Abstract
Pancreatic cancer is the fourth leading cause of cancer death in the United States, with 4% survival 5 years after diagnosis. Biomarkers are desperately needed to improve earlier, more curable cancer diagnosis and to develop new effective therapeutic targets. The development of quantitative proteomics technologies in recent years offers great promise for understanding the complex molecular events of tumorigenesis at the protein level, and has stimulated great interest in applying the technology for pancreatic cancer studies. Proteomic studies of pancreatic tissues, juice, serum/plasma, and cell lines have recently attempted to identify differentially expressed proteins in pancreatic cancer to dissect the abnormal signaling pathways underlying oncogenesis, and to detect new biomarkers. It can be expected that the continuing evolution of proteomics technology with better resolution and sensitivity will greatly enhance our capability in combating pancreatic cancer.
Keywords: pancreatic cancer, proteomics, biomarker, mass spectrometry, ICAT
Introduction
Pancreatic cancer is a highly lethal disease that has seen little progress in diagnosis and treatment for many years. One of the major obstacles for improving the outcome in this highly aggressive tumor arises from the difficulty in diagnosing the disease at an early stage, a goal that could markedly improve the survival rate[1]. Unfortunately, the current methods for diagnosing pancreatic cancer are ineffective and/or impractical for identifying smaller, potentially curable lesions in the general population. Therefore, the use of biomarkers for the early detection of pancreatic cancer would be of invaluable clinical benefit. Because of the relative low prevalence of pancreatic cancer in the general population, a biomarker test would have to have extremely high specificity, in order to be feasible and cost-effective.
Many genetic alterations in pancreatic cancer have been discovered [2;3]. For example, Kras point mutations occur in over 90% of pancreatic cancers[2]. Tumor suppressor genes, such as p53, p16, and DPC4 are frequently inactivated[3], while epidermal growth factor receptor HER-2/neu becomes activated [4–6]. More recently, a palladin mutation was linked with certain familial pancreatic cancer[7–9]. However, the significant progress in the identification and characterization of cancer-related gene abnormalities has not translated into useable biomarkers for the general population (or even moderately elevated risk population). The emerging technology of quantitative proteomics has stimulated great interest to apply the technique to investigate the proteome of diseased samples, with the goals of identifying biomarkers and revealing the pathogenesis of disease mechanisms. While different approaches of disease-associated protein discovery are now available, the typical scheme starts with the comparison of the proteome of diseased and normal samples in a global scale to identify differentially expressed proteins. Such a comparative approach for protein profiling greatly facilitates the identification of dysregulated proteins associated with a specific biological condition. Further more, the quantitative measurement or comparison of a protein that is mechanistically informative for a disease may be essential in revealing the role of the protein in the pathogenesis of the disease. For example, if protein X is discovered in cancer and in normal tissue—the significance of the protein for the disease state (cancer) can be inferred if the protein is significantly down or up-regulated compared to the normal tissue. As proof of this principle, recent investigations using mass spectrometry-based quantitative proteomics for large scale protein profiling of tumors, tumor cells, or bodily fluids in comparison of pancreatic cancer patients and normal controls have identified many important biomarker candidates, and provide new hypotheses to further elucidate molecular pathways of cancer[10;11]. With this report, we hope to provide an update on the recent progress in proteomic studies of pancreatic cancer.
Overview of current proteomics technology
Quantitative proteomics aims to systematically and quantitatively compare two or more proteomes to assess static- or perturbation-induced changes in protein profiles. At present, no single technique in a one-step operation can provide the identification and quantification of all proteins in a complex system, such as body fluids, cell lysate or tissue extracts. To reach that ambitious goal, concerted approaches, including sample preparation, protein or peptide separation, mass spectrometric analysis, as well as bioinformatic tools for database search and quantification, are typically integrated for such applications, and need to be further improved. In general, most of the quantitative proteomics methods can be categorized into two different approaches: the 2-dimensional electrophoresis (2DE) based approach and the mass spectrometry (MS) based approach.
In the 2DE approach, the proteins from a sample are separated based on their isoelectric point and molecular weight. The in-gel separated proteins can be excised and in-gel digested for protein identification using either peptide mass-mapping by matrix-assisted laser desorption/ionization (MALDI) time-of-flight (TOF) mass spectrometer, or peptide sequencing by electrospray ionization (ESI) or MALDI tandem mass spectrometers. The quantification of the stained protein spots is achieved assessing the staining intensity, and the 2DE patterns can be compared to contrast the characteristic spots or patterns between different samples using pattern-matching algorithms. A quantitative comparison of multiple samples in a single gel under similar experimental condition can also be accomplished using more sophisticated fluorescence difference gel electrophoresis (DIGE) technique[12;13]. While 2DE is one of the most versatile technologies for protein separation, the major disadvantage for 2DE is the limited dynamic range. The recent technical advancements in this field, including higher resolving gels[14;15] and more sensitive staining methods[13;16] have aimed to improve its separation efficiency and alleviated other inherent limitations.
Currently, the most widely used MS based approach for quantitative proteomics is based on the bottom-up or “shot-gun” proteomics strategy[14], in which the identification and quantification of proteins relies on the sequential and quantitative analysis of corresponding peptides. The protein mixtures from a sample are digested, separated by one or multiple dimensional liquid chromatography and introduced into tandem mass spectrometer for automated analysis. In general, such an approach can be divided into four components, each emphasizing a different technical aspect: separation, mass spectrometry, peptide/protein identification and quantification.
First, most of the clinical samples, such as tissue and body fluids, are highly complex with substantial dynamic range in protein concentration. One-dimensional reversed phase liquid chromatography (RP LC), which is typically used prior to MS analysis, does not provide adequate separation power to resolve the complex protein tryptic digest, and consequently, many of low abundance peptides can be missed. The most common approach at the present time is the two-dimensional chromatographic separation, combining strong cation exchange (SCX) and RP LC, which significantly enhances the resolution power and peak capacity for MS analysis. In addition, a variety of purification or enrichment techniques, including organic fractionation, size exclusion, chemical extraction, affinity binding and immunochemical precipitation, have been developed to enrich a sub-proteome or sub-group of proteins with specific properties or posttranslational modifications (PTMs) prior to proteomic analytical flow.
