Abstract
Acute promyelocytic leukemia (APL) is associated with reciprocal chromosomal translocations involving the retinoic acid receptor α (RARα) locus on chromosome 17. In the majority of cases, RARα translocates and fuses with the promyelocytic leukemia (PML) gene located on chromosome 15. The resulting fusion genes encode the two structurally unique PML/RARα and RARα/PML fusion proteins as well as aberrant PML gene products, the respective pathogenetic roles of which have not been elucidated. We have generated transgenic mice in which the PML/RARα fusion protein is specifically expressed in the myeloid-promyelocytic lineage. During their first year of life, all the PML/RARα transgenic mice have an abnormal hematopoiesis that can best be described as a myeloproliferative disorder. Between 12 and 14 months of age, 10% of them develop a form of acute leukemia with a differentiation block at the promyelocytic stage that closely mimics human APL even in its response to retinoic acid. Our results are conclusive in vivo evidence that PML/RARα plays a crucial role in the pathogenesis of APL.
Acute promyelocytic leukemia (APL), the M3 subtype of the French–American–British classification of acute myeloid leukemias (1), is characterized by three distinct and unique features: (i) the accumulation in the bone marrow (BM) of tumor cells with promyelocytic features; (ii) the invariable association with specific translocations that always involve chromosome 17 (reviewed in ref. 2); and (iii) the exquisite sensitivity of APL blasts to the differentiating action of retinoic acid (RA) (reviewed in refs. 3 and 4). From this point of view, APL has become the paradigm for therapeutic approaches using differentiating agents. In the vast majority of patients, APL is associated with a reciprocal translocation between chromosomes 15 and 17 (5–8). At the molecular level, the breakpoint on chromosome 17 lies within the RA receptor α (RARα) locus. The breakpoints on chromosome 15 cluster within the PML locus [for promyelocytic leukemia (9–12)]. In rare cases, the translocation involves chromosome 11 instead of chromosome 15, and a newly identified gene named promyelocytic leukemia zinc finger (PLZF) instead of PML (13). In only four cases does the translocation involve chromosome 5 and the nucleophosmin gene (NPM) or chromosome 11 and the NUMA gene (refs. 14 and 15 and unpublished results). The common feature of the four translocations is the involvement of chromosome 17 and of the RARα locus (2). The 15;17 translocation results in the production of two fusion genes that encode a PML/RARα fusion protein (9–11), an RARα/PML fusion protein (16), and an aberrant PML protein that lacks the PML C-terminal end and its phosphorylation site (12). Because the PML/RARα hybrid retains most of the functional domains of its parental proteins and can heterodimerize with PML and retinoid-X-receptor (RXR), the chimeric proteins might act as doubly dominant negative oncogenic products, interfering with both PML and RAR/RXR pathways (17, 18). This expectation has been corroborated by the finding that, while PML is normally concentrated within discrete speckled nuclear structures called PML nuclear bodies (NBs) [also called Kremer Bodies, ND10, or PODs (for PML oncogenic domains)], this normal localization pattern is abolished in promyelocytes from APL patients and in the promyelocytic cell line NB4, due to the presence of PML/RARα. In these cases, aberrant structures, within which PML, RARα, RXRα, and the PML/RARα fusion protein colocalize, become apparent (19–21). PML is specifically up-regulated at the transcriptional level by interferons, and its overexpression is accompanied by growth suppression (22, 23). All these observations taken together would suggest that the deregulation of the PML pathway plays a crucial role in promyelocytic leukemogenesis and that the disruption of this activity by PML/RARα could result in the expansion of the APL blasts. On the other hand, the possible role of RARα in the control of myeloid terminal differentiation (24–26) suggests that the concomitant interference of PML/RARα with RARα/RXRα function could lead to the block of maturation at the promyelocytic stage observed in APL. Indeed, the PML/RARα protein is capable of blocking the terminal differentiation of myeloid precursor cells such as U937, HL60, or K562 induced by certain stimuli, including vitamin D3 and transforming growth factor type β. However, the information available on the function of the APL-specific chimeric products relies thus far only on data derived from leukemic cell lines, which are obviously not the ideal system for the study of oncogenic transformation (27–29). In fact, there is no direct proof that the PML/RARα and RARα/PML fusion proteins, and the aberrant PML protein, can cause APL, and their individual relative contributions to producing the leukemic phenotype have not been elucidated. Here we provide evidence that the myeloid-promyelocytic restricted expression of the PML/RARα fusion protein in the transgenic mouse results in acute leukemia characterized by the specific differentiation block at the promyelocytic stage that defines human promyelocytic leukemia.
MATERIALS AND METHODS
Construction of the Transgene.
To generate the human cathepsin-G (hCG) minigene expression vector, we screened a human genomic placenta library with a hCG full-length cDNA probe generated by reverse transcriptase (RT)-PCR and obtained four overlapping phages which spanned the hCG locus. A 3.3-kb BamHI–EcoRI fragment that contains the hCG 5′ flanking region and its promoter and a 6-kb SalI–BamHI fragment that contains the coding and the 3′ flanking regions were subcloned into Bluescript plasmids (Stratagene) and named pT2BE3 and pT4BS6, respectively. Two pairs of PCR primers (AF–HR and SalI BF–LR; AF, CTTGCTTTGCTGGAGTATTCTG; HR, TTTCCTGAAAGGCTGCCCAG; SalI BF, GTCGACAGCCACTCCTGCTTCTGCTG; LR, GTCCAATTCAAGACCACGTGGGACT; see ref. 30) were used to amplify a 0.3-kb fragment ending one base before the ATG and a 0.4-kb fragment starting one base after the ATG of hCG gene with an artificial SalI site at its 5′ end. The two PCR products were cloned into Bluescript, sequenced to exclude PCR amplification errors, and named pBST and pB15, respectively. The 3.3-kb 5′ BamHI–EcoRI fragment from pT2BE3 was fused to the pBST fragment to reconstruct the 5′ of the gene just before the ATG and named HB33. A BamHI–SalI fragment from HB33 was cloned at a SalI site into the plasmid pBSNN, into which a Tk–Neo cassette had been cloned for the purpose of selection. In parallel, a 3-kb PmlI–BamHI fragment containing the coding and 3′ flanking regions of the hCG excised from pT4BS6 was fused at the 3′ of pB15 at a PmlI site to reconstruct the 3′ of the gene just after the ATG. A ClaI–BamHI fragment from this vector was cloned into pBSNN at the ClaI site, thus reconstructing a cathepsin-G minigene in which the ATG was replaced by two unique cloning sites, ClaI and SalI (hCG-GAG vector). This plasmid vector is 11 kb long and can be linearized before transfection with use of a unique BamHI site. Subsequently, a blunt-ended 3.15-kb NotI–EcoRI fragment spanning the PML/RARα fusion cDNA (9), and the Lac-Z cDNA were cloned into the blunt-ended artificial SalI site of the hCG-GAG vector (hCG–PML/RARα). The resulting mRNAs are processed and stabilized by the splicing events occurring between the various hCG exons, and are polyadenylylated. Finally, BamHI–NotI fragments from these constructs were purified for egg injection according to standard procedures (31).
DNA and RNA Analysis.
Genomic DNA extracted from mouse tails was digested with EcoRI and HindIII and hybridized with probe CT, a 250-bp HindIII–XbaI hCG genomic fragment, and probe HS1, a 1-kb hCG genomic fragment (see Fig. 1A). A murine PML EcoRI–ApaI genomic fragment (PML probe A) (unpublished work) was used to normalize and determine the number of copies of the transgene with the help of a PhosphorImager densitometer (Bio-Rad). For Northern blot analysis, total RNA was prepared from mouse BM and spleen using the guanidinium thiocyanate method as previously described (32). Denatured RNA (20 μg) was hybridized with the human RARα probe IT (5) following standard procedures (33). RT-PCR was also used to analyze the expression of the transgene (34) using the primers M4-R7 for the first round of amplification and M2-Rj for the nested round, or RAR(d)-Cat(b) for first and F1-B1 for nested (see Fig. 1A). Primers: M4, CTGGAGGCTGTGGACGCGCGGTA; R7, TCTTCTGGATGCTGCGGCGGAA; M2, AGTGTACGCCTTCTCCATCAAAG; RJ, CTTGCAGCCCTCACAGGCGCT; RAR(d), CAGCTCCAACAGAAGCAGC; Cat(b), CAATGAGCTGCTGTCAGCA; F1, TCTCTGCCTTTCTACCGACC; B1, CTGGATCTGAAGATACGCC.
Figure 1.
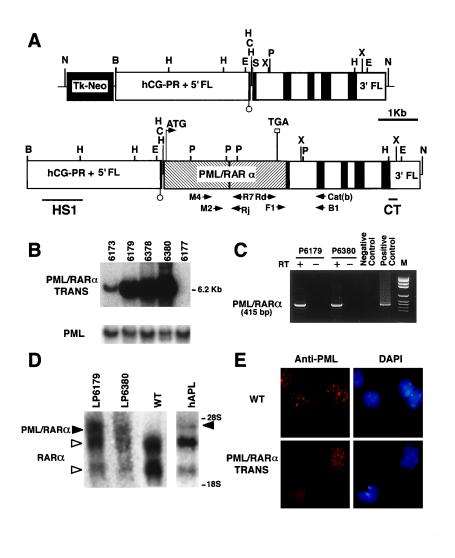
Structure, integration, and expression of the hCG–PML/RARα transgene. (A) Structure of the hCG minigene expression vector (Upper) and the hCG–PML/RARα transgene fragment used for injection (Lower). The exons of hCG gene are designated by solid boxes. The promoter and the 5′ flanking region (hCG–PR + 5′ FL) as well as the 3′ flanking region (3′ FL) of the hCG gene are indicated. The PML/RARα cDNA was cloned into hCG exon 1. Restriction endonuclease sites used in construction and Southern blotting are as follows: N, NotI; B, BamHI; E, EcoRI; C, ClaI; S, SalI; P, PmlI; X, XbaI; H, HindIII. The location of probes for Southern blotting and primers for RT-PCR are shown underneath the structure of the injected fragment. (B) Southern blot of mouse genomic tail DNA from the four transgenic founders and one control digested with EcoRI and hybridized with probe CT reveals the expected band of 6.2 kb. PML probe A (see Materials and Methods) was used as an internal control probe to normalize DNA loading. Lanes: 6173, 6179, 6378, and 6380, four founders; 6177, nontransgenic mouse. (C) RT-PCR analysis of PML/RARα fusion mRNA expression in BM cells from progeny from founders 6179 (P6179) and 6380 (P6380). Representative result of nested PCR is shown. Negative control, RNA from a nontransgenic mouse; positive control, RNA from an APL patient with t(15;17); M, pGEM DNA markers (Promega). (D) Northern blot on total RNA from leukemic progeny from lines 6179 (LP6179) and 6380 (LP6380) hybridized with the human RARα probe IT (5). WT, nontransgenic mouse; hAPL, APL patient with t(15;17) [PML/RARα of bcr1 type (ref. 12)]. Expected 4.0-kb transcript for hCG–PML/RARα transgene (left solid arrowhead) and 4.4-kb transcript for human PML/RARα chimeric mRNA (right solid arrowhead) are indicated. Open arrowheads indicate the positions of the two RARα transcripts. The position of the 28S and the 18S ribosomal RNA transcripts are also indicated. (E) Immunofluorescence staining of BM cells from control mice (WT) and leukemic hCG–PML/RARα transgenic mice. PML is localized within the NBs in nontransgenic mouse (Upper), whereas it is in the form of microspeckles in transgenic mouse (Lower). 4′,6-Diamidino-2-phenylindole (DAPI) staining was used to visualize cell nuclei.
