A vault from rat liver was crystallized in space group C2. Rotational symmetry searches indicated that the particle has 39-fold dihedral symmetry.
Keywords: vault, ribonucleoproteins
Abstract
Vault is a 12.9 MDa ribonucleoprotein particle with a barrel-like shape, two protruding caