Summary
The vacuolar (H+)-ATPases (V-ATPases) are ATP-dependent proton pumps responsible for both acidification of intracellular compartments and, for certain cell types, proton transport across the plasma membrane. Intracellular V-ATPases function in both endocytic and intracellular membrane traffic, processing and degradation of macromolecules in secretory and digestive compartments, coupled transport of small molecules such as neurotransmitters and ATP and in the entry of pathogenic agents, including envelope viruses and bacterial toxins. V-ATPases are present in the plasma membrane of renal cells, osteoclasts, macrophages, epididymal cells and certain tumor cells where they are important for urinary acidification, bone resorption, pH homeostasis, sperm maturation and tumor cell invasion, respectively.
The V-ATPases are composed of a peripheral domain (V1) that carries out ATP hydrolysis and an integral domain (V0) responsible for proton transport. V1 contains eight subunits (A-H) while V0 contains six subunits (a,c,c’,c”,d and e). V-ATPases operate by a rotary mechanism in which ATP hydrolysis within V1 drives rotation of a central rotary domain, that includes a ring of proteolipid subunits (c,c’ and c”), relative to the remainder of the complex. Rotation of the proteolipid ring relative to subunit a within V0 drives active transport of protons across the membrane. Two important mechanisms of regulating V-ATPase activity in vivo are reversible dissociation of the V1 and V0 domains and changes in coupling efficiency of proton transport and ATP hydrolysis. This review focuses on recent advances in our lab in understanding the structure and regulation of the V-ATPases.
Function of V-ATPases
V-ATPases function as ATP-driven proton pumps in a wide variety of cellular membranes, including endosomes, lysosomes, Golgi-derived vesicles, secretory vesicles and the plasma membrane of various cell types [1]. Acidification of endosomes facilitates the dissociation of internalized ligand-receptor complexes and allows unoccupied receptors to recycle to the plasma membrane [2]. Receptors that follow this pathway include, among others, those for the cholesterol carrier low density lipoprotein (LDL), asialoglycoproteins and peptide hormones, such as insulin. A similar acid-activated dissociation occurs in Golgi-derived vesicles and is involved in the delivery of newly synthesized lysosomal enzymes from the trans-Golgi network to lysosomes utilizing the mannos-6-phosphate receptor [3]. Endosomal acidification is also required for the budding of endosomal carrier vesicles that move cargo proteins from early to late endosomes [4]. Exposure of various envelope viruses (like influenza virus) and toxins (like anthrax toxin) to the acidic environment of the endosome facilitates the entry of the cytotoxic portions of these agents into cells [5]. A low pH within lysosomes activates degradative enzymes present within the lysosome lumen and provides a driving force for the coupled transport of small molecules and ions across the lysosomal membrane. Similarly, acidification of secretory vesicles, like synaptic vesicles, drives the uptake of small molecules, such as neurotransmitters, coupled either to the proton gradient or the positive interior membrane potential generated by the V-ATPase. The low pH within secretory vesicles is also required for the activity of proteolytic enzymes that process precursor proteins, such as proinsulin, to their mature forms [6].
Plasma membrane V-ATPases function in both normal and disease processes. V-ATPases in the apical membrane of renal intercalated cells of the distal tubule and collecting duct serve to secrete protons into the urine, thus participating in the regulation of plasma pH [7]. Defects in this process lead to the human genetic disorder renal tubule acidosis [8]. Plasma membrane V-ATPases in osteoclasts are essential for the ability of these cells to degrade bone, with mutations in the isoform responsible for plasma membrane targeting in osteoclasts leading to osteopetrosis [9]. In macrophages and neutrophils, V-ATPases at the cell surface partcipate in pH homeostasis [10] whereas in the epididymus and vas deferens, V-ATPases function in sperm maturation and storage [11]. V-ATPases have been identified at the plasma membrane of both vascular endothelial cells and certain tumor cells where they are thought to participate in the invasive properties of these cells [12, 13]. V-ATPases are thus being investigated as a potential target in the treatment of a variety of human diseases, including osteoporosis, diabetes and cancer.
Structure and mechanism of the V-ATPases
The V-ATPases are large, multi-subunit complexes organized into two domains (Fig. 1a) [1]. The peripheral V1 domain is responsible for ATP hydrolysis whereas the integral V0 domain carries out proton translocation. V1 is composed of eight different subunits (A-H) of molecular mass 70-10 kDa that are present in a stoichiometry of A3B3C1D1E2F1G2H1-2 [14-16]. The three A and three B subunits are arranged in an alternating hexamer with the nucleotide binding sites located at the interface of the A and B subunits. The three catalytic sites are composed principally of residues contributed by the A subunits, although certain residues on the B subunits are also present [17-20]. Similarly, the three “non-catalytic” sites, whose function is proposed to be regulatory, are located principally on the B subunits [17, 19, 21, 22]. The remaining V1 subunits are organized into one of two types of stalks, referred to as the central and peripheral stalks, connecting the V1 and V0 domains. We have localized subunits to the central and peripheral stalks of the V-ATPase complex using both electron microscopy and cysteine-mediated cross-linking in yeast [23-28]. In the latter approach, unique cysteine residues are introduced into structurally defined sites of a subunit that has been otherwise engineered to remove endogenous cysteine residues. For subunits C and H, we have employed available high resolution crystal structures [29, 30] to place unique cysteine residues over the surface of the protein [25, 28]. For subunit B, a molecular model based on sequence homology with the F-ATPase and a high resolution structure of the F1 domain was employed [23, 24, 26, 31]. Results from both cross-linking and electron microscopy have shown that subunit D is present in the central stalk whereas subunits C, E, G and H together with the amino-terminal domain of subunit a are present in the peripheral stalk [23-28]. Based upon the formation of tight heterodimeric complexes of subunits D and F [32], we have also assigned subunit F to the central stalk.
Figure 1. Structure and mechanism of the V-ATPase.