Immunofluorescence.
BM and spleen cells were fixed with 4% paraformaldehyde for 10 min and permeabilized with methanol at −20°C for 7 min. The cells were subsequently incubated with a primary α-PML antibody [kindly provided by P. Freemont, Imperial Cancer Research Fund (ICRF), London].
Hematology, Histopathology, and Immunohistochemistry.
White blood cell (WBC), hemoglobin, and platelet counts in peripheral blood (PB) were determined using an automated counter (Technicon-H2). Differential counts of PB and BM were performed microscopically on Wright-Giemsa stained smears. A minimum of 100 cells for PB or 300–500 cells for BM were examined and reported as a percentage of each kind of cell in the nucleated cells. Sections (4 μm) from various hematopoietic organs, including BM, spleen, liver, thymus, and lymph nodes, were stained with hematoxylin and eosin and examined. Myeloperoxidase (MPO; Dako), B220 (CD45R), and CD3 antibodies (PharMingen) were used for the immunohistochemistry staining on BM and spleen sections as described (35).
Flow Cytometry.
PB, BM, and spleen cells (106 cells) were incubated with Mac-1 (CD11b) antibody and Gr-1 (myeloid differentiation antigen) or c-Kit (CD117) antibodies after being blocked with an unlabeled FcγR (CD16/32) antibody. All antibodies were obtained from PharMingen. At least 2 × 104 events were analyzed from each sample on a FACScan flow cytometer (Becton Dickinson).
Clonogenic Assays.
BM and spleen cells were seeded in 0.9% methylcellulose medium containing 30% FBS, 1% BSA, 10−4 M 2-mercaptoethanol, 2 mM l-glutamine, 2% pokeweed mitogen-stimulated spleen cell conditioned media, and 3 units/ml erythropoietin (StemCell Technologies, Vancouver). CFU-E were scored on day 2, and BFU-E, CFU-GM, and CFU-GEMM were scored on day 10.
RA Differentiation Assays.
Mononuclear cells were isolated from BM and spleen suspensions by Ficoll-hypaque density gradient centrifugation. Cultures were established at a cell concentration of 106 per ml in α-MEM supplemented with 10% fetal calf serum, 2 mM l-glutamine, and antibiotics in a 24-well-plate with or without RA at a final concentration of 10−6 M. RA-sensitive NB4 cells were used as a positive control for differentiation. At days 0, 3, 5, and 7, cells were harvested from culture for quantitative evaluation of differentiation induction by applying the nitro blue tetrazolium (NBT) test (see below), and morphological differential count. The NBT-positive percentage was obtained by microscopic scoring of 100 cells.
RESULTS
Production of PML/RARα Transgenic Mice.
In our attempt to generate PML/RARα transgenic mice, we noticed that transgene expression driven by housekeeping promoters such as β-actin resulted in fetal lethality. We presume this is due to high expression of a mutant RARα interfering with the RAR/RXR pathway. To avoid the potential toxicity of the fusion proteins for the developing embryo, we decided to generate transgenic mice in which the APL-specific fusion genes (cDNAs) are under the control of the regulatory elements of the myeloid/promyelocytic specific cathepsin-G gene (36). To this end, we generated a cathepsin-G minigene expression vector by cloning and modifying a 6.7-kb BamHI–BamHI genomic fragment of the hCG gene that is larger than a fragment which has previously been shown to retain promyelocytic specificity in transgenic mice (Fig. 1A and ref. 36). We subcloned the PML/RARα cDNA as well as a Lac-Z cDNA into this vector using the unique SalI restriction enzyme cloning site (Fig. 1A and data not shown). The hCG–Lac-Z construct was injected into mouse eggs, and embryos were killed at 12.5, 14.5, and 16.5 days post coitus and stained for β-galactosidase activity to test the activity of the hCG expression vector. Positive staining was observed in the liver (12.5 and 14.5 days post coitus) and in the BM (16.5 days post coitus) (data not shown). In addition, we observed variable staining in other organs, indicating that the expression of the minigene is susceptible to position-dependent effects (data not shown). The progeny obtained from injection of the hCG–PML/RARα was analyzed by Southern blotting on DNA obtained from mouse tails and digested with EcoRI, using a human-specific cathepsin-G probe (CT) which does not cross-react with the endogenous mouse cathepsin-G gene (Fig. 1A). Using this screening method, we identified four independent transgenic founders carrying the hCG–PML/RARα transgene. Southern blot analysis of offspring from the various founders with two distinct probes (HS1 and CT) and using two different restriction enzyme digestions (EcoRI and HindIII) showed integration of between 2 and 32 intact copies of the transgene in a head-to-tail and/or tail-to-tail arrangement (Fig. 1 A and B and Materials and Methods).
Analysis of PML/RARα Expression Driven by the hCG Minigene.
The expression of the PML/RARα fusion gene was initially studied by RT-PCR analysis on RNA extracted from BM and spleen of the progeny of the four founders following the nested RT-PCR approach that is presently used in our lab to detect minimal residual disease in APL patients (see Materials and Methods, Fig. 1 A and C, and ref. 34). Progeny from founders 6173, 6179, and 6380 were found to be positive. Lines 6179 and 6380 were further expanded and characterized. To make sure that the hCG/PML/RARα fusion cDNA was not contaminated by genomic transgene sequences, we took several precautions. (i) We performed the RT-PCR in parallel on RNA samples with and without RT (Fig. 1C). (ii) We also used two pairs of oligonucleotides [Rd and Cat(b), and F1 and B1] that span cathepsin-G intron 1, thus allowing us to distinguish the unprocessed gene from the processed cDNA (Fig. 1A and Materials and Methods). The expression of the transgene was subsequently confirmed by Northern blotting and immunofluorescence analysis on BM samples obtained from mice that had developed overt leukemia. Northern blot analysis confirmed the expression of the expected hCG/PML/RARα fusion transcript of approximately 4 kb in both transgenic lines (Fig. 1D).
Immunofluorescence analysis carried out with a PML antibody that cross-reacts with both human and murine PML protein as well as with the PML/RARα fusion protein revealed a striking microspeckled PML distribution in the BM cells from PML/RARα transgenic mice when compared with the classical PML accumulation in the NBs observed in the cells from wild-type control mice (Fig. 1E). Therefore, these data confirm the expression of the transgene and at the same time demonstrate that, as with human APL, the presence of PML/RARα results in delocalization of PML from the NBs.
Aberrant Hematopoiesis and Leukemia in PML/RARα Transgenic Mice.
The hematopoiesis of progeny from the two expressing lines was systematically analyzed in the following manner over a 14-month follow-up period. (i) In one group, the mice were bled on a monthly basis, together with appropriate littermate controls of the same sex and age (25 transgenic mice and 25 control mice per line). Differential counts, as well as morphological analysis of PB cells, were performed on each sample to monitor the incidence of leukemia. (ii) In the second group, 4 transgenic mice and 4 controls were killed every 2 months, at 2, 4, 6, etc., months of age, for gross and microscopic examination of all organs and in particular PB, BM, spleen, lymph nodes, and thymus. These organs and their cell populations were analyzed morphologically on touch preparation, smears, and cytospin preparations stained with Wright-Giemsa.
This analysis revealed a profound disturbance of myeloid hematopoiesis in both transgenic lines that can be divided into two distinct phases.
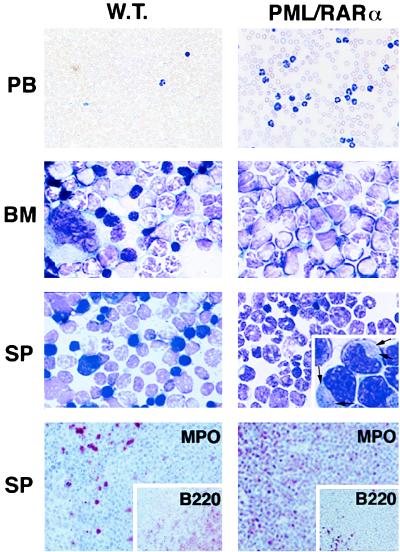
(i) The first phase is a long asymptomatic preleukemia phase in which the transgenic mice develop what in human would be described as a myeloproliferative disorder. In this phase, myeloid precursors, including promyelocytes (up to 10%; see Table 1), progressively accumulate in the BM, and infiltrate the spleen, the lymph nodes, and the thymus (data not shown). These cells partially retain the ability to terminally differentiate toward mature granulocytes (data not shown). During this phase, the PB remains essentially normal, except, in some cases, for an increased granulocyte/lymphocyte ratio (Table 1). By 12 months of age, all the mice analyzed had developed this myeloproliferative disorder to variable degrees of severity, but no leukemia was observed before this age.
Table 1.
Automated and differential counts of PB and BM from transgenic and wild-type mice
| Mice | PB
|
BM
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hb, g/dl | Plt, 109/liter | WBC, 109/liter | M/L | Pro+blasts, % | M/L | M/E | Poly, % | Mye+meta, % | Pro+blasts, % | |
| Wild type | 15.1 ± 0.9 | 1,278 ± 201 | 11.0 ± 4.0 | 0.23 ± 0.1 | 0 | 3.54 ± 2.6 | 1.92 ± 0.8 | 34.1 ± 11.1 | 9.9 ± 5.2 | 3.0 ± 1.0 |
| Preleukemia | 17.7 ± 2.1 | 990 ± 299 | 9.1 ± 3.0 | 0.82 ± 0.7 | 0 | 11.38 ± 1.7 | 7.15 ± 5.2 | 43.6 ± 6.2 | 24.9 ± 10.0 | 7.9 ± 0.9 |
| Leukemia | 10.0 ± 5.7 | 280 ± 100 | 148.4 ± 114.4 | 7.75 ± 5.7 | 6.8 ± 4.9 | 38.2 ± 14.5 | 47.8 ± 16.3 | 35.2 ± 23.4 | 12.0 ± 8.8 | 46.3 ± 15.0 |
Preleukemia, transgenic mice showing some abnormal signs in PB, BM, or spleen but not having reached leukemic criteria; Hb, hemoglobin; Plt, platelet; M/L, ratio of various differentiation stage granulocytes (M) to lymphocytes (L); M/E, ratio of various differentiation stage granulocytes (M) to nucleated erythrocytes (E); Poly, polymorphonuclear granulocytes; Mye+meta, myelocytes plus metamyelocytes; Pro+blasts, promyelocytes plus blasts. The number of mice used in this analysis is as follows: 4 leukemia, 4 preleukemia, 5 wild-type for BM, and 10 wild-type for PB.
(ii) After mice reached 1 year of age, approximately 10% of them developed overt acute leukemia and died or were killed. Leukemia was diagnosed according to the postmortem analysis (infiltrates of all organs analyzed by the myeloid blast population) and to hematological parameters (promyelocytes and blasts percentage in the BM and PB). This leukemia is characterized by the accumulation of promyelocytes in the BM, PB, and spleen, which is distinctive of APL (Table 1 and Fig. 2), with some promyelocytes showing the presence of pathognomonic cytoplasmic Auer’s bodies (Fig. 2), and by white cell counts in the PB (WBC) higher than in classical human APL (Table 1 and ref. 1). As in the myeloproliferative phase, the leukemic cellular population partially retains the ability to terminally differentiate toward mature granulocytes (Table 1 and Fig. 2). The remaining 90% of the mice are still leukemia-free after a 14-month follow-up period.
Figure 2.
Morphology and immunohistochemistry of PB, BM, and spleen (SP) cells from wild-type (W.T.) and leukemic hCG–PML/RARα transgenic mice. The smears of PB or imprints of BM and spleen were stained with Wright-Giemsa stain. Magnified detail of promyelocyte morphology is shown in the spleen imprint from the leukemic hCG–PML/RARα mouse. Auer’s bodies are visible in the cytoplasm of the leukemic promyelocytes as indicated by the arrows. The serial spleen sections shown in the bottom panel were immunohistochemically stained with MPO (myeloid precursors and mature granulocytes) and B220 (pan-B cells, Inset) antibodies. (×400 for PB; ×1,000 for BM and spleen; ×128 for immunohistochemical staining.) Note that in the leukemic mice, the number of WBC in PB is much increased; the normal BM components, including erythrocytes and megakaryocytes, are replaced by myeloid cells at various maturation stages; the spleen is heavily infiltrated by MPO-positive cells.
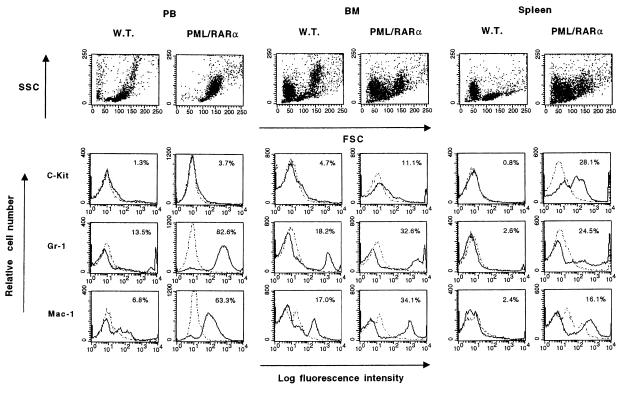
We next subjected the PB, BM, and spleen cells from leukemic mice to a flow cytometric analysis with antibodies against the following cell surface markers: c-Kit (totipotent stem cells), Gr-1 (mature granulocytes and monocytes), and Mac-1 (mature monocytes and granulocytes) (see Fig. 3 and Materials and Methods). This analysis confirmed the myeloid nature of the leukemia revealed by the dramatic accumulation of myeloid cells (Mac-1- and Gr-1-positive) in the PB and spleen of the leukemic mice, as well as the aforementioned observation that the leukemic cellular population partially retains the ability to terminally differentiate. It is also interesting to note the marked accumulation of c-Kit-positive hematopoietic precursors in the spleen of the leukemic mice.
Figure 3.
Flow cytometric analysis of PB, BM, and spleen cells from leukemic hCG–PML/RARα transgenic and wild-type mice. The cells were analyzed with antibodies against the cell surface markers, c-Kit, Gr-1, and Mac-1, conjugated with fluorescein isothiocyanate (Mac-1) or R-phycoerythrin (others) (solid lines), and with control antibodies of the same isotype (dotted lines). Before Gr-1 and Mac-1 staining, cells were incubated with CD16/32 antibody to block non-antigen-specific binding of antibodies to the murine FcγII/III receptors. Values given in upper right corner of each histogram indicate the positive population for each antibody.
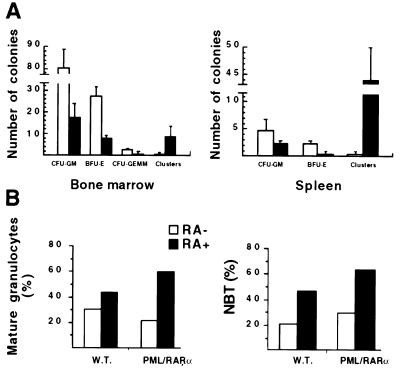
In vitro cultures of BM and spleen cells from leukemic mice and control littermates were also performed using standard conditions (see Materials and Methods and Fig. 4A). CFU-E were scored on day 2, and BFU-E, CFU-GM, and CFU-GEMM were scored on day 10. The clonogenic capacity of hematopoietic precursors from the transgenic mice was markedly reduced, particularly the number of BFU-E, in agreement with the almost complete replacement of the normal hematopoietic precursors by the neoplastic population in the BM and spleen of the leukemic transgenic mice (Fig. 4A). On the contrary, the number of clusters (small colonies, containing 8–20 cells), characteristically observed in BM cultures from acute myeloid leukemia patients (37), is clearly increased in cultures from the BM, and dramatically increased in the cultures from the spleen of leukemic mice.
Figure 4.