By function, mass spectrometers consist of three major modules: an ion source, a mass analyzer and an ion detector. Based on the ionization method, ESI[17] and MALDI[18;19] are the most commonly used soft-ionization techniques to volatilize and ionize peptides and proteins. In ESI, analytes in an acidic solution are spayed directly into the inlet of a mass spectrometer by generating an electrically generated fine mist, producing ions with multiple charges. In MALDI, analytes are entrapped in an UV-absorbing compound, and dissociated and ionized by laser in a vacuum environment, generating predominately single charge ions. A variety of mass analyzers or combination of mass analyzers can be coupled with either ESI or MALDI ion sources. The most widely used mass analyzers for proteomics application include ion trap (three-dimensional trap, linear trap, orbitrap), quadrupole, time-of-flight (TOF) and Fourier transform ion cyclotron (FT-MS), based on the mechanism to control the ion motion. FT-MS is basically an ion-trap instrument equipped with high magnetic field to achieve high mass accuracy. More details in discussing mass spectrometers for proteomics applications can be found in many reviews[14;20].
The most common proteomics approach for peptide and protein identification is based on peptide sequencing using MS/MS spectra. The MS/MS spectra are generated by collision induced dissociation (CID) or post-source decay (PSD) and searched against the theoretical spectra in a database using search algorithm, such as SEQUEST[21] or MASCOT[22]. More recently, electron transfer dissociation (ETD), an emerging fragmentation method, was introduced to more effectively sequence peptides with PTMs[23]. The matching of empirical data and theoretical spectra leads to the identification of peptides. The assignment of peptide sequences and consequently proteins are typically validated with statistically based approaches, such as PeptideProphet[24] and ProteinProphet[25], or a decoy database search[26].
Lastly, quantification is one of the most important aspects of quantitative proteomics, relevant to many clinical related applications. The most widely used and versatile approaches utilizes stable isotope labeling to introduce mass tags on proteins or peptides in different samples, allowing mass spectrometry to distinguish and quantitatively compare a peptide with identical sequence but from different origins. Such an approach allows peptides with different isotopic labels to be separated under the same chromatographic conditions, ionized, separated and analyzed under the same conditions in a mass spectrometer, thus, providing a reliable quantitative comparison. In general, two main methods are applied to introduce stable isotope labeling: chemical reaction and biological incorporation. The chemical reaction method is a post-isolation method providing great flexibility for quantitative proteomics analysis. The methods include the widely used isotope coded affinity tag (ICAT)[27], isotope tags for relative and absolute quantification (iTRAQ)[28] and global internal standard technology (GIST)[29;30]. The biological incorporation approaches include metabolic stable isotope labeling[31–35], such as stable isotope labeling by amino acids in cell culture (SILAC)[31–34], and stable isotope incorporation via enzyme reaction[36;37]. In recent years, targeted quantitative proteomics has been introduced, which utilizes synthetic reference peptides with stable isotope labeling as signature markers for targeted identification and quantification, providing a complementary approach for protein and peptide validation in a complex system[38;39]. Other quantitative / semi-quantitative proteomics approaches including label free approach[40;41] and SELDI (surface-enhanced laser desorption ionization) [42–45] have also been reported.
Proteomics study of pancreatic tissue
The typical sample types available for proteomics studies in pancreatic cancer include pancreatic tissue, pancreatic juice, serum or plasma, and pancreatic cell lines. Global profiling of tumor tissue has been used to successfully identify novel tumor-associated biomarkers in a number of cancers[11;46–48]. Study of the tumor proteome can lead to understanding of abnormally regulated signal pathways underlying tumorigenesis, providing new targets for cancer diagnosis and intervention. Moreover, biomarkers identified through proteomic profiling of tumor tissue can provide candidates for further biomarker development in serum or plasma.
2DE study of pancreatic cancer tissue
A direct comparison of tissues from cancer and normal control represents one common proteomics approach to discover differentially expressed proteins in tumor. 2DE was used to compare 12 pancreatic cancer samples to the matching adjacent unaffected pancreatic tissues, and successfully identified 70 over-expressed proteins in cancer [49]. Two over-expressed proteins identified by the 2DE comparison were selected for validation by immunohistochemistry (IHC) analysis. Fascin was observed to be positively stained in 13 out of 21 cancer samples by IHC. More interestingly, the study found that the expression of fascin positively co-related with the degree of differentiation of pancreatic cancer. Another over-expressed protein identified by 2DE, cathepsin D, also showed increased staining in the cancerous tissue in 12 of the 21 cases tested using IHC analysis. In a different 2-DE study of pancreatic cancer[50], the authors identified 40 differentially expressed proteins, five of them (including α-amylase; copper zinc superoxide dismutase; protein disulfide isomerase, pancreatic; tropomyosin 2; and galectin-1) had previously been associated with pancreatic disease in gene expression studies, indicating that the over-expression of these five genes at the RNA level can be reflected into protein levels. Although similar in the experimental approach, the two 2DE studies did not identify common proteins with the exception of cathepsin D, suggesting the biologically variability and experimental design may both play important roles in the outcome of the investigations.
2DE study of microdissected pancreatitc cancer cells
Pancreatic ductal adenocarcinoma is characterized by a strong stromal presence, with 30–90% of tumor cells surrounded and interspersed by the fibroblastic stroma. There is therefore a good reason to isolate cancer cells or normal epithelial cells from the stroma for expression analysis. The use of enriched cancer cells may facilitate the discovery of very low abundant proteins derived from the cancer cells. Different approaches have recently been applied to obtain enriched cancer cells and epithelial cells, including Dynal bead based epithelial enrichment, short-term cultures of pancreatic ductal cells, and laser capture microdissection (LCM)[11]. LCM is usually the method of choice and has been shown to be effective in microdissection of epithelial cells from cancers, providing enriched populations of target cells. One limitation is the relatively low number of cells that can be obtained from the capture. However, in a study using LCM to enrich for both normal and malignant pancreatic ductal epithelial cells, investigators have managed to obtain sufficient material for 2-DE analysis[51]. About 800 spots were successfully detected with as little as approximately 50,000 microdissected cells. It is not surprising that the study found that the protein profiles from unmicrodissected normal pancreas and LCM isolated nonmalignant ductal cells were different. Comparison of protein profiles from nonmalignant and malignant ductal cells revealed nine protein spots that were consistently differentially regulated. One of these spots was identified as S100A6, and this protein was further showed to be over-expressed in moderately or poorly differentiated pancreatic cancers by IHC analysis. LCM, while labor intensive, may provide a solution for qualitative and quantitative proteomic analysis of enriched cancer cells.
2D DIGE study of pancreatic cancer tissue
One challenge in quantitative studies employing 2DE is the technical variations between different gels, which require steps such as replicates and normalization. To overcome this, 2D DIGE was introduced, enabling analysis of multiple samples in one gel[12;13]. This was accomplished by the labeling of the proteins with structurally similar, but spectrally different, fluorophores. In the improved version of 2D DIGE with saturation labeling dyes, the sensitivity has been improved, facilitating simultaneous analysis of multiple samples with limited amount of proteins[52]. When 2D DIGE was applied to the study of pancreatic cancer, proteins extracted from about 1000 LCM microdissected cells resulted in a high resolution of up to 2500 protein spots[53]. Using protein lysates from the bulk pancreatic cancer tissue as a reference proteome, the authors were able to demonstrate that 92% of the spots of the microdissected sample map could be matched to bulk cancer tissue proteome. Eight differentially expressed proteins were identified in PanIN lesions (pancreatic intra-epithelial neoplasias), the precursor lesions of pancreatic cancer. Interestingly, among the differentially expressed proteins identified, in addition to actin itself, three of them were actin filament-associated proteins, including transgelin, vimentin and MRCL3, suggesting a relevant role of the actin cytoskeleton during pancreatic tumor progression.
ICAT MS/MS based quantitative proteomics study of pancreatic cancer tissue
With the development of mass spectrometry and stable isotope tagging methods, quantitative proteomics can now be effectively applied to study clinical samples, despite the fact that complete proteome analysis of complex samples is still challenging. Among the various stable isotope tagging methods, the ICAT method was the first introduced and widely used approach for quantitative proteomic analysis[27]. This methodology has demonstrated a significant improvement over gel-based methods in identifying low abundance proteins[54]. The approach of ICAT labeling coupled with LC MS/MS was employed to perform quantitative protein profiling of pancreatic cancer tissues compared to normal pancreas [46]. Identification and quantification of the proteins was accomplished by differentially labeling the proteins in cancer with heavy ICAT reagents and the normal comparator proteins with light ICAT reagents. The isotopically labeled proteins were then combined, purified and followed by liquid chromatography (LC) tandem mass spectrometry (MS/MS) analysis. The study identified 151 differential regulated proteins in cancer samples, most of which were involved in signal transduction, cell growth and/or maintenance, metabolism, and cellular physiological processes[11;46]. Validation of the discovered proteins from ICAT analysis, was performed using Western blotting, IHC and tissue microarray analysis. This study identified a number of unique proteins that have not been associated with pancreatic cancer or other cancers before, providing potential new targets and biological hypotheses for future development of biomarkers for early diagnosis and therapy. Furthermore, the study revealed a group of proteins that were involved in orchestrating a complex relationship between the tumor cells, the extracellular matrix and the immune system. These proteins include the upregulation of: a) proteins that destroy the extracellular matrix (annexin A2, cathepsin B and matrix metalloproteinase inducer); b) proteins that promote invasion and metastases (versican and lumican); c) angiogenesis (periostin and endothelial actin-binding protein); d) cancer cell migration (integrin β1, thrombospondin-1, fibronectin, and migration stimulation factor FN70); and e) proteins that allow tumor cell evasion (neutrophil defensin and annexin A1)[46]. The data demonstrated the important interaction and cooperation between ductal epithelium (cancer) and the extracellular matrix. These newly discovered proteins provide valuable insights in dissecting the process of pancreatic tumorigenesis and towards devising strategies for new interventions.
There is currently a vast amount of information on the differential gene expression of pancreatic cancer at the mRNA level, including RNA expression arrays, DNA microarrays, differential display experiments, and SAGE analysis of pancreatic cancer[55]. Compared to the gene expression profiling data, about 80% of the differentially regulated proteins reported in the proteomics study described above[46] have not been found to be differentially expressed by previous mRNA studies. It is well known that the dysregulation at gene level does not always reflect at protein level. The results reflected the complementary nature of proteomics versus genomics approaches.
Proteomics study of pancreatic juice
Pancreatic juice is rich in proteins that are secreted directly from the pancreatic duct, where pancreatic cancers arise. Cancer cells are preferentially shed into the ductal lumen, making juice a rich source of cancer-specific proteins. While pancreatic juice may not be as easily accessible as serum, it is a more specific source for searching biologically significant proteins associated with pancreatic cancer due to the proximity to the tumor.
1DE and 2DE study of the pancreatic juice proteome
Pancreatic juice was extensively studied in late 1970s and 1980s, primarily by early 2-DE analyses, which led to the discovery and description of several pancreatic enzymes [56]’[57–60]. With recent progress in 2DE and mass spectrometry, several groups have reported proteomic studies on pancreatic juice. A recent investigation employed 1DE and LC MS/MS to comprehensively categorize the pancreatic juice proteome [61]. Pancreatic juice obtained from pancreatic cancer patients was first separated in 1DE, cut into several gel slices and subsequently analyzed by LC MS/MS for protein identification. A total of 170 unique proteins were identified, including several proteins that were previously found to be over-expressed in pancreatic cancer or known pancreatic cancer tumor markers, including carcinoembryonic antigen-related cell adhesion molecules, mucin 1, hepatocarcinoma-intestine-pancreas/pancreatitis-associated protein and lipocalin 2. The study represented a comprehensive study of pancreatic juice proteome. The approach was aimed at identifying the protein constituents of pancreatic juice, and therefore, was non-quantitative.
ICAT MS/MS based quantitative proteomics study of pancreatic juice
In an effort to search for biomarkers associated with pancreatic cancer the ICAT-based method was applied to analyze the proteome of pancreatic juice and to quantify proteins that are differentially expressed in pancreatic cancer compared to normal controls[62]. Proteins from pancreatic juice samples from cancer patients and from normal controls were labeled with isotopically heavy and light ICAT reagents, respectively, for quantitative analysis. To reduce sample complexity and enhance protein identification and quantification, two-dimensional LC separation, including SCX and RP LC, was applied for fractionation of the samples before mass spectrometry analysis. To that end, 105 proteins were identified and quantified in the pancreatic juice, of which 30 proteins showed abundance changes of at least two-fold in pancreatic cancer juice compared to normal controls. Many of these proteins have been externally validated to be associated with pancreatic cancer, providing additional candidates for diagnostic biomarkers. As proof of principle, that the identified cancer specific proteins could be used as a foundation for biomarker development, one of the over-expressed proteins in cancer pancreatic juice identified by ICAT analysis, insulin-like growth factor binding protein 2 (IGFBP-2), was further validated and showed to be consistently over-expressed in pancreatic cancer tissue by western blotting[62]. The study demonstrated a comprehensive strategy for quantitative protein profiling of pancreatic juice and its potential for future biomarker development.
Pancreatitis is an inflammatory condition of the pancreas. However, it often shares many molecular features with pancreatic cancer. Biomarker candidates present in pancreatic cancer frequently occur in the setting of pancreatitis as well. The efforts to develop diagnostic biomarkers for pancreatic cancer have thus been complicated by the false positive involvement of pancreatitis. Moreover, chronic pancreatitis is a risk factor for eventual neoplastic progression. Therefore, understanding the proteins involved in both diseases may yield valuable insights into the mechanisms that link these events, while helping to minimizing false positive biomarker candidates. In a comparative proteomics study of pancreatic juices from patients with chronic pancreatitis and controls[63], nine proteins (hemoglobin, fibrinogen, trypsin i, trypsin ii, chymotrypsinogen b, Ig alpha-1 chain c region, Ig mu chain c region, ribonuclease, and human serum albumin) that were previously shown to be up-regulated in pancreatic cancer juice, were also differentially expressed in pancreatitis juice. Furthermore, among the 27 differentially expressed proteins identified in pancreatitis juice, several proteins, including plasminogen, neural cell adhesion molecule L1, and caldecrin, have not been previously associated with pancreatitis, nor identified in pancreatic cancer juice; thus they may serve as novel biomarker candidates for chronic pancreatitis[63].
Proteomics study of serum/plasma
Plasma/serum represents an ideal diagnostic specimen for clinical tests due to its easy and inexpensive accessibility. Unfortunately, it is technically difficult to study low abundance proteins in blood because of its enormous complexity and depth of proteome. Extensive separation at both protein and peptide level using electrophoresis and multidimensional LC is one way to enhance the identification of low abundant proteins in serum/plasma. Several alternative approaches have also been developed to reduce the abundant proteins or the complexity of the sample. Immunoaffinity methods are now available to deplete albumin and other high abundance proteins from serum, allowing for the detection of lower abundance proteins[64]. While depletion of high abundant proteins can increase the detection of lower abundance proteins, the major concern for this approach is the potential variable and selective losses of other proteins along with the immunoglobulins and albumin. A different approach to reduce the complexity of serum/plasma proteins focuses on the in-depth analysis of biological sub-proteomes to minimize the repeated analyses of abundantly expressed proteins. For example, isolation of glycoproteins or glycopeptides, and enrichment of phosphoproteins from serum/plasma for comparative analysis may reduce the interference of albumin and other highly abundant proteins and allow a more specific search of biomarker candidates that have undergone post-translation modification.
2DE and 2D DIGE study of serum
In a large-scale 2DE proteomic analysis of serum samples, 62 serum samples obtained from 32 patients with pancreatic cancer and 30 healthy volunteers were used to create protein profiles[65]. The study identified 154 differentially expressed protein spots which could reliably discriminate pancreatic cancer serum samples from normal using a PCA model. The study found that nine selected protein spots were sufficient to discriminate 100% pancreatic cancer samples and 94% of normal controls using cross-validation. One of the 9 spot was further identified as fibrinogen gamma, and was over-expressed in 67% of pancreatic cancers by IHC analysis. The finding of over-expression of fibrinogen in the sera of pancreatic cancer patients suggests a link between pancreatic cancer and the coagulation cascade.
In another 2DE based study[66], the highly abundant proteins including albumin, immunoglobulins, transferrin, haptoglobin, and antitrypsin were first depleted by immunoaffinity HPLC columns. The depleted serum was then subjected to 2D DIGE analysis. The study identified 24 unique proteins that were increased and 17 unique proteins that were decreased in cancer serum samples. Some of the proteins were validated in an independent series of serum samples from 20 patients with pancreatic cancer and 14 controls. The increased levels of apolipoprotein E, α-1-antichymotrypsin, and inter-α-trypsin inhibitor were found to be associated with pancreatic cancer.
Glycoprotein enrichment of serum proteins
Malignant transformation is associated with abnormal glycosylation, leading to the accumulation of tumor-specific glycoproteins actively involved in tumor progression and metastasis[67]. This could explain why many of the tumor markers are glycoproteins. Comparative studies of glycoproteins from cancer cells and corresponding normal cells can lead to identification of biomarkers that are useful for diagnosis, prognosis, and therapy.
Mass spectrometry is a powerful tool to evaluate glycosylation sites and elucidate oligosaccharide structures[68;69]. A method for comprehensive analysis of N-glycans and glycosylation sites in human pancreatic cancer serum was recently reported[70]. N-glycopeptides were first captured by double lectin column, and enzymatically cleaved to separate the glycan and peptides for analysis. The observation of a 1 Da shift on asparagines, as well as the consensus sequence for N-linked glycosylation was then used for identification of the N-glycosylation sites. A capillary hydrophilic interaction column was applied to fractionate the un-derivatized oligosaccharide mixtures before online detection by ESI-TOF MS. Using this approach, 45 oligosaccharides were found altered in pancreatic cancer serum, of which 44 were distinct in the cancer sample. Although demonstrated as a proof of principle, this approach may become a useful tool for finding protein glycosylation sites and oligosaccharides specific to cancer cells.
Comparative study of serum before and after resection of pancreatic cancer
A challenge in biomedical research is the biological variability that can obscure the true differences or changes induced by a diseased state. To minimize the biological variability, a study compared samples of plasma from the same patient at different time points during diagnosis and treatment of pancreatic cancer[71]. 2D DIGE was applied to quantitatively analyze plasma samples longitudinally collected along with the disease course of 10 individual pancreatic cancer patients to identify the plasma proteome profile patterns associated with clinical prognosis and response to surgical therapy. To this end, the authors found several protein spots consistently associated with tumor across ten patients studied[71]. Some of these protein spots were identified, including complement factors, hemopexin, and apolipoprotein A IV. The authors were able to further identify 14 proteins which were differentially expressed between patients who had no recurrence of disease versus those who exhibited recurrence or had died. Although the study unavoidably suffered with the technical difficulties associated with plasma analysis, it demonstrated an experimental model to develop therapeutic response biomarkers for pancreatic cancer-- such biomarkers, if effective, could potentially change the strategy for patient management[71].
Proteomics study of pancreatic cancer cell lines
Cancer cell lines are important model systems to study cancer that would not have been feasible with tumor/tissue/body fluid. One unique feature of studying in vitro cell models is the ability to perform perturbations/treatments on cells and investigate the effects, providing useful model systems for the investigation of cancer therapeutic response, including drug effectiveness, toxicity, drug resistance etc.