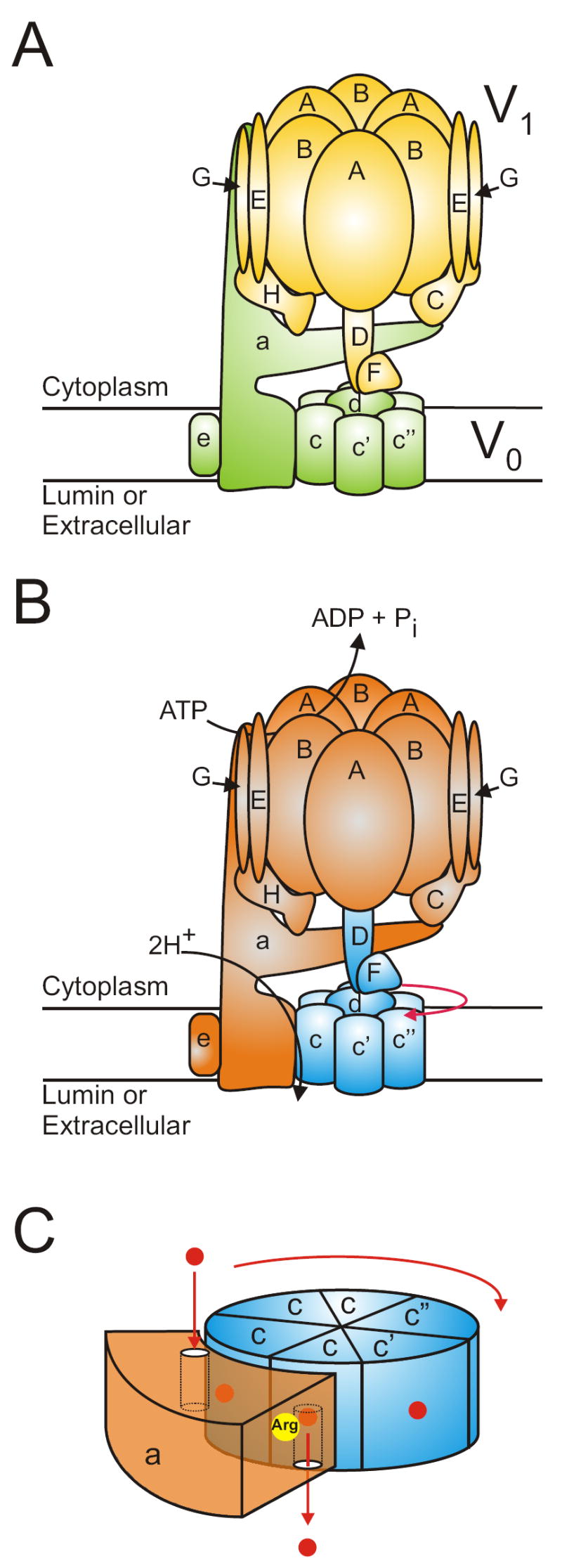
A, subunit model of the V-ATPase highlighting the V1 (yellow) and V0 (green) domains. The model is based on the homology to F-ATPase, the known crystal structure of isolated V-ATPase subunits, electron microscopy and cysteine mediated crosslinking studies. B, the rotary mechanism of V-ATPase. The rotary subunits are highlighted in blue, while the stationary subunits are shown in orange. Hydrolysis of ATP causes conformational changes in the A subunits which drives the rotation of the rotor in the direction shown by the red arrow. C, the mechansim of proton transport through V0. Shown is a schematic representation of the C-terminal domain of a, and the proteolipid ring. Protons (shown as red dots) enter the V0 complex through the cytoplasmic hemi-channel on a and bind to the essential glutamic acid residue on the proteolipid subunit that is aligned with the hemichannel. As the proteolipids rotate, the protonated form of the glutamic acid travels through the lipid bilayer and eventually returns to the a subunit and aligns with the luminal hemichannel. Residue Arg-735 (shown in yellow) on a promotes the deprotonation of the glutamate, and the proton is released through the hemichannel into the lumen.
V0 (in yeast) is composed of six different subunits of molecular mass 100-9 kDa that are present in a stoichiometry of a1d1enc4-5c’1c”1 [14, 33]. Subunits c, c’ and c” are highly hydrophobic proteins that share sequence homology to each other and to the F-ATPase subunit c and are referred to as proteolipid subunits [34]. Subunits c and c’ contain four transmembrane helices whereas subunit c” contains five transmembrane helices [35]. Each proteolipid subunit contains a single buried carboxyl group present in TM4 (for c and c’) or TM3 (for c”) that is essential for proton translocation. These buried carboxyl groups are located near the middle of the bilayer and are thought to undergo reversible protonation during proton translocation, although it is possible that protons may be transported as hydronium ions, as proposed for the F-ATPases [36]. The proteolipid subunits form a ring containing single copies of subunit c’ and c” and 4-5 copies of subunit c [14, 33]. Subunit d is a hydrophilic protein that binds tightly to the cytoplasmic surface of the proteolipid ring [37] and serves as the binding site for the V1 subunits (D and F) of the central stalk complex. The proteolipid ring, together with subunit d of V0 and subunits D and F of V1, is also referred to as the rotary complex (see below).
Subunit a is a 100 kDa transmembrane protein containing a 50 kDa amino-terminal cytoplasmic domain and a 50 kDa carboxy-terminal hydrophobic domain containing 8-9 transmembrane helices [38]. Subunit a contains a buried arginine residue located in TM7 which, like the carboxyl groups of the proteolipid subunits, is essential for proton translocation through V0 [39]. Subunit a is also thought to form two hemi-channels (one oriented towards the cytoplasmic side of the membrane and one oriented towards the luminal side) which allow protons to reach and to leave the buried carboxyl groups on the proteolipid subunits [1].
