactive GIF bound to YT cells with high affinity (Fig. 1). The number of the high-affinity receptors (Kd = 20 pM) on YT cells was 1,400 per cell.
We determined whether the cytosolic inactive GIF would bind to the 12H5 cells. The GIF protein isolated from the cytosol of 2FH2 cells failed to show the bioactivity at the level of 1 μg/ml. The protein was radiolabeled with 125I and incubated with 12H5 cells; however, specific binding of the protein was not detectable. To confirm this finding, we determined the ability of the cytosolic GIF to block the high-affinity binding of bioactive GIF. A constant amount of radiolabeled bioactive GIF was mixed with either the 2FH2-derived cytosolic GIF or bioactive GIF of various concentrations, and the mixtures were added to YT cells. Final concentration of the radiolabeled GIF in the mixtures was 1 nM. As expected, bioactive GIF inhibited the binding of radiolabeled GIF in a dose-dependent manner, whereas even a 75-fold excess of the cytosolic GIF failed to affect the binding of radiolabeled bioactive GIF (results not shown).
Bind Ability of rGIF and Its Derivatives to Target Cells.
Previous experiments have shown that E. coli-derived rhGIF did not show bioactivity at the level of 1 μg/ml (6), but the activity was generated by the Cys-57 → Ala replacement. Thus we determined the affinity of the wild-type rGIF and mutated rGIF for target cells. Bioactivity of the mutated GIF employed in the present experiments is shown in Table 1. Radiolabeled wild-type rhGIF failed to show any specific binding to either 12H5 or YT cells (data not shown); however, C57A/N106S bound to both cell types with high affinity. The binding data shown in Fig. 2 indicated that Kd values between C57A/N106S and the high-affinity binding sites on 12H5 and YT cells were 10 pM and 40 pM, respectively. The numbers of the high-affinity binding sites on 12H5 and YT cells were 200 and 500 per cell, respectively. The murine Th1 clone AD10 also expressed high-affinity binding sites (Kd = 300 pM for 1,400 sites per cell) and low-affinity binding sites (Kd = 130 nM for 400 × 103 sites per cell) (Fig. 2). The high-affinity binding sites for C57A/N106S were detected on the Th2 clone A1C6 cells as well.
Table 1.
Increase in bioactivity of rGIF by mutation and chemical modifications of the protein
| Mutated GIF | Chemical modification
|
||
|---|---|---|---|
| None | DTNB | Iodoacetate | |
| Wild-type GIF | >1,000 | 8 | 120 |
| C57A | 100 | 3 | 10 |
| C57A/N106S | 100 | 3 | ND |
| N106S | >1,000 | ND | ND |
Data are the minimum concentration (ng/ml) of GIF species required for the detection of bioactivity. Mutated GIF was prepared by site-directed mutagenesis. ND, not done.
Figure 2.
Scatchard plot analysis of the binding of 125I-labeled C57A/N106S to 12H5 cells, YT cells, and AD10 cells. Similar results were obtained in five separate experiments.
Recent experiments have shown that chemical modification of Cys-60 in the wild-type rGIF with iodoacetate or DTNB resulted in the generation of bioactivity (7). Thus, we treated both wild-type rGIF and C57A with a sulfhydryl reagent and compared the affinity of the derivatives for 12H5 and YT cells through their ability to inhibit the binding of 1 nM 125I-labeled C57A/N106S. As shown in Fig. 3A, wild-type rhGIF could not inhibit the specific binding of radiolabeled C57A/N106S even with a 100-fold excess, but modification of wild-type rGIF with DTNB generated the ability to inhibit the binding of the radiolabeled C57A/N106S. As expected, C57A inhibited the binding in a dose-dependent manner, and treatment of C57A with either iodoacetate or DTNB markedly increased the ability of inhibiting the binding of 125I-labeled C57A/N106S (Fig. 3B). Indeed, C57A-DTNB inhibited the binding of 125I-labeled C57A/N106S as effectively as unlabeled C57A/N106S. Conversely, C57A/N106S inhibited the specific binding of 1 nM 125I-labeled C57A-DTNB to 12H5 and YT cells as effectively as unlabeled C57A-DTNB (data not shown). Furthermore, Scatchard analysis of the binding between 125I-labeled C57A-DTNB and the target cells indicated that Kd values for the binding between the C57A-DTNB and high-affinity receptors on YT and 12H5 cells were 20 pM and 10 pM, respectively.
Figure 3.