Study of drug response in cancer cell lines
Recent studies have been carried out in proteomics prospects to investigate the effects of drug treatments, combination therapy, the mechanisms of drug resistance and chemoresistance using pancreatic cancer cell lines as model systems. The most commonly used drug for chemotherapy of pancreatic cancer is gemcitabine, which is a nucleoside analogue that can prevent DNA repair and cause apoptotic cell death when incorporated into the cells. Other anti-cancer drugs can act on epigenetic events to suppress tumor growth by inhibiting DNA methylation (such as 5-aza-2’-deoxycytidine) or histone deacetylation (such as trichostatin A). One study used 2DE to investigate the protein profile in pancreatic cancer cell lines treated with trichostatin A and/or gemcitabine in single or combined treatment[72]. Among the differentially expressed proteins identified, most were involved in two major biological processes: apoptotic cell death and proliferation, consistent with the modes of drug action. Moreover, the study revealed that trichostatin A enhances the anti-tumoral effect of gemcitabine by promoting growth inhibition and apoptosis. Further pathway analysis suggested that the combined treatment is able to compensate for the p53 loss of function occurring in most of pancreatic cancers. These findings provide useful information for future therapeutic strategies.
Resistance to chemotherapy is usually a major cause of cancer treatment failure. The outcomes for many cancers could be improved by understanding the mechanisms of drug resistance, developing biomarkers capable of identifying resistant tumors, and better therapies for treating them. A 2DE study of proteomics profiles of chemo-resistant pancreatic cancer cell lines identified three novel proteins, including E-FABP, cofilin and 14-3-4-sigma. The study suggested that these proteins might be involved in the development of chemoresistance[73], providing new insights and hypothesis into the mechanism of developing chemoresistance.
Study of signaling pathway via siRNA silencing
RNAi (RNA interference) technology is a powerful tool to study gene silencing. Proteomics study of RNAi mediated gene silencing provides the opportunity to study the global changes of protein expression associated with the loss of a specific gene function. Smad4 is a tumor –suppressor gene that is lost or mutated in 50% of pancreatic cancer[74]. Its tumor suppressor function likely acts through TGF-β (transforming growth factor- β) induced growth inhibition. Analysis of the proteomic profile of the Smad4-knockdown pancreatic cancer cell line revealed 10 novel targets for TGF- β, suggesting a novel TGF- β signal pathway independent of Smad4[74]. The study demonstrated the combined and complementary approach of proteomics and RNAi for examining novel target molecules in unknown pathways.
Study of cancer cell lines secretome
Body fluid based biomarkers are often secreted proteins or proteins that may come from necrotic and apoptotic cells. Secreted proteins (the secretome) from cancer cells are therefore among the candidates that have the greatest potential for translation to fluidbased biomarkers for cancer detection. In a recent study, multidimensional protein identification technology (MudPIT) was applied to identify proteins released by pancreatic cancer cells in cultured systems[75]. Although the approach is nonquantitative in nature, the study revealed multiple proteins that have not been previously associated with pancreatic cancer. In addition to the identification of novel proteins released by pancreatic cancer cells, the study also suggested that pancreatic cancer cells secreted a series of proteoglycans, including versican, perlecan, syndecan 1 and 4, challenging the common view that fibroblasts of tumor stroma are the sole source of these molecules[75].
In a quantitative proteomics study investigating secreted proteins from a pancreatic cancer cell line, SILAC was used to identify differentially expressed proteins in the culture media of a pancreatic cancer cell line in comparison with a non-neoplastic pancreatic ductal cell line[76]. A total of 145 differentially secreted proteins (>1.5-fold change) were identified, several of which were previously reported as differentially regulated in pancreatic cancer, confirming the validity of the approach. When the protein expression data from SILAC were compared with mRNA expression data obtained using gene expression microarrays, a correlation coefficient of 0.28 was obtained, confirming previously reported poor associations between RNA and protein expression studies[76].
Summary
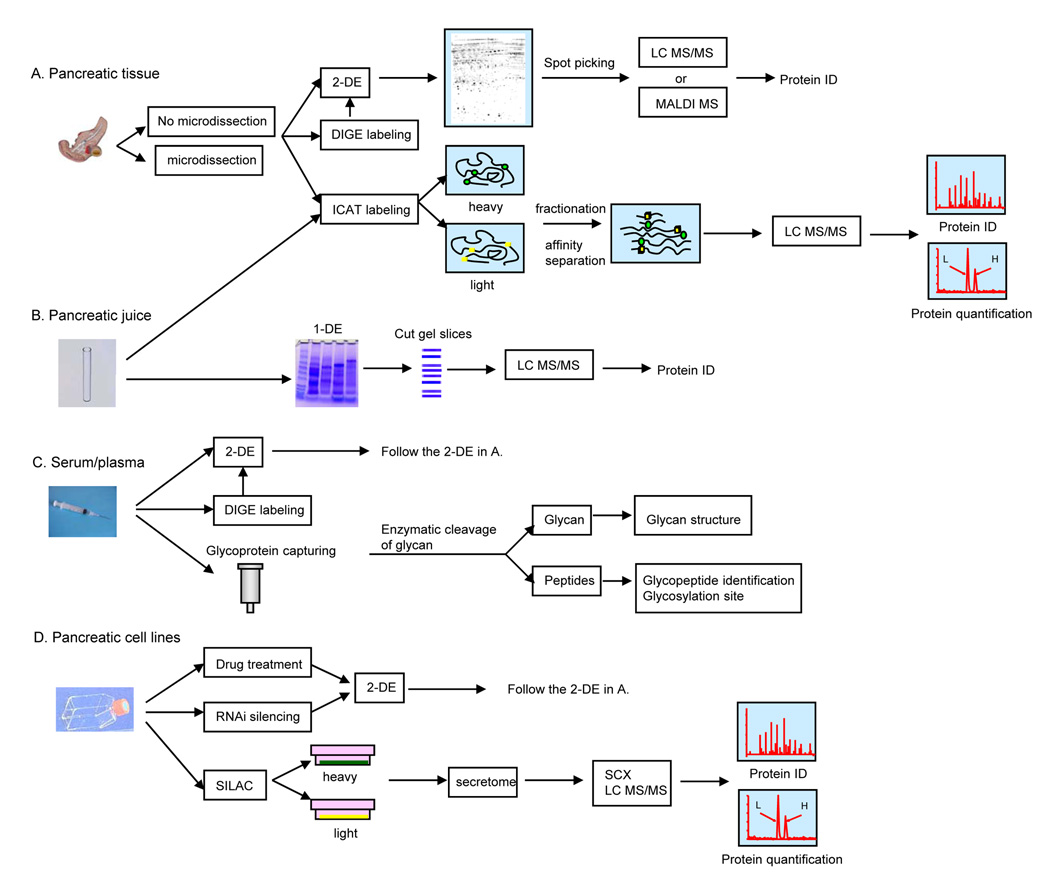
With the development of proteomic technologies and different working platforms, global measurement of the perturbed state induced by pancreatic cancer is now feasible at the protein level. Various sample types, including pancreatic tissue, pancreatic juice, serum/plasma and cancer cell lines have been studied in searching of candidate markers for early diagnosis, biomarkers of treatment response, and new targets for therapy (Figure 1), Due to their proximity to the tumor, pancreatic tissue and pancreatic juice represent the most direct and effective sources for proteomics investigation aimed at biomarker discovery or an increased mechanistic understanding of pancreatic cancer. In contrast, serum/plasma remains as a better material for developing clinical assays. Because proteins carry most of the biological activities, the identification of novel proteins associated with pancreatic cancer is expected to provide new hypotheses and help to reveal abnormal signaling pathways to understand the mechanisms of pancreatic oncogenesis.