Like the related family of F-ATPases (or ATP synthases), the V-ATPases operate by a rotary mechanism (Fig. 1b and c) [40, 41]. ATP hydrolysis within the V1 domain drives rotation of the rotary complex (subunits D, F, d and the proteolipid ring). The proteolipid ring rotates relative to subunit a, which is held fixed relative to the A3B3 hexameric head of V1 by the peripheral stalk(s), or stator(s), which include subunits C, E, G, H and the amino terminal domain of subunit a. The mechanism of proton translocation, based upon the mechanism first proposed for the F-ATPases [42], is envisioned as follows (Fig. 1c) [1]. A proton enters from the cytoplasmic side of the membrane by the cytoplasmic hemi-channel in subunit a and protonates a buried carboxyl group on one of the proteolipid subunits. This proton remains attached to the carboxyl group as the proteolipid ring is forced to rotate because of the hydrophobic environment in which this group is located. As the protonated carboxyl group reaches the luminal hemi-channel, interaction with the buried Arg residue on subunit a stabilizes the carboxyl group in its charged form, thus releasing the proton into the luminal hemi-channel. Continued rotation of the charged carboxyl group past the Arg residue of subunit a places it once again in contact with the cytoplasmic hemi-channel, where it is again available for protonation.
Evidence for close interaction between TM7 of subunit a containing the critical arginine residue and TM 4 of subunit c’ containing the essential glutamic acid residue has come from zero-length cross-linking studies between engineered cysteine residues in each protein [43]. TM7 of subunit a also shows disulfide-mediated cross-linking to TM3 but not TM5 of subunit c” [44]. Both TM3 and TM5 of subunit c” contain buried glutamic acid residues, but only that in TM3 is essential for proton translocation [35]. Interestingly, cross-linking of subunit a to both subunits c’ and c” shows evidence for helical swiveling in both subunit a and the proteolipid subunits [43, 44]. That is, zero-length cross-linking can be observed between residues that would require considerable radial mobility in both helices. It is tempting to speculate that such helical swiveling in subunit a may help to control proton accessibility through the postulated hemi-channels that participate in proton translocation. The specific and highly potent inhibitors bafilomycin and concanamycin have been shown to bind at helical interfaces within the proteolipid ring [45] and it is possible that they inhibit activity by preventing helical swiveling within the ring. Subunit a may also participate to a lesser degree in conferring sensitivity to these inhibitors [46].
We have also recently shown that the three proteolipid subunits in the yeast V-ATPase adopt a particular arrangement in functional V-ATPase complexes [47]. Subunit c’ must be adjacent to and counterclockwise from subunit c” as viewed from the luminal side of the membrane. The reason that only this arrangement gives rise to a functional V-ATPase is not known, but one possibility is that, because subunit c’ serves as the docking site for the dedicated ER chaperone protein Vma21p [48], it may be that only this arrangement leads to fully assembled V0 domains.
Regulation of V-ATPase activity
Different intracellular compartments are known to be maintained at different pH values, with lysosomes more acidic than late endosomes which are in turn more acidic than early endosomes [49]. Similarly, late endosomes are more acidic than the trans-Golgi network, which is also more acidic than the other Golgi compartments [50]. In many cases, cells also need to dramatically change proton translocation across the plasma membrane without changing acidification of intracellular compartments [7]. To accomplish this differential regulation of proton transport, V-ATPase activity is tightly controlled in vivo using a variety of cellular mechanisms.
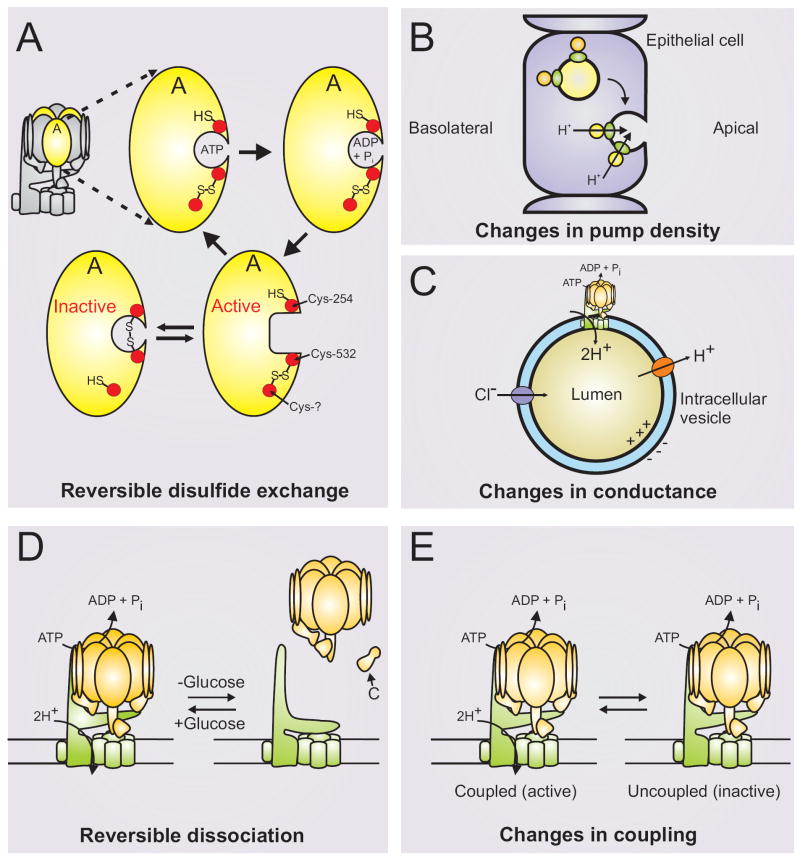
One early identified mechanism of controlling V-ATPase activity involves reversible disulfide bond formation between conserved cysteine residues at the catalytic site of the V-ATPase [51-53]. When a disulfide bond is formed between a highly conserved cysteine in the Walker A sequence of the catalytic A subunit (Cys254 of the bovine potein) and a second highly conserved cysteine residue (Cys532) located in the C-terminal domain of the same subunit, the V-ATPase activity is reversibly inhibited. This results from a covalent locking of the catalytic site in a closed conformation which is unable to undergo the opening required for product release and ATP re-binding [31]. Cleavage of this disulfide bond occurs principally by a thio-disulfide exchange involving an additional cysteine residue in subunit A, although nitric oxide is also able to induce disulfide bond formation leading to reversible inhibition of V-ATPase activity [54]. Estimates of the fraction of V-ATPase existing in the reversibly disulfide bonded state in vivo suggest that this number is approximately 50% in clathrin-coated vesicles in brain [53].