Inhibition of the binding of 125I-labeled C57A/N106S to 12H5 and YT cells by derivatives of rGIF. Aliquots of the radiolabeled C57A/N106S were mixed with a 6.25- to 100-fold excess of unlabeled GIF of various species, and the mixtures were incubated with 1 × 106 cells. The final concentration of 125I-labeled C57A/N106S in the cell suspensions was 1 nM. Cell-bound cpm were determined in duplicate, and nonspecifically bound cpm were subtracted. The ordinate represents the ratio between specifically bound radioactivity in the presence of unlabeled GIF and that in the absence of GIF. (A) Effect of rGIF (○), DTNB-derivative of rGIF (•), N106S (□), or C57A/N106S (▪). (B) Effect of rGIF (○), C57A (•), C57A-CM (▵), C57A-DTNB (▴), or C57A/N106S (▪). Cell-bound radioactivity in the absence of unlabeled GIF was 1,421 cpm for 12H5 and 1,600 cpm for YT (A) and 2,252 cpm for 12H5 or 1,481 cpm for YT (B). Three experiments of the same design gave similar results.
Determination of bioactivity of mutated GIFs and their derivatives indicated that modification of Cys-60 in C57A by a sulfhydryl reagent resulted in parallel increase in its bioactivity and affinity for 12H5 cells (Table 1 and Fig. 3B). However, replacement of Asn-106 in C57A with Ser markedly increased the affinity for 12H5 cells without affecting the bioactivity. To evaluate the discrepancy between the bioactivity and affinity for the target cells, we determined the effect of the replacement of Asn-106 with Ser in the wild-type rGIF. N106S, the mutated GIF, did not show bioactivity at the level of 1 μg/ml (Table 1) but significantly blocked the specific binding of 125I-labeled C57A/N106S to 12H5 cells (Fig. 3A).
High-affinity binding of bioactive GIF and GIF derivatives to Th clones raised a question as to whether 125I-labeled C57A/N106S may bind to unprimed lymphocytes. As shown in Fig. 4, the mutated GIF did not bind to either naive T cells or Ia+ cells but did bind to NK1.1+ cells. The number of high-affinity binding sites (Kd = 110 pM) on NK1.1+ cells was approximately 100 per cell. As expected, wild-type rGIF failed to bind to any of the unprimed lymphocyte populations.
Figure 4.
Scatchard plot analysis of the binding of 125I-labeled C57A/N106S to freshly purified naive T cells, Ia+ cells, and NK1.1+ cells. Ninety percent of the total cells in the naive T cell fraction were CD4+ CD62Lhigh, 80% of the cells in Ia+ cell fraction were I-Ad+, and 92% of the cells in NK1.1+ fraction were NK1.1+, as determined by flow cytometry. Similar results were obtained in three experiments.
Failure of GIF and Its Derivatives to Stimulate Macrophages.
In view of previous reports that deduced amino acid sequence of hGIF is identical to the sequence of MIF except for one amino acid (4, 8), we determined the binding of radiolabeled rhGIF to murine macrophage cell lines P388D1 and RAW264.7 and human monocyte cell line U937. However, specific binding of rhGIF to any of these cells was not detectable. Similar experiments with 125I-labeled C57A/N106S demonstrated that the binding of the protein to the cells was one order of magnitude less than that to the 12H5 cells, and that high-affinity binding was not detectable between the mutated rhGIF and the macrophages or monocytes (Fig. 5).
Figure 5.
Scatchard plot analysis of the binding of 125I-labeled C57A/N106S to 12H5, P388D1, U937, or RAW264.7 cells. Similar results were obtained in three experiments.
Lack of high-affinity binding between macrophages and rGIF or its mutant is in conflict with previous observations that rMIF induced RAW264.7 cells for the formation of TNF-α (9, 10). To test the possibility that bioactive or inactive GIF may induce the formation of TNF-α, aliquots of RAW264.7 cells were cultured with serial dilutions (0.1 μg/ml to 10 μg/ml) of wild-type rGIF, DTNB-treated rGIF, C57A/N106S, or C57A-DTNB. As controls, the same cells were cultured in the presence of LPS at 10 ng/ml to 10 μg/ml. Measurement of TNF-α in culture supernatants indicated that LPS induced the formation of TNF-α between 11 and 45 ng/ml in a dose-dependent manner, whereas none of the GIF species induced the formation of a detectable amount (0.2 ng/ml) of TNF-α at any of the concentrations examined. It is known that TNF-α is expressed on the cell surface as a precursor of the secreted form and that expression of membrane form TNF-α is inducible by various stimuli (21, 22). Thus, RAW264.7 cells were cultured for 12 hr in the presence of various concentrations of LPS, rGIF, or bioactive rGIF derivatives, and the level of TNF-α expression was determined by flow cytometry analysis. As shown in Fig. 6, LPS at 0.1 μg/ml was sufficient for maximal expression of TNF-α on RAW264.7 cells. In contrast, rGIF at even 10 μg/ml or its bioactive derivatives failed to increase the expression of the membrane form of TNF-α. It is evident that neither rGIF nor its bioactive derivative can induce the formation of TNF-α by macrophages.
Figure 6.
Failure of GIF derivatives to induce cell surface TNF-α on RAW264.7 cells. RAW264.7 cells were incubated with 0 (bold solid line), 0.1 μg/ml (dashed line), 1.0 μg/ml (dashed and dotted line), or 10 μg/ml (dotted line) of LPS (a), rGIF (b), C57A/N106S (c), or C57A-DTNB (d) for 12 hr. Cell-surface TNF-α was stained and analyzed by flow cytometry. The thin solid line represents background immunofluorescence. The ordinate and abscissa represent the cell number and the fluorescence intensity on a logarithmic scale, respectively. Data presented are representative of comparable results from three experiments.
DISCUSSION
The present experiments demonstrated that bioactive GIF secreted from murine Ts hybridoma bound to Th hybridomas, Th clones, and NK cell line cells with high affinity, whereas inactive GIF in cytosol and E. coli-derived rhGIF failed to do so. A Scatchard plot of the binding data indicates that these target cells have two types of receptors for bioactive GIF and that inactive GIF bound to neither receptor. It was noted that the Kd values between the bioactive GIF and high-affinity receptors were different from one experiment to another, in the range of 20 pM to 900 pM. Such inconsistency in Kd values is partly due to the presence of both bioactive GIF and inactive GIF in culture supernatant of 2FH2 cells (6). After removal of the majority of inactive GIF from the 2FH2 culture supernatant, the minimum concentration of the 13-kDa protein for the detection of GIF bioactivity was 3–5 ng/ml (6). For the GIF sample used for the binding to 12H5 cells in Fig. 1, the minimum concentration for the bioactivity was 20 ng/ml, indicating that bioactive GIF in the sample represents only 10–20% of total GIF. Considering this factor, the actual Kd value between bioactive GIF and high-affinity receptors appears to be in the range of 10 pM to 100 pM. Possible relationships between high-affinity receptors and low-affinity receptors are not known. Nevertheless, the Kd value between bioactive GIF and low-affinity receptors suggests that receptors of this type do not have a biological role.
An identical amino acid sequence shared by the bioactive GIF and inactive GIF (6) and lack of an intra-chain disulfide bond in these molecules (7, 23) collectively suggest that the specific binding of GIF molecules to target cells depends on their tertiary structure. No difference was detected by SDS/PAGE among the bioactive GIF, inactive GIF in cytosol, and E. coli-derived rGIF (5, 6). One may speculate that heterogeneity of GIF in bioactivity and affinity for target cells is due to conformational transition of the same peptide. The crystal structure of GIF showed that the GIF monomer consists of one β-sheet and two α-helices, which are located on the same side of the β-sheet. Although the distance between the center axes of the two helices is 12 Å in the crystal, analysis of the structure indicated conformational flexibility in the two helices and adjacent loop regions (23). Subsequent experiments showed that SH group of Cys-60, which is located on the β-strand and sticking out between the two α-helices, was easily carboxymethylated or reacted with DTNB, and such chemical modifications of rGIF resulted in generation of bioactivity (7). The results suggested that binding of a chemical group to the sulfur atom in Cys-60 increased the distance between the two helices and caused some conformational changes in the molecule and that such conformational changes might be responsible for the generation of bioactivity. Replacement of Cys-57, located in the loop region, with Ala also generated the bioactivity. The present experiments showed that all of the chemical modification of Cys-60, the replacement of Cys-57 with Ala, and the replacement of Asn-106 with Ser resulted in the generation of affinity for target cells and that such independent chemical changes in the molecule had synergistic effects on increasing the affinity of rGIF for target cells. Our more recent experiments have shown that kinetics of the reaction of DTNB to Cys-60 in N106S and C57A was significantly faster than the kinetics of the reaction between DTNB and Cys-60 in wild-type rGIF, suggesting that both the Asn-106 → Ser substitution and Cys-57 → Ala substitution may have increased the distance between the two α-helices, rendering the SH group of Cys-60 more accessible to DTNB (data not shown). These findings collectively indicate that the receptors on target cells recognize conformational structures in bioactive GIF, which are lacking in inactive homologue, and that high-affinity binding of the molecule is an essential step for manifestation of bioactivity. It should be noted that the Kd value between C57A-DTNB and high-affinity receptors on 12H5 and YT cells was 10–20 pM and that the minimum concentration of this GIF derivative required for the detection of bioactivity was comparable to that of highly purified Ts-derived GIF (6).
However, the present experiments also indicate that the affinity of GIF derivatives for 12H5 cells does not parallel their bioactivity. It was found that substitution of Ser for Asn-106 in rGIF resulted in the generation of affinity for target cells but not the GIF bioactivity. The same substitution in C57A markedly increased the affinity of the molecule for target cells without increasing its bioactivity (see Fig. 3B and Table 1). Structural basis for the lack of parallelism between the bioactivity and cell affinity is not known. Nevertheless, the results suggest that the binding of GIF molecule to target cells may not be sufficient for signal transduction. This finding may not be surprising. It is known that TNF-α and lymphotoxin are structurally related and bind to the same receptors with comparable affinity; however, TNF-α was 200- to 300-fold more effective than lymphotoxin in inhibiting the growth of tumor cell line cells (24).
Present experiments show that bioactive derivatives of rGIF bind to both Th1 and Th2 clones with high affinity but do not bind to unprimed naive T cells or Ia+ cells, which include normal B cells. Evidence has been presented that bioactive GIF is a subunit of antigen-specific Ts factors (25, 26). The high affinity of bioactive GIF for antigen-primed Th cells suggests that the Ts factors may bind to Th cells through GIF and exert their immunoregulatory effects. The present experiments also showed that bioactive GIF and GIF derivatives bind to NK cells with high affinity. It is known that NK cells are the major source of interferon γ (27, 28). Indeed, we have recently observed that bioactive GIF from 2FH2 cells and some GIF derivatives enhanced NK activity in vivo and induced interferon γ production in vitro. NK1.1+ cells are implicated not only in the production of cytokines and cytotoxicity but also in immunosuppression (29). It is possible that NK1.1+ cells may be involved in the immunoregulatory effect of GIF.
We have reported (4) that affinity-purified bioactive GIF does not inhibit the migration of human monocytes. The present experiments actually showed that bioactive rGIF derivatives that had high affinity for Th cells and NK cells failed to bind to macrophage/monocyte lines. Neither the bioactive derivatives nor inactive rGIF induced TNF-α production in RAW264.7 cells. Failure of rGIF for specific binding to the cells is in agreement with previous findings by Herriott et al. (30) that rMIF/GIF at up to 5 μg/ml failed to inhibit migration of macrophages and eliminates the possibility that GIF directly acts on the cells.
Acknowledgments
We express our great appreciation to Dr. Carl Ware for advice in determining TNF-α production by macrophages. This paper is publication 174 from the La Jolla Institute for Allergy and Immunology. The work was supported in part by Research Grant AI-14784 from the U.S. Department of Human and Health Services.
ABBREVIATIONS
- GIF
glycosylation-inhibiting factor
- IgE-BF
IgE-binding factor
- Ts
suppressor T
- Th
helper T
- DTNB
5,5′-dithiobis(2-nitrobenzoic acid)
- MIF
macrophage migration inhibitory factor
- TNF-α
tumor necrosis factor α
- r
recombinant
- h
human
- OVA
ovalbumin
- LPS
lipopolysaccharide
References
- 1.Ishizaka K. Annu Rev Immunol. 1984;2:159–182. doi: 10.1146/annurev.iy.02.040184.001111. [DOI] [PubMed] [Google Scholar]
- 2.Tagaya Y, Mori A, Ishizaka K. Proc Natl Acad Sci USA. 1991;88:9117–9121. doi: 10.1073/pnas.88.20.9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jardieu P, Akasaki M, Ishizaka K. J Immunol. 1987;138:1494–1501. [PubMed] [Google Scholar]
- 4.Mikayama T, Nakano T, Gomi H, Nakagawa Y, Liu Y-C, Sato M, Iwamatsu A, Ishii Y, Weiser W Y, Ishizaka K. Proc Natl Acad Sci USA. 1993;90:10056–10060. doi: 10.1073/pnas.90.21.10056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y-C, Nakano T, Elly C, Ishizaka K. Proc Natl Acad Sci USA. 1994;91:11227–11231. doi: 10.1073/pnas.91.23.11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakano T, Liu Y-C, Mikayama T, Watarai H, Taniguchi M, Ishizaka K. Proc Natl Acad Sci USA. 1995;92:9196–9200. doi: 10.1073/pnas.92.20.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakano T, Watarai H, Liu Y-C, Oyama Y, Mikayama T, Ishizaka K. Proc Natl Acad Sci USA. 1997;94:202–207. doi: 10.1073/pnas.94.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiser W Y, Temple P A, Witek-Giannotti J S, Remold H G, Clark S C, David J R. Proc Natl Acad Sci USA. 1989;86:7522–7526. doi: 10.1073/pnas.86.19.7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calandra T, Bernhagen J, Mitchell R A, Bucala R. J Exp Med. 1994;179:1895–1902. doi: 10.1084/jem.179.6.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernhagen J, Mitchell R A, Calandra T, Voelter W, Cerami A, Bucala R. Biochemistry. 1994;33:14144–14155. doi: 10.1021/bi00251a025. [DOI] [PubMed] [Google Scholar]
- 11.Iwata M, Adachi M, Ishizaka K. J Immunol. 1988;140:2534–2542. [PubMed] [Google Scholar]
- 12.Hedrick S M, Engel I, Fink P J, Hsu M-L, Hansburg D, Matis L A. Science. 1988;239:1541–1544. doi: 10.1126/science.2832942. [DOI] [PubMed] [Google Scholar]
- 13.Gajewski T F, Joyce J, Fitch F W. J Immunol. 1989;143:15–22. [PubMed] [Google Scholar]
- 14.Yodoi J, Teshigawara K, Nikaido T, Fukui K, Noma T, Honjo T, Takigawa M, Sasaki M S, Minato N, Tsudo M, Uchiyama T, Maeda M. J Immunol. 1985;134:1623–1630. [PubMed] [Google Scholar]
- 15.Murphy K M, Heimberger A B, Loh D Y. Science. 1990;250:1720–1722. doi: 10.1126/science.2125367. [DOI] [PubMed] [Google Scholar]
- 16.Thomas P, Gomi H, Takeuchi T, Carini C, Tagaya Y, Ishizaka K. J Immunol. 1992;148:729–737. [PubMed] [Google Scholar]
- 17.Wang H-M, Smith K A. J Exp Med. 1987;166:1055–1069. doi: 10.1084/jem.166.4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugie K, Nakamura Y, Tagaya Y, Koyasu S, Yahara I, Takakura K, Kumagai S, Imura H, Yodoi J. Int Immunol. 1990;2:391–397. doi: 10.1093/intimm/2.5.391. [DOI] [PubMed] [Google Scholar]
- 19.Munson P J, Rodbard D. Anal Biochem. 1980;107:220–239. doi: 10.1016/0003-2697(80)90515-1. [DOI] [PubMed] [Google Scholar]
- 20.Iwata M, Ishizaka K. J Immunol. 1988;141:3270–3277. [PubMed] [Google Scholar]
- 21.Ware C, Crowe P D, Grayson M H, Androlewicz M J, Browning J L. J Immunol. 1992;149:3881–3888. [PubMed] [Google Scholar]
- 22.Kriegler M, Perez C, DeFay K, Albert I, Lu S D. Cell. 1988;53:45–53. doi: 10.1016/0092-8674(88)90486-2. [DOI] [PubMed] [Google Scholar]
- 23.Kato U, Muto T, Tomura T, Tsumura H, Watarai H, Mikayama T, Ishizaka K, Kuroki R. Proc Natl Acad Sci USA. 1996;93:3007–3010. doi: 10.1073/pnas.93.7.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Browning J, Ribolini A. J Immunol. 1989;143:1859–1867. [PubMed] [Google Scholar]
- 25.Nakano T, Ishii Y, Ishizaka K. J Immunol. 1996;156:1728–1734. [PubMed] [Google Scholar]
- 26.Ishii Y, Nakano T, Ishizaka K. J Immunol. 1996;156:1735–1742. [PubMed] [Google Scholar]
- 27.Young H A, Ortaldo J R. J Immunol. 1987;139:724–727. [PubMed] [Google Scholar]
- 28.Scharton T M, Scott P. J Exp Med. 1993;178:567–577. doi: 10.1084/jem.178.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vicari A P, Zlotnik A. Immunol Today. 1996;17:71–76. doi: 10.1016/0167-5699(96)80582-2. [DOI] [PubMed] [Google Scholar]
- 30.Herriott M J, Jiang H, Stewart C A, Fast D J, Leu R W. J Immunol. 1993;150:4524–4531. [PubMed] [Google Scholar]