(A) Colony formation assay on BM and spleen cells from hCG–PML/RARα leukemic mice (solid bars) and sex- and age-matched wild-type littermates (open bars). Cells (3 × 104) were plated per dish in 0.9% methylcellulose medium in triplicate. Colony numbers shown here are from one representative experiment (mean ± SD from the triplicate). CFU-GM includes CFU-G, -M, and -GM. Clusters are clones with <20 cells, often observed in assays performed on cells from acute leukemia and thought to represent colonies of leukemic origin (37). Note that colony formation from all lineages decreased and the number of clusters often observed in cultures from acute leukemias dramatically increased in BM and spleen cultures from leukemic transgenic mice. (B) In vitro response of BM cells to RA. BM cells from transgenic (PML/RARα) and wild-type (W.T.) mice were incubated with or without RA. Percentages of mature granulocytes and NBT-positive cells on day 5 are shown from two independent experiments. Note that both the percentage of mature granulocytes and the percentage of NBT-positive cells from leukemic transgenic mice are increased by 2-fold in the presence of RA.
Response to RA of Leukemic Cells from PML/RARα Transgenic Mice.
We studied the capacity of the blasts from the leukemic transgenic mice to differentiate upon exposure to RA at the same concentration (10−6 M) used to trigger the differentiation of human APL blasts (38). To this end, mononuclear cells were isolated from BM and spleen suspensions and cultured for a week with or without RA (see Materials and Methods). We evaluated the induction of differentiation with the help of the NBT test, and scored by morphological differential count for the percent of terminal maturation toward the granulocytic compartment. In control mice, RA increased the differentiation of the hematopoietic precursors to some extent, whereas it markedly increased the terminal differentiation (up to 60%) of the leukemic cells (Fig. 4B). These results demonstrate that, as with human APL, the leukemic blasts accumulating in the PML/RARα transgenic mice are sensitive to the differentiating action of RA and that the maturation block at the promyelocytic stage can be reverted by this agent.
DISCUSSION
In APL, RARα variably fuses to PML, PLZF, NPM, and NUMA (X proteins). In terms of structure, what the various RARα partners contribute to the chimeric products is dramatically different; and at the first glance, it appears that the only common feature among the fusion proteins is the presence of the B to F domains of RARα (2). However, in spite of their diversity, the various fusion molecules share one common feature, their ability to heterodimerize with X proteins, because the regions which can mediate X protein homodimerization and oligomerization are retained in the fusion moiety (13–15, 17, 18). Similarly, the RARα portion can heterodimerize with RXR (17, 18), and consequently, the various fusion products have the ability to interfere with both X and RARα/RXR pathways. Therefore, X/RARα proteins can dominantly interfere with the aforementioned activity of X proteins as well as with their and RARα’s possible involvement in modulating normal hemopoietic processes, with the cooperation of RARα/X and aberrant X products. The importance of the RARα/PML fusion product in APL leukemogenesis has been recently highlighted by the identification of APL cases with nonreciprocal translocations resulting in the expression of the RARα/PML fusion gene but lacking the PML/RARα fusion gene (39). We decided to test the above hypothesis in vivo by a direct genetic approach and on a comparative basis by generating transgenic mice in which the expression of all these various molecules is restricted to the hemopoietic/myeloid cellular compartment.
The development of a suitable vector to direct the expression of the transgene to the hemopoietic/myeloid compartment was dictated on one hand by the fact that our attempt to generate PML/RARα transgenic mice using ubiquitous promoters was unsuccessful because of embryonic lethality, and on the other hand, by our aim to target the transgene expression specifically to promyelocytes, to possibly elicit the promyelocytic block observed in APL. We therefore generated a cathepsin-G minigene expression vector from the human locus into which we cloned cDNAs from all the various APL-associated products as well as the Lac-Z reporter gene (Fig. 1A and data not shown).
Transgenic mice expressing the PML/RARα fusion gene develop a severe myeloproliferative disorder with a dramatic accumulation of immature myeloid precursors in the BM and lymphoid organs. After a long latency period (>12 months), approximately 10% of the transgenic mice from two independent lines develop an acute myeloid leukemia characterized by features that are distinctive of APL: (i) a dramatic accumulation of promyelocytes in the BM (>30%); (ii) the presence of Auer’s bodies within the cytoplasm of the promyelocytic blasts; and (iii) the response of the leukemic blast to the differentiating activity of RA (Figs. 2 and 4 and Table 1).
However, in this leukemia, the white cell counts in the PB (WBC) were higher than those in classical APL and reminiscent of the human “variant” APL (APL-M3v in the French–American–British classification; see Table 1 and ref. 1). In addition, both in the myeloproliferative and the leukemic phase the infiltrating cellular population partially retains the ability to terminally differentiate toward mature granulocytes (Table 1 and Figs. 2 and 3).
The observation that hCG–PML/RARα mice are not born with leukemias, and in fact present a long preleukemic incubation period, suggests two possible interpretations that are not mutually exclusive. (i) PML/RARα is necessary, but not sufficient, to cause full-blown APL in humans also; additional mutations are needed, which in humans may consist of the expression of RARα/PML and aberrant PML. (ii) The relative level of expression of the PML/RARα transgene with respect to the PML and RARα genes, and/or other genes, is not sufficient to confer to the PML/RARα fusion protein the ability to dominantly interfere with their functions. From this point of view, it is important to remember that the 15;17 translocation in humans reduces the normal PML and RARα genes to heterozygosity, whereas the hCG–PML/RARα transgenic mice still carry two normal copies of these genes.
The second interpretation is unlikely because the expression of the PML/RARα transgene is not negligible, as evaluated by Northern blot analysis in the leukemic phase, and high enough to dislocate PML from the NBs, an event observed in the blasts of APL patients and thought to be pathognomonic for the disease (Fig. 1 D and E).
On the other hand, PML/RARα seems sufficient to cause the myeloproliferative disorder and the block of maturation because this is observed in 100% of transgenic mice. The expansion of the promyelocytic cellular compartment would then favor the occurrence of additional transforming events (40).