A second important mechanism of controlling V-ATPase activity involves modulation of pump density. This is particularly important in controlling the amount of V-ATPase-dependent proton transport occurring across the plasma membrane of epithelial cells. Thus proton transport across the apical membrane of both renal alpha intercalated cells and epididymal clear cells is controlled by reversible fusion with the apical membrane of intracellular vesicles containing a high density of V-ATPases [7, 11]. In the case of clear cells this fusion has been shown to be activated by a bicarbonate-sensitive adenylyl cyclase.
A third mechanism of regulating V-ATPase activity in vivo involves reversible dissociation of the V1 and V0 domains. In yeast and in insect cells dissociation has been shown to be activated by nutrient depletion [55, 56], presumably as a way of conserving cellular stores of ATP, although changes in assembly of the V-ATPase have also been reported in renal cells and dendritic cells involved in antigen processing [57, 58]. In yeast, dissociation occurs rapidly, reversibly and does not require new protein synthesis [59]. It also does not involve a variety of signaling pathways which are altered in response to glucose depletion [60]. It has been reported by both our lab and the Kane laboratory that dissociation requires catalytic activity [20, 60], although we have recently found that dissociation is not absolutely dependent upon activity [61]. We have also found that dissociation, but not reassembly, is dependent upon an intact microtubule network [62], suggesting that dissociation and reassembly are independently controlled processes. Consistent with this idea is the observation that glucose-dependent reassembly of the complex (but not dissociation) is dependent upon a novel heterotrimeric complex termed RAVE (regulator of acidification of vacuoles and endosomes) [63]. RAVE is composed of two novel proteins (Rav1p and Rav2p) and a ubiquitin-ligase subunit (Skp1) and promotes both glucose-dependent and normal biosynthetic assembly of the V-ATPase complex [64].
We have investigated the dependence of in vivo dissociation of the V-ATPase on isoforms of subunit a and cellular environment. Targeting of V-ATPases to different cellular membranes is controlled by isoforms of subunit a [1]. In yeast there are two isoforms of subunit a (Vph1p and Stv1p), with Vph1p targeting to the vacuole and Stv1p targeting to a late Golgi compartment [65, 66]. If Stv1p is overexpressed in a strain deleted in both genes, however, Stv1p also appears in the vacuole. We have found that V-ATPases containing Vph1p present in the vacuole dissociate upon removal of glucose from the media whereas Stv1p-containing complexes localized to the Golgi do not [65]. If Stv1p-containing complexes are re-directed to the vacuole, however, dissociation upon glucose depletion is observed. We have employed vps (vacuolar protein sorting) mutants of yeast that are disrupted in particular steps in the trafficking of proteins from the Golgi to the vacuole to further probe the dependence of dissociation on cellular environment [61, 65]. Vps21 mutants are disrupted in trafficking from a post-Golgi compartment to a prevacuolar compartment whereas vps27 mutants are interrupted in movement of proteins from the prevacuolar compartment to the vacuole. We have found that V-ATPases targeted to the same intracellular compartment behave similarly with respect to glucose-dependent dissociation, with dissociation being most complete in the vacuole, followed by the post-Golgi and prevacuolar compartments and lastly by the Golgi, which shows no significant dissociation [61]. These results suggest that cellular environment rather than a subunit isoform is the primary determinant in controlling the degree of in vivo dissociation. Although the precise environmental factors important in controlling dissociation are not known, the lumenal pH appears to be one such parameter, with a sufficiently low luminal pH being permissive for in vivo dissociation [67].
We have recently demonstrated that a unique domain of the catalytic A subunit, termed the non-homologous domain, appears to play a role in controlling in vivo dissociation [67, 68]. This 90 amino acid domain located about one third of the way from the N-terminus of subunit A is highly conserved among V-ATPase sequences from different species but is unique to the V-ATPases, being absent from the homologous beta subunit of the F-ATPases (hence the name). We have found that certain mutations in the non-homologous domain lead to complete inhibition of glucose-dependent dissociation but give rise to V-ATPase complexes with full catalytic activity [68]. Moreover, we have found that the separate non-homologous domain expressed as an HA-tagged protein is able to bind to the V0 domain in the absence of the other V1 subunits in a glucose-dependent manner (i.e., removing glucose from the media induces in vivo dissociation) [67]. Interestingly, upon release from V0 the non-homologous domain appears to bind to another as yet unidentified protein. These results suggest that changes in the non-homologous domain of subunit A may help to activate dissociation upon glucose depletion.
An important question concerning reversible dissociation of the V-ATPase complex is how ATPase activity of the free V1 domain and passive proton conductance by the free V0 domain are silenced upon dissociation in vivo. Earlier data demonstrated that the free V1 and V0 domains are indeed inhibited in their respective functions and results from the Kane laboratory showed that subunit H plays a crucial role in inhibiting ATP hydrolysis by free V1 [69]. Using cysteine-mediated cross-linking of complexes containing subunit H mutants engineered to contain single cysteine residues, we have demonstrated that subunit H comes into close proximity to subunit F (a rotor subunit) in free V1 but not the intact V1V0 complex [28]. Because subunit H has been shown to form part of the stator complex [26], these results suggest that subunit H inhibits the activity of free V1 by bridging the rotor and stator complexes and physically preventing ATP-driven rotation.
A fourth mechanism of controlling V-ATPase activity involves changes in the coupling of proton transport and ATP hydrolysis. The enzyme appears poised to alter the efficiency of coupling based upon the observation that a variety of changes, including limited proteolysis [70], elevated ATP concentrations [71] and mutations in a number of subunits, including subunit D [72] and the non-homologous region of subunit A [68], cause a partial uncoupling of proton transport from ATP hydrolysis. Perhaps most interesting is the finding that certain mutations in the non-homologous region actually cause an increase in the coupling efficiency [68], suggesting that the wild type V-ATPase is not optimally coupled. Although the normal physiological signals that control coupling have not yet been identified, we have observed that V-ATPase complexes containing the Vph1p isoform of subunit a are 4-5 fold more tightly coupled than those containing the Stv1p isoform [65]. This, combined with the observation that Vph1p-containing V0 domains show about 10-fold better assembly with V1 than Stv1p-containing V0 domains [65], helps to explain why the vacuole (which normally contains Vph1p-containing complexes) is significantly more acidic in vivo than the Golgi (which contains Stv1p-containing complexes). Thus the a subunit isoform plays an important role not only in controlling intracellular targeting of the V-ATPase but also in controlling assembly and coupling efficiency of the complex. Interestingly, information to control targeting and dissociation appears to reside in the N-terminal domain of subunit a whereas information controlling coupling resides in the C-terminal domain [73].
A final mechanism of regulating vacuolar acidification in vivo involves changes in transporters other than the V-ATPase. Because the V-ATPase is electrogenic, it establishes a luminal positive membrane potential during transport. As a result, little proton transport can occur in intracellular compartments unless accompanied by a compensating charge flux. In vivo this appears to primarily involve intracellular chloride channels that allow chloride influx to accompany ATP-driven proton transport, thus dissipating the luminal positive membrane potential. Moreover, chloride channel activity has been shown to be modulated by PKA [74]. Because the final pH of an intracellular compartment is a balance between active proton transport and passive proton leakage, organellar pH may also be controlled by regulating passive proton conductance. Recent data suggest that intracellular Na/H antiporters may be playing this role in yeast [75].
Figure 2. Regulation of the V-ATPase.
V-ATPases are regulated by a number of mechanisms including: A, reversible disulfide bond formation between conserved cysteine residues that prevent the catalytic site from cycling between the open and closed conformations required by the binding change mechanism [31, 76]; B, changes in pump density through fusion of vesicles containing a high number of V-ATPase; C, changes in either Cl- or H+ conductance through distinct channels; D, reversible dissociation into inactive V1 and V0 domains; and E, changes in coupling efficiency.
Acknowledgments
This work was supported by National Institutes of Health Grant GM34478 (to M.F.), a Canadian Institutes of Health Research Postdoctoral Fellowship (to D.J.C), a Postdoctoral Fellowship from the Northeast Affiliate of the American Heart Association (to Y.W.), a TEACRS Fellowship from the NIH (to A.H.), and NIH Training Grant DK07542 (to K.C.J., and S.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 2.Nishi T, Forgac M. The vacuolar (H+)-ATPases--nature’s most versatile proton pumps. Nat Rev Mol Cell Biol. 2002;3:94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh P, Dahms NM, Kornfeld S. Mannose 6-phosphate receptors: new twists in the tale. Nature reviews. 2003;4:202–212. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- 4.Gu F, Gruenberg J. ARF1 regulates pH-dependent COP functions in the early endocytic pathway. J Biol Chem. 2000;275:8154–8160. doi: 10.1074/jbc.275.11.8154. [DOI] [PubMed] [Google Scholar]
- 5.Abrami L, Lindsay M, Parton RG, Leppla SH, van der Goot FG. Membrane insertion of anthrax protective antigen and cytoplasmic delivery of lethal factor occur at different stages of the endocytic pathway. J Cell Biol. 2004;166:645–651. doi: 10.1083/jcb.200312072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun-Wada GH, Toyomura T, Murata Y, Yamamoto A, Futai M, Wada Y. The a3 isoform of V-ATPase regulates insulin secretion from pancreatic beta-cells. J Cell Sci. 2006;119:4531–4540. doi: 10.1242/jcs.03234. [DOI] [PubMed] [Google Scholar]
- 7.Wagner CA, Finberg KE, Breton S, Marshansky V, Brown D, Geibel JP. Renal vacuolar H+-ATPase. Physiol Rev. 2004;84:1263–1314. doi: 10.1152/physrev.00045.2003. [DOI] [PubMed] [Google Scholar]
- 8.Karet FE, Finberg KE, Nelson RD, Nayir A, Mocan H, Sanjad SA, Rodriguez-Soriano J, Santos F, Cremers CW, Di Pietro A, Hoffbrand BI, Winiarski J, Bakkaloglu A, Ozen S, Dusunsel R, Goodyer P, Hulton SA, Wu DK, Skvorak AB, Morton CC, Cunningham MJ, Jha V, Lifton RP. Mutations in the gene encoding B1 subunit of H+-ATPase cause renal tubular acidosis with sensorineural deafness. Nat Genet. 1999;21:84–90. doi: 10.1038/5022. [DOI] [PubMed] [Google Scholar]
- 9.Frattini A, Orchard PJ, Sobacchi C, Giliani S, Abinun M, Mattsson JP, Keeling DJ, Andersson AK, Wallbrandt P, Zecca L, Notarangelo LD, Vezzoni P, Villa A. Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nat Genet. 2000;25:343–346. doi: 10.1038/77131. [DOI] [PubMed] [Google Scholar]
- 10.Nanda A, Brumell JH, Nordstrom T, Kjeldsen L, Sengelov H, Borregaard N, Rotstein OD, Grinstein S. Activation of proton pumping in human neutrophils occurs by exocytosis of vesicles bearing vacuolar-type H+-ATPases. J Biol Chem. 1996;271:15963–15970. doi: 10.1074/jbc.271.27.15963. [DOI] [PubMed] [Google Scholar]
- 11.Pietrement C, Sun-Wada GH, Silva ND, McKee M, Marshansky V, Brown D, Futai M, Breton S. Distinct expression patterns of different subunit isoforms of the V-ATPase in the rat epididymis. Biol Reprod. 2006;74:185–194. doi: 10.1095/biolreprod.105.043752. [DOI] [PubMed] [Google Scholar]
- 12.Rojas JD, Sennoune SR, Martinez GM, Bakunts K, Meininger CJ, Wu G, Wesson DE, Seftor EA, Hendrix MJ, Martinez-Zaguilan R. Plasmalemmal vacuolar H+-ATPase is decreased in microvascular endothelial cells from a diabetic model. Journal of cellular physiology. 2004;201:190–200. doi: 10.1002/jcp.20059. [DOI] [PubMed] [Google Scholar]
- 13.Sennoune SR, Bakunts K, Martinez GM, Chua-Tuan JL, Kebir Y, Attaya MN, Martinez-Zaguilan R. Vacuolar H+-ATPase in human breast cancer cells with distinct metastatic potential: distribution and functional activity. Am J Physiol Cell Physiol. 2004;286:C1443–1452. doi: 10.1152/ajpcell.00407.2003. [DOI] [PubMed] [Google Scholar]
- 14.Arai H, Terres G, Pink S, Forgac M. Topography and subunit stoichiometry of the coated vesicle proton pump. J Biol Chem. 1988;263:8796–8802. [PubMed] [Google Scholar]
- 15.Xu T, Vasilyeva E, Forgac M. Subunit interactions in the clathrin-coated vesicle vacuolar (H(+))-ATPase complex. J Biol Chem. 1999;274:28909–28915. doi: 10.1074/jbc.274.41.28909. [DOI] [PubMed] [Google Scholar]
- 16.Ohira M, Smardon AM, Charsky CM, Liu J, Tarsio M, Kane PM. The E and G subunits of the yeast V-ATPase interact tightly and are both present at more than one copy per V1 complex. J Biol Chem. 2006;281:22752–22760. doi: 10.1074/jbc.M601441200. [DOI] [PubMed] [Google Scholar]
- 17.Liu Q, Kane PM, Newman PR, Forgac M. Site-directed mutagenesis of the yeast V-ATPase B subunit (Vma2p) J Biol Chem. 1996;271:2018–2022. doi: 10.1074/jbc.271.4.2018. [DOI] [PubMed] [Google Scholar]
- 18.Liu Q, Leng XH, Newman PR, Vasilyeva E, Kane PM, Forgac M. Site-directed mutagenesis of the yeast V-ATPase A subunit. J Biol Chem. 1997;272:11750–11756. doi: 10.1074/jbc.272.18.11750. [DOI] [PubMed] [Google Scholar]
- 19.MacLeod KJ, Vasilyeva E, Baleja JD, Forgac M. Mutational analysis of the nucleotide binding sites of the yeast vacuolar proton-translocating ATPase. J Biol Chem. 1998;273:150–156. doi: 10.1074/jbc.273.1.150. [DOI] [PubMed] [Google Scholar]
- 20.MacLeod KJ, Vasilyeva E, Merdek K, Vogel PD, Forgac M. Photoaffinity labeling of wild-type and mutant forms of the yeast V-ATPase A subunit by 2-azido-[(32)P]ADP. J Biol Chem. 1999;274:32869–32874. doi: 10.1074/jbc.274.46.32869. [DOI] [PubMed] [Google Scholar]
- 21.Vasilyeva E, Forgac M. 3’-O-(4-Benzoyl)benzoyladenosine 5’-triphosphate inhibits activity of the vacuolar (H+)-ATPase from bovine brain clathrin-coated vesicles by modification of a rapidly exchangeable, noncatalytic nucleotide binding site on the B subunit. J Biol Chem. 1996;271:12775–12782. doi: 10.1074/jbc.271.22.12775. [DOI] [PubMed] [Google Scholar]
- 22.Vasilyeva E, Liu Q, MacLeod KJ, Baleja JD, Forgac M. Cysteine scanning mutagenesis of the noncatalytic nucleotide binding site of the yeast V-ATPase. J Biol Chem. 2000;275:255–260. doi: 10.1074/jbc.275.1.255. [DOI] [PubMed] [Google Scholar]
- 23.Arata Y, Baleja JD, Forgac M. Localization of subunits D, E, and G in the yeast V-ATPase complex using cysteine-mediated cross-linking to subunit B. Biochemistry. 2002;41:11301–11307. doi: 10.1021/bi0262449. [DOI] [PubMed] [Google Scholar]
- 24.Arata Y, Baleja JD, Forgac M. Cysteine-directed cross-linking to subunit B suggests that subunit E forms part of the peripheral stalk of the vacuolar H+-ATPase. J Biol Chem. 2002;277:3357–3363. doi: 10.1074/jbc.M109967200. [DOI] [PubMed] [Google Scholar]
- 25.Inoue T, Forgac M. Cysteine-mediated cross-linking indicates that subunit C of the V-ATPase is in close proximity to subunits E and G of the V1 domain and subunit a of the V0 domain. J Biol Chem. 2005;280:27896–27903. doi: 10.1074/jbc.M504890200. [DOI] [PubMed] [Google Scholar]
- 26.Wilkens S, Inoue T, Forgac M. Three-dimensional structure of the vacuolar ATPase. Localization of subunit H by difference imaging and chemical cross-linking. J Biol Chem. 2004;279:41942–41949. doi: 10.1074/jbc.M407821200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, Inoue T, Forgac M, Wilkens S. Localization of subunit C (Vma5p) in the yeast vacuolar ATPase by immuno electron microscopy. FEBS Lett. 2006;580:2006–2010. doi: 10.1016/j.febslet.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Jefferies KC, Forgac M. Subunit H of the V-ATPase inhibits ATP hydrolysis by the free V1 domain by interaction with the rotary subunit F. J Biol Chem. 2007 doi: 10.1074/jbc.M707144200. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sagermann M, Stevens TH, Matthews BW. Crystal structure of the regulatory subunit H of the V-type ATPase of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2001;98:7134–7139. doi: 10.1073/pnas.131192798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drory O, Frolow F, Nelson N. Crystal structure of yeast V-ATPase subunit C reveals its stator function. EMBO reports. 2004;5:1148–1152. doi: 10.1038/sj.embor.7400294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 32.Tomashek JJ, Graham LA, Hutchins MU, Stevens TH, Klionsky DJ. V1-situated stalk subunits of the yeast vacuolar proton-translocating ATPase. J Biol Chem. 1997;272:26787–26793. doi: 10.1074/jbc.272.42.26787. [DOI] [PubMed] [Google Scholar]
- 33.Powell B, Graham LA, Stevens TH. Molecular characterization of the yeast vacuolar H+-ATPase proton pore. J Biol Chem. 2000;275:23654–23660. doi: 10.1074/jbc.M004440200. [DOI] [PubMed] [Google Scholar]
- 34.Mandel M, Moriyama Y, Hulmes JD, Pan YC, Nelson H, Nelson N. cDNA sequence encoding the 16-kDa proteolipid of chromaffin granules implies gene duplication in the evolution of H+-ATPases. Proc Natl Acad Sci USA. 1988;85:5521–5524. doi: 10.1073/pnas.85.15.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirata R, Graham LA, Takatsuki A, Stevens TH, Anraku Y. VMA11 and VMA16 encode second and third proteolipid subunits of the Saccharomyces cerevisiae vacuolar membrane H+-ATPase. J Biol Chem. 1997;272:4795–4803. doi: 10.1074/jbc.272.8.4795. [DOI] [PubMed] [Google Scholar]
- 36.von Ballmoos C, Dimroth P. Two distinct proton binding sites in the ATP synthase family. Biochemistry. 2007;46:11800–11809. doi: 10.1021/bi701083v. [DOI] [PubMed] [Google Scholar]
- 37.Iwata M, Imamura H, Stambouli E, Ikeda C, Tamakoshi M, Nagata K, Makyio H, Hankamer B, Barber J, Yoshida M, Yokoyama K, Iwata S. Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase. Proc Natl Acad Sci USA. 2004;101:59–64. doi: 10.1073/pnas.0305165101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leng XH, Nishi T, Forgac M. Transmembrane topography of the 100-kDa a subunit (Vph1p) of the yeast vacuolar proton-translocating ATPase. J Biol Chem. 1999;274:14655–14661. doi: 10.1074/jbc.274.21.14655. [DOI] [PubMed] [Google Scholar]
- 39.Kawasaki-Nishi S, Nishi T, Forgac M. Arg-735 of the 100-kDa subunit a of the yeast V-ATPase is essential for proton translocation. Proc Natl Acad Sci USA. 2001;98:12397–12402. doi: 10.1073/pnas.221291798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imamura H, Nakano M, Noji H, Muneyuki E, Ohkuma S, Yoshida M, Yokoyama K. Evidence for rotation of V1-ATPase. Proc Natl Acad Sci USA. 2003;100:2312–2315. doi: 10.1073/pnas.0436796100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirata T, Iwamoto-Kihara A, Sun-Wada GH, Okajima T, Wada Y, Futai M. Subunit rotation of vacuolar-type proton pumping ATPase: relative rotation of the G and C subunits. J Biol Chem. 2003;278:23714–23719. doi: 10.1074/jbc.M302756200. [DOI] [PubMed] [Google Scholar]
- 42.Vik SB, Long JC, Wada T, Zhang D. A model for the structure of subunit a of the Escherichia coli ATP synthase and its role in proton translocation. Biochim Biophys Acta. 2000;1458:457–466. doi: 10.1016/s0005-2728(00)00094-3. [DOI] [PubMed] [Google Scholar]
- 43.Kawasaki-Nishi S, Nishi T, Forgac M. Interacting helical surfaces of the transmembrane segments of subunits a and c’ of the yeast V-ATPase defined by disulfide-mediated cross-linking. J Biol Chem. 2003;278:41908–41913. doi: 10.1074/jbc.M308026200. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Inoue T, Forgac M. TM2 but not TM4 of subunit c” interacts with TM7 of subunit a of the yeast V-ATPase as defined by disulfide-mediated cross-linking. J Biol Chem. 2004;279:44628–44638. doi: 10.1074/jbc.M407345200. [DOI] [PubMed] [Google Scholar]
- 45.Bowman BJ, McCall ME, Baertsch R, Bowman EJ. A model for the proteolipid ring and bafilomycin/concanamycin-binding site in the vacuolar ATPase of Neurospora crassa. J Biol Chem. 2006;281:31885–31893. doi: 10.1074/jbc.M605532200. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Inoue T, Forgac M. Subunit a of the yeast V-ATPase participates in binding of bafilomycin. J Biol Chem. 2005;280:40481–40488. doi: 10.1074/jbc.M509106200. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Cipriano DJ, Forgac M. Arrangement of subunits in the proteolipid ring of the V-ATPase. J Biol Chem. 2007;282:34058–34065. doi: 10.1074/jbc.M704331200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malkus P, Graham LA, Stevens TH, Schekman R. Role of Vma21p in assembly and transport of the yeast vacuolar ATPase. Mol Biol Cell. 2004;15:5075–5091. doi: 10.1091/mbc.E04-06-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tycko B, Maxfield FR. Rapid acidification of endocytic vesicles containing alpha 2-macroglobulin. Cell. 1982;28:643–651. doi: 10.1016/0092-8674(82)90219-7. [DOI] [PubMed] [Google Scholar]
- 50.Kim JH, Lingwood CA, Williams DB, Furuya W, Manolson MF, Grinstein S. Dynamic measurement of the pH of the Golgi complex in living cells using retrograde transport of the verotoxin receptor. J Cell Biol. 1996;134:1387–1399. doi: 10.1083/jcb.134.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feng Y, Forgac M. A novel mechanism for regulation of vacuolar acidification. J Biol Chem. 1992;267:19769–19772. [PubMed] [Google Scholar]
- 52.Feng Y, Forgac M. Cysteine 254 of the 73-kDa A subunit is responsible for inhibition of the coated vesicle (H+)-ATPase upon modification by sulfhydryl reagents. J Biol Chem. 1992;267:5817–5822. [PubMed] [Google Scholar]
- 53.Feng Y, Forgac M. Inhibition of vacuolar H(+)-ATPase by disulfide bond formation between cysteine 254 and cysteine 532 in subunit A. J Biol Chem. 1994;269:13224–13230. [PubMed] [Google Scholar]
- 54.Forgac M. The vacuolar H+-ATPase of clathrin-coated vesicles is reversibly inhibited by S-nitrosoglutathione. J Biol Chem. 1999;274:1301–1305. doi: 10.1074/jbc.274.3.1301. [DOI] [PubMed] [Google Scholar]
- 55.Kane PM. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase, Microbiol. Mol Biol Rev. 2006;70:177–191. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beyenbach KW, Wieczorek H. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J Exp Biol. 2006;209:577–589. doi: 10.1242/jeb.02014. [DOI] [PubMed] [Google Scholar]
- 57.Sautin YY, Lu M, Gaugler A, Zhang L, Gluck SL. Phosphatidylinositol 3-kinase-mediated effects of glucose on vacuolar H+-ATPase assembly, translocation, and acidification of intracellular compartments in renal epithelial cells. Mol Cell Biol. 2005;25:575–589. doi: 10.1128/MCB.25.2.575-589.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. Activation of lysosomal function during dendritic cell maturation. Science. 2003;299:1400–1403. doi: 10.1126/science.1080106. [DOI] [PubMed] [Google Scholar]
- 59.Kane PM. Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J Biol Chem. 1995;270:17025–17032. [PubMed] [Google Scholar]
- 60.Parra KJ, Kane PM. Reversible association between the V1 and V0 domains of yeast vacuolar H+-ATPase is an unconventional glucose-induced effect. Mol Cell Biol. 1998;18:7064–7074. doi: 10.1128/mcb.18.12.7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qi J, Forgac M. Cellular environment is important in controlling V-ATPase dissociation and its dependence on activity. J Biol Chem. 2007;282:24743–24751. doi: 10.1074/jbc.M700663200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu T, Forgac M. Microtubules are involved in glucose-dependent dissociation of the yeast vacuolar [H+]-ATPase in vivo. J Biol Chem. 2001;276:24855–24861. doi: 10.1074/jbc.M100637200. [DOI] [PubMed] [Google Scholar]
- 63.Seol JH, Shevchenko A, Shevchenko A, Deshaies RJ. Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat Cell Biol. 2001;3:384–391. doi: 10.1038/35070067. [DOI] [PubMed] [Google Scholar]
- 64.Smardon AM, Tarsio M, Kane PM. The RAVE complex is essential for stable assembly of the yeast V-ATPase. J Biol Chem. 2002;277:13831–13839. doi: 10.1074/jbc.M200682200. [DOI] [PubMed] [Google Scholar]
- 65.Kawasaki-Nishi S, Nishi T, Forgac M. Yeast V-ATPase complexes containing different isoforms of the 100-kDa a-subunit differ in coupling efficiency and in vivo dissociation. J Biol Chem. 2001;276:17941–17948. doi: 10.1074/jbc.M010790200. [DOI] [PubMed] [Google Scholar]
- 66.Manolson MF, Wu B, Proteau D, Taillon BE, Roberts BT, Hoyt MA, Jones EW. STV1 gene encodes functional homologue of 95-kDa yeast vacuolar H(+)-ATPase subunit Vph1p. J Biol Chem. 1994;269:14064–14074. [PubMed] [Google Scholar]
- 67.Shao E, Forgac M. Involvement of the nonhomologous region of subunit A of the yeast V-ATPase in coupling and in vivo dissociation. J Biol Chem. 2004;279:48663–48670. doi: 10.1074/jbc.M408278200. [DOI] [PubMed] [Google Scholar]
- 68.Shao E, Nishi T, Kawasaki-Nishi S, Forgac M. Mutational analysis of the non-homologous region of subunit A of the yeast V-ATPase. J Biol Chem. 2003;278:12985–12991. doi: 10.1074/jbc.M212096200. [DOI] [PubMed] [Google Scholar]
- 69.Parra KJ, Keenan KL, Kane PM. The H subunit (Vma13p) of the yeast V-ATPase inhibits the ATPase activity of cytosolic V1 complexes. J Biol Chem. 2000;275:21761–21767. doi: 10.1074/jbc.M002305200. [DOI] [PubMed] [Google Scholar]
- 70.Adachi I, Arai H, Pimental R, Forgac M. Proteolysis and orientation on reconstitution of the coated vesicle proton pump. J Biol Chem. 1990;265:960–966. [PubMed] [Google Scholar]
- 71.Arai H, Pink S, Forgac M. Interaction of anions and ATP with the coated vesicle proton pump. Biochemistry. 1989;28:3075–3082. doi: 10.1021/bi00433a051. [DOI] [PubMed] [Google Scholar]
- 72.Xu T, Forgac M. Subunit D (Vma8p) of the yeast vacuolar H+-ATPase plays a role in coupling of proton transport and ATP hydrolysis. J Biol Chem. 2000;275:22075–22081. doi: 10.1074/jbc.M002983200. [DOI] [PubMed] [Google Scholar]
- 73.Kawasaki-Nishi S, Bowers K, Nishi T, Forgac M, Stevens TH. The amino-terminal domain of the vacuolar proton-translocating ATPase a subunit controls targeting and in vivo dissociation, and the carboxyl-terminal domain affects coupling of proton transport and ATP hydrolysis. J Biol Chem. 2001;276:47411–47420. doi: 10.1074/jbc.M108310200. [DOI] [PubMed] [Google Scholar]
- 74.Mulberg AE, Tulk BM, Forgac M. Modulation of coated vesicle chloride channel activity and acidification by reversible protein kinase A-dependent phosphorylation. J Biol Chem. 1991;266:20590–20593. [PubMed] [Google Scholar]
- 75.Brett CL, Tukaye DN, Mukherjee S, Rao R. The yeast endosomal Na+K+/H+ exchanger Nhx1 regulates cellular pH to control vesicle trafficking. Mol Biol Cell. 2005;16:1396–1405. doi: 10.1091/mbc.E04-11-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boyer PD. The binding change mechanism for ATP synthase--some probabilities and possibilities. Biochim Biophys Acta. 1993;1140:215–250. doi: 10.1016/0005-2728(93)90063-l. [DOI] [PubMed] [Google Scholar]