Figure 1. Proteomic approaches in the studies of pancreatic cancer.
Different experimental approaches have been applied to study different sample types. (A) Pancreatic tissue; (B) Pancreatic juice; (C) Serum/plasma; (D) Pancreatic cell lines.
One of the major technical challenges for current proteomics studies is the validation of biomarker candidates across different proteomics platforms and laboratories. A main obstacle is to the large biological variation, such as gender, age, genetic factors, dietary, environmental, and drug treatment; and the fact that none of the current platform technologies can provide a non-biased and complete protein identification and quantification in a complex biological sample. As an evolving technology, many technical aspects, including sample handling, separation methods, mass spectrometry, search algorithms and databases used, can all impact the final outcomes of the experiments. While the results of many investigations have suffered from a lack of sensitivity and reproducibility of the methods used, the discoveries in the past few years have been encouraging and informative, and laid important groundwork for future studies. Many of the current limitations may be transient as the technologies in this field are quickly evolving. It can be anticipated that proteomics technologies with better resolution, sensitivity, quantification and reproducibility will greatly enhance our effort in revealing low abundant protein markers, and provide more consistent platforms for validating marker candidates.
As most of the current proteomics studies in pancreatic cancer research were designed and performed from a discovery perspective with global approaches, the targeted validation of identified marker candidates is becoming an important part for biomarker development for pancreatic cancer. Targeted quantitative proteomics aims to analyze specific proteins with biological significance; and not only provides technical advantages, such as absolute quantification, enhanced sensitivity and higher throughput, but also facilitates in-depth characterization of a protein, such as different isoforms and PTMs. The combination of global profiling with targeted validation techniques, such as tissue microarray, ELISA, protein array or MS based targeted proteomics, provides a more systematic approach and is expected to greatly facilitate the identification of specific candidate biomarkers for cancer diagnosis, prognosis and therapy.
Acknowledgements
This work has been supported by NCI NIH grants 1K07CA11629601A2, 1R01CA10720901A2 and funds from the Canary Foundation, Gene and Mary Ann Walters Pancreatic Cancer Foundation, the AACR-PanCAN Career Development Award for Pancreatic Cancer Research, and Federal funds from the National Heart, Lung and Blood Institute NIH, under contract # NOI-HV-28179.
Abbreviations
- ICAT
isotope coded affinity tag
- IHC
immunohistochemistry
- LCM
laser capture microdissection
- SCX
strong cation exchange
- ETD
electron transfer dissociation
Reference List
- 1.Brand R. Cancer J. 2001;7:287–297. [PubMed] [Google Scholar]
- 2.Hruban RH, van Mansfeld AD, Offerhaus GJ, van Weering DH, Allison DC, Goodman SN, Kensler TW, Bose KK, Cameron JL, Bos JL. Am.J.Pathol. 1993;143:545–554. [PMC free article] [PubMed] [Google Scholar]
- 3.Hruban RH, Offerhaus GJ, Kern SE, Goggins M, Wilentz RE, Yeo CJ. J.Hepatobiliary.Pancreat.Surg. 1998;5:383–391. doi: 10.1007/s005340050062. [DOI] [PubMed] [Google Scholar]
- 4.Day JD, Digiuseppe JA, Yeo C, Lai-Goldman M, Anderson SM, Goodman SN, Kern SE, Hruban RH. Hum.Pathol. 1996;27:119–124. doi: 10.1016/s0046-8177(96)90364-0. [DOI] [PubMed] [Google Scholar]
- 5.Hall PA, Hughes CM, Staddon SL, Richman PI, Gullick WJ, Lemoine NR. J.Pathol. 1990;161:195–200. doi: 10.1002/path.1711610305. [DOI] [PubMed] [Google Scholar]
- 6.Satoh K, Sasano H, Shimosegawa T, Koizumi M, Yamazaki T, Mochizuki F, Kobayashi N, Okano T, Toyota T, Sawai T. Cancer. 1993;72:51–56. doi: 10.1002/1097-0142(19930701)72:1<51::aid-cncr2820720112>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 7.Pogue-Geile KL, Chen R, Bronner MP, Crnogorac-Jurcevic T, Moyes KW, Dowen S, Otey CA, Crispin DA, George RD, Whitcomb DC, Brentnall TA. PLoS.Med. 2006;3:e516. doi: 10.1371/journal.pmed.0030516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slater E, Amrillaeva V, Fendrich V, Bartsch D, Earl J, Vitone LJ, Neoptolemos JP, Greenhalf W. PLoS.Med. 2007;4:e164. doi: 10.1371/journal.pmed.0040164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zogopoulous G, Rothenmund H, Eppel A, Ash C, Akbari MR, Hedley D, Narod SA, Gallinger S. Hum.Genet. 2007;121:635–637. doi: 10.1007/s00439-007-0361-z. [DOI] [PubMed] [Google Scholar]
- 10.Alaiya A, Al Mohanna M, Linder S. J.Proteome.Res. 2005;4:1213–1222. doi: 10.1021/pr050149f. [DOI] [PubMed] [Google Scholar]