The interbreeding of PML/RARα, RARα/PML, and aberrant PML transgenic mice will allow us to understand if all these molecules are concomitantly necessary to trigger full-blown leukemia. In addition, to test if the lack of PML and RARα, and therefore the relative levels of expression of genes and fusion genes, potentiates the leukemogenic activity of the APL-specific fusion molecules, we have interbred the PML/RARα transgenic mice with PML and RARα knockout mice.
Transgenic mice for the PML/RARα fusion gene under the control of the myeloid CD11b promoter did not develop leukemia over the course of a more than 2-year follow-up period and total WBC and differential counts of myeloid subpopulations did not show appreciable differences between PML/RARα and control mice (41). Appropriate timing of PML/RARα expression in early myeloid progenitors certainly plays a crucial role with respect to its leukemogenic potential and its ability to interfere with normal hematopoiesis, thus possibly explaining the difference observed with our mice. In addition, retrovirally mediated expression of a mutated version of PML/RARα in the chicken has been shown to cause leukemias, but the cells exhibit features and membrane antigen patterns of very immature cells (42). The reasons why the chicken leukemic cells are different from the human APL cells are unknown, although the specific phenotype might be a result of the two point mutations present in the PML/RARα fusion protein that was used in those studies (42). On the contrary, in our transgenic mice the expression of the PML/RARα fusion protein in the myeloid/promyelocytic compartment resulted in profound disturbances in myeloid hematopoiesis and in myeloid leukemias characterized by a block of differentiation at the promyelocytic stage that defines the APL phenotype. Thus, our data conclusively demonstrate in vivo that PML/RARα plays a crucial role in APL pathogenesis.
The comparative analysis of PML/RARα, PLZF/RARα, NPM/RARα, and NUMA/RARα transgenic mice will tell us how the various fusion molecules contribute to the leukemic phenotype, and whether they cause a more severe or benign phenotype, also with respect to their response to RA treatment.
Acknowledgments
We thank P. Freemont for the PML antibody; L. Luzzatto for critical reviewing of the manuscript; N. Hawe and V. Petronis for help with the editing of the manuscript; D. Gandini, R. Notaro, and V. Rosti for help with some experiments and critical support; and Jia Hui Dong for assistance with the management of the mice. R.R. was partially supported by the Associazione Italiana per la Ricerca sul Cancro. G.C. is a Leukemia Society of America Special Fellow. This work was supported by the Sloan-Kettering Institute and the National Institutes of Health (P.P.P.).
ABBREVIATIONS
- APL
acute promyelocytic leukemia
- RA
retinoic acid
- RARα
RA receptor α
- PML
promyelocytic leukemia
- PLZF
promyelocytic leukemia zinc finger
- NPM
nucleophosmin
- NBs
nuclear bodies
- hCG
human cathepsin-G
- RT
reverse transcriptase
- WBC
white blood cell(s)
- PB
peripheral blood
- BM
bone marrow
- MPO
myeloperoxidase
- BFU-E
burst-forming units-erythroid
- CFU-E
-GM, and -GEMM, colony-forming units-erythroid, -granulocyte/macrophage, -granulocyte/erythroid/macrophage/megakaryocyte, respectively
- NBT
nitro blue tetrazolium
- RXR
retinoid-X-receptor
References
- 1.Bennett J M, Catovsky D, Daniel D, Flandrin G, Galton D A G, Gralnick H R, Sultan C. Br J Haematol. 1976;33:451–458. doi: 10.1111/j.1365-2141.1976.tb03563.x. [DOI] [PubMed] [Google Scholar]
- 2.Pandolfi P P. Haematologica. 1996;81:472–482. [PubMed] [Google Scholar]
- 3.Grignani F, Fagioli M, Alcalay M, Longo L, Pandolfi P P, Donti E, Biondi A, Lo Coco F, Pelicci P G. Blood. 1994;83:10–25. [PubMed] [Google Scholar]
- 4.Warrell R P, Jr, de Thé H, Wang Z Y, Degos L. N Engl J Med. 1993;329:177–189. doi: 10.1056/NEJM199307153290307. [DOI] [PubMed] [Google Scholar]
- 5.Longo L, Pandolfi P P, Biondi A, Rambaldi A, Mencarelli A, Lo Coco F, Diverio D, Pegoraro L, Avanzi G, Tabilio A, Zangrilli D, Alcalay M, Donti E, Grignani F, Pelicci P G. J Exp Med. 1990;172:1571–1575. doi: 10.1084/jem.172.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcalay M, Zangrilli D, Pandolfi P P, Longo L, Mencarelli A, Giacomucci A, Rocchi M, Biondi A, Rambaldi A, Lo Coco F, Diverio D, Donti E, Grignani F, Pelicci P G. Proc Natl Acad Sci USA. 1991;88:1977–1981. doi: 10.1073/pnas.88.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrow J, Goddard A D, Sheer D, Solomon E. Science. 1990;249:1577–1580. doi: 10.1126/science.2218500. [DOI] [PubMed] [Google Scholar]
- 8.de Thé H, Chomienne C, Lanotte M, Degos L, Dejean A. Nature (London) 1990;347:558–561. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- 9.Pandolfi P P, Grignani F, Alcalay M, Mencarelli A, Biondi A, Lo Coco F, Pelicci P G. Oncogene. 1991;6:1285–1292. [PubMed] [Google Scholar]
- 10.Kakizuka A, Miller W H, Jr, Umesono K, Warrell R P, Jr, Frankel S R, Murty V V V S, Dmitrovsky E, Evans R M. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 11.de Thé H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 12.Pandolfi P P, Alcalay M, Fagioli M, Zangrilli D, Mencarelli A, Diverio D, Biondi A, Lo Coco F, Rambaldi A, Grignani F, Rochette-Egly C, Gaub M-P, Chambon P, Pelicci P G. EMBO J. 1992;11:1397–1407. doi: 10.1002/j.1460-2075.1992.tb05185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z, Brand N J, Chen A, Chen S-J, Tong J-H, Wang Z-Y, Waxman S, Zelent A. EMBO J. 1995;12:1161–1167. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Redner R L, Rush E A, Faas S, Rudert W A, Corey S J. Blood. 1996;3:882–886. [PubMed] [Google Scholar]
- 15.Wells R A, Kamel-Reid S. Blood. 1996;88:365. (abstr.). [Google Scholar]
- 16.Alcalay M, Zangrilli D, Fagioli M, Pandolfi P P, Mencarelli A, Lo Coco F, Biondi A, Pelicci P G. Proc Natl Acad Sci USA. 1992;89:4840–4844. doi: 10.1073/pnas.89.11.4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kastner P, Perez A, Lutz Y, Rochette-Egly C, Gaub M P, Durand B, Lanotte M, Berger R, Chambon P. EMBO J. 1992;11:629–642. doi: 10.1002/j.1460-2075.1992.tb05095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez A, Kastner P, Sethi S, Lutz Y, Reibel C, Chambon P. EMBO J. 1993;12:3171–3182. doi: 10.1002/j.1460-2075.1993.tb05986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyck J, Maul G G, Miller W H, Jr, Chen J D, Kakizuka A, Evans R M. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 20.Koken M H M, Puvion-Dutilleul F, Guillemin M C, Viron A, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, Degos L, Puvion E, de Thé H. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 22.Mu Z-M, Chin K-V, Liu J-H, Lozano G, Chang K-S. Mol Cell Biol. 1994;14:6858–6867. doi: 10.1128/mcb.14.10.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koken M H M, Linares-Cruz G, Quignon F, Viron A, Chelbi-Alix M K, Sobczak-Thepot J, Juhlin L, Degos L, Calvo F, de Thé H. Oncogene. 1995;10:1315–1324. [PubMed] [Google Scholar]
- 24.Collins S J, Robertson K, Mueller L. Mol Cell Biol. 1990;10:2154–2163. doi: 10.1128/mcb.10.5.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robertson K A, Emami B, Collins S J. Blood. 1992;80:1885–1889. [PubMed] [Google Scholar]
- 26.Tsai S, Bartelmez S, Heyman R, Damm K, Evans R M, Collins S J. Genes Dev. 1992;6:2258–2269. doi: 10.1101/gad.6.12a.2258. [DOI] [PubMed] [Google Scholar]
- 27.Grignani F, Ferrucci P F, Testa U, Talamo G, Fagioli M, Alcalay M, Mencarelli A, Peschle C, Nicoletti I, Pelicci P G. Cell. 1993;74:423–431. doi: 10.1016/0092-8674(93)80044-f. [DOI] [PubMed] [Google Scholar]
- 28.Grignani F, Testa U, Fagioli M, Barberi T, Masciulli R, Mariani G, Peschle C, Pelicci P G. Cancer Res. 1995;55:440–443. [PubMed] [Google Scholar]
- 29.Rousselot P, Hardas B, Patel A, Guidez F, Gäken J, Castaigne S, Dejean A, de Thé H, Degos L, Farzaneh F, Chomienne C. Oncogene. 1994;9:545–551. [PubMed] [Google Scholar]
- 30.Hohn P A, Popescu N C, Hanson R D, Salvesen G, Ley T J. J Biol Chem. 1989;264:13412–13419. [PubMed] [Google Scholar]
- 31.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 226–250. [Google Scholar]
- 32.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 34.Seale J R C, Varma S, Swirsky D, Pandolfi P P, Goldman J M, Cross N C P. Br J Haematol. 1996;95:95–101. doi: 10.1046/j.1365-2141.1996.d01-1881.x. [DOI] [PubMed] [Google Scholar]
- 35.Cattoretti G, Chang C-C, Cechova K, Zhang J, Ye B H, Falini B, Louie D C, Offit K, Chaganti R S K, Dalla-Favera R. Blood. 1995;86:45–53. [PubMed] [Google Scholar]
- 36.Grisolano J L, Sclar G M, Ley T J. Proc Natl Acad Sci USA. 1994;91:8989–8993. doi: 10.1073/pnas.91.19.8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore M A, Spitzer G, Williams N, Metcalf D, Buckley J. Blood. 1974;44:1–18. [PubMed] [Google Scholar]
- 38.Castaigne S, Chomienne C, Daniel M T, Berger R, Fenaux P, Degos L. Blood. 1990;76:1704–1709. [PubMed] [Google Scholar]
- 39.Lafage-Pochitaloff M, Alcalay M, Brunnel V, Longo L, Sainty D, Simonetti J, Birg F, Pelicci P G. Blood. 1995;85:1169–1174. [PubMed] [Google Scholar]
- 40.Luzzatto L, Pandolfi P P. Br Med J. 1993;307:579–580. doi: 10.1136/bmj.307.6904.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Early E, Moore M A S, Kakizuka A, Nason-Burchenal K, Martin P, Evans R M, Dmitrovsky E. Proc Natl Acad Sci USA. 1996;93:7900–7904. doi: 10.1073/pnas.93.15.7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altabef M, Garcia M, Lavau C, Bae S-C, Dejean A, Samarut J. EMBO J. 1996;15:2707–2716. [PMC free article] [PubMed] [Google Scholar]