- 11.Chen R, Pan S, Brentnall TA, Aebersold R. Mol.Cell Proteomics. 2005;4:523–533. doi: 10.1074/mcp.R500004-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Tonge R, Shaw J, Middleton B, Rowlinson R, Rayner S, Young J, Pognan F, Hawkins E, Currie I, Davison M. Proteomics. 2001;1:377–396. doi: 10.1002/1615-9861(200103)1:3<377::AID-PROT377>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 13.Unlu M, Morgan ME, Minden JS. Electrophoresis. 1997;18:2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- 14.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 15.Gauss C, Kalkum M, Lowe M, Lehrach H, Klose J. Electrophoresis. 1999;20:575–600. doi: 10.1002/(SICI)1522-2683(19990301)20:3<575::AID-ELPS575>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 16.Rabilloud T. Proteomics. 2002;2:3–10. [PubMed] [Google Scholar]
- 17.Whitehouse CM, Dreyer RN, Yamashita M, Fenn JB. Anal.Chem. 1985;57:675–679. doi: 10.1021/ac00280a023. [DOI] [PubMed] [Google Scholar]
- 18.Karas M, Hillenkamp F. Anal.Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka K, Waki H, Waki H, Akita S, Yoshida Y, Yoshida T. Rapid Commun.Mass Spectrom. 1988;2:151–153. [Google Scholar]
- 20.Aebersold R, Goodlett DR. Chem.Rev. 2001;101:269–295. doi: 10.1021/cr990076h. [DOI] [PubMed] [Google Scholar]
- 21.Eng J, McCormack AL, Yates JR. J.Am.Soc.Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 22.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc.Natl.Acad.Sci.U.S.A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Analytical Chemistry. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 25.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. Anal.Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 26.Elias JE, Gygi SP. Nat.Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 27.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Nat.Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 28.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Mol.Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Chakraborty A, Regnier FE. J.Chromatogr.A. 2002;949:173–184. doi: 10.1016/s0021-9673(02)00047-x. [DOI] [PubMed] [Google Scholar]
- 30.Regnier FE, Julka S. Proteomics. 2006 [Google Scholar]
- 31.Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M. Nat.Biotechnol. 2003;21:315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 32.Foster LJ, de Hoog CL, Mann M. Proc.Natl.Acad.Sci.U.S.A. 2003;100:5813–5818. doi: 10.1073/pnas.0631608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ibarrola N, Kalume DE, Gronborg M, Iwahori A, Pandey A. Anal.Chem. 2003;75:6043–6049. doi: 10.1021/ac034931f. [DOI] [PubMed] [Google Scholar]
- 34.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Mol.Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 35.Veenstra TD, Martinovic S, Anderson GA, Pasa-Tolic L, Smith RD. J.Am.Soc.Mass Spectrom. 2000;11:78–82. doi: 10.1016/S1044-0305(99)00120-8. [DOI] [PubMed] [Google Scholar]
- 36.Mirgorodskaya OA, Kozmin YP, Titov MI, Korner R, Sonksen CP, Roepstorff P. Rapid Commun.Mass Spectrom. 2000;14:1226–1232. doi: 10.1002/1097-0231(20000730)14:14<1226::AID-RCM14>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 37.Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Anal.Chem. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 38.Anderson L, Hunter CL. Mol.Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Pan S, Zhang H, Rush J, Eng J, Zhang N, Patterson D, Comb MJ, Aebersold RH. Mol.Cell.Proteomics. 2004;4:182–190. doi: 10.1074/mcp.M400161-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Fang R, Elias DA, Monroe ME, Shen Y, McIntosh M, Wang P, Goddard CD, Callister SJ, Moore RJ, Gorby YA, Adkins JN, Fredrickson JK, Lipton MS, Smith RD. Mol.Cell Proteomics. 2006;5:714–725. doi: 10.1074/mcp.M500301-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, Norton S, Kumar P, Anderle M, Becker CH. Anal.Chem. 2003;75:4818–4826. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- 42.Diamandis EP. Mol.Cell Proteomics. 2004;3:367–378. doi: 10.1074/mcp.R400007-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Diamandis EP. J.Natl.Cancer Inst. 2004;96:353–356. doi: 10.1093/jnci/djh056. [DOI] [PubMed] [Google Scholar]
- 44.Issaq HJ, Veenstra TD, Conrads TP, Felschow D. Biochem.Biophys.Res.Commun. 2002;292:587–592. doi: 10.1006/bbrc.2002.6678. [DOI] [PubMed] [Google Scholar]
- 45.Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Mills GB, Simone C, Fishman DA, Kohn EC, Liotta LA. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 46.Chen R, Yi EC, Donohoe D, Pan S, Eng J, Crispin DA, Lane Z, Goodlett DA, Bronner MP, Aebersold R, Brentnall TA. Gastroenterology. 2005;129:1187–1197. doi: 10.1053/j.gastro.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 47.DeSouza L, Diehl G, Rodrigues MJ, Guo J, Romaschin AD, Colgan TJ, Siu KW. J.Proteome.Res. 2005;4:377–386. doi: 10.1021/pr049821j. [DOI] [PubMed] [Google Scholar]
- 48.Li C, Hong Y, Tan YX, Zhou H, Ai JH, Li SJ, Zhang L, Xia QC, Wu JR, Wang HY, Zeng R. Mol.Cell Proteomics. 2004;3:399–409. doi: 10.1074/mcp.M300133-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Lu Z, Hu L, Evers S, Chen J, Shen Y. Proteomics. 2004;4:3975–3988. doi: 10.1002/pmic.200300863. [DOI] [PubMed] [Google Scholar]
- 50.Shen J, Person MD, Zhu J, Abbruzzese JL, Li D. Cancer Res. 2004;64:9018–9026. doi: 10.1158/0008-5472.CAN-04-3262. [DOI] [PubMed] [Google Scholar]
- 51.Shekouh AR, Thompson CC, Prime W, Campbell F, Hamlett J, Herrington CS, Lemoine NR, Crnogorac-Jurcevic T, Buechler MW, Friess H, Neoptolemos JP, Pennington SR, Costello E. Proteomics. 2003;3:1988–2001. doi: 10.1002/pmic.200300466. [DOI] [PubMed] [Google Scholar]
- 52.Kondo T, Seike M, Mori Y, Fujii K, Yamada T, Hirohashi S. Proteomics. 2003;3:1758–1766. doi: 10.1002/pmic.200300531. [DOI] [PubMed] [Google Scholar]
- 53.Sitek B, Luttges J, Marcus K, Kloppel G, Schmiegel W, Meyer HE, Hahn SA, Stuhler K. Proteomics. 2005;5:2665–2679. doi: 10.1002/pmic.200401298. [DOI] [PubMed] [Google Scholar]
- 54.Gygi SP, Rist B, Griffin TJ, Eng J, Aebersold R. Journal of Proteome Research. 2002;1:47–54. doi: 10.1021/pr015509n. [DOI] [PubMed] [Google Scholar]
- 55.Chen R, Pan S, Crispin DA, Brentnall TA. Cancer Genomics and Proteomics. 2006;3:1–10. [PubMed] [Google Scholar]
- 56.Lohr M, Faissner R. Pancreatology. 2004;4:67–75. doi: 10.1159/000077212. [DOI] [PubMed] [Google Scholar]
- 57.Goke B, Keim V, Dagorn JC, Arnold R, Adler G. Pancreas. 1990;5:261–266. doi: 10.1097/00006676-199005000-00004. [DOI] [PubMed] [Google Scholar]
- 58.Keim V, Iovanna JL, Rohr G, Usadel KH, Dagorn JC. Gastroenterology. 1991;100:775–782. doi: 10.1016/0016-5085(91)80025-5. [DOI] [PubMed] [Google Scholar]
- 59.Scheele GA. J.Biol.Chem. 1975;250:5375–5385. [PubMed] [Google Scholar]
- 60.Scheele GA, Palade GE. J.Biol.Chem. 1975;250:2660–2670. [PubMed] [Google Scholar]
- 61.Gronborg M, Bunkenborg J, Kristiansen TZ, Jensen ON, Yeo CJ, Hruban RH, Maitra A, Goggins MG, Pandey A. J.Proteome Res. 2004;3:1042–1055. doi: 10.1021/pr0499085. [DOI] [PubMed] [Google Scholar]
- 62.Chen R, Pan S, Yi EC, Donohoe S, Bronner MP, Potter JD, Goodlett DR, Aebersold R, Brentnall TA. Proteomics. 2006;6:3871–3879. doi: 10.1002/pmic.200500702. [DOI] [PubMed] [Google Scholar]
- 63.Chen R, Pan S, Cooke K, Moyes KW, Bronner MP, Goodlett DR, Aebersold R, Brentnall TA. Pancreas. 2007;34:70–79. doi: 10.1097/01.mpa.0000240615.20474.fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Whiteaker JR, Zhang H, Eng JK, Fang R, Piening BD, Feng LC, Lorentzen TD, Schoenherr RM, Keane JF, Holzman T, Fitzgibbon M, Lin C, Zhang H, Cooke K, Liu T, Camp DG, Anderson L, Watts J, Smith RD, McIntosh MW, Paulovich AG. J.Proteome.Res. 2006;6:828–836. doi: 10.1021/pr0604920. [DOI] [PubMed] [Google Scholar]
- 65.Bloomston M, Zhou JX, Rosemurgy AS, Frankel W, Muro-Cacho CA, Yeatman TJ. Cancer Res. 2006;66:2592–2599. doi: 10.1158/0008-5472.CAN-05-3659. [DOI] [PubMed] [Google Scholar]
- 66.Yu KH, Rustgi AK, Blair IA. J.Proteome.Res. 2005;4:1742–1751. doi: 10.1021/pr050174l. [DOI] [PubMed] [Google Scholar]
- 67.Ono M, Hakomori S. Glycoconj.J. 2004;20:71–78. doi: 10.1023/B:GLYC.0000018019.22070.7d. [DOI] [PubMed] [Google Scholar]
- 68.Wada Y, Tajiri M, Yoshida S. Anal.Chem. 2004;76:6560–6565. doi: 10.1021/ac049062o. [DOI] [PubMed] [Google Scholar]
- 69.Wada Y, Azadi P, Costello CE, Dell A, Dwek RA, Geyer H, Geyer R, Kakehi K, Karlsson NG, Kato K, Kawasaki N, Khoo KH, Kim S, Kondo A, Lattova E, Mechref Y, Miyoshi E, Nakamura K, Narimatsu H, Novotny MV, Packer NH, Perreault H, Peter-Katalinic J, Pohlentz G, Reinhold VN, Rudd PM, Suzuki A, Taniguchi N. Glycobiology. 2007;17:411–422. doi: 10.1093/glycob/cwl086. [DOI] [PubMed] [Google Scholar]
- 70.Zhao J, Qiu W, Simeone DM, Lubman DM. J.Proteome.Res. 2007;6:1126–1138. doi: 10.1021/pr0604458. [DOI] [PubMed] [Google Scholar]
- 71.Lin Y, Goedegebuure PS, Tan MC, Gross J, Malone JP, Feng S, Larson J, Phommaly C, Trinkaus K, Townsend RR, Linehan DC. J.Proteome.Res. 2006;5:2126–2176. doi: 10.1021/pr0600374. [DOI] [PubMed] [Google Scholar]
- 72.Cecconi D, Donadelli M, Scarpa A, Milli A, Palmieri M, Hamdan M, Areces LB, Rappsilber J, Righetti PG. J.Proteome.Res. 2005;4:1909–1916. doi: 10.1021/pr050154j. [DOI] [PubMed] [Google Scholar]
- 73.Sinha P, Hutter G, Kottgen E, Dietel M, Schadendorf D, Lage H. Electrophoresis. 1999;20:2952–5960. doi: 10.1002/(SICI)1522-2683(19991001)20:14<2952::AID-ELPS2952>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 74.Imamura T, Kanai F, Kawakami T, Amarsanaa J, Ijichi H, Hoshida Y, Tanaka Y, Ikenoue T, Tateishi K, Kawabe T, Arakawa Y, Miyagishi M, Taira K, Yokosuka O, Omata M. Biochem.Biophys.Res.Commun. 2004;318:289–296. doi: 10.1016/j.bbrc.2004.04.029. [DOI] [PubMed] [Google Scholar]
- 75.Mauri P, Scarpa A, Nascimbeni AC, Benazzi L, Parmagnani E, Mafficini A, Della PM, Bassi C, Miyazaki K, Sorio C. FASEB J. 2005;19:1125–1127. doi: 10.1096/fj.04-3000fje. [DOI] [PubMed] [Google Scholar]
- 76.Gronborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, Sato N, Molina H, Jensen ON, Hruban RH, Goggins MG, Maitra A, Pandey A. Mol.Cell Proteomics. 2006;5:157–171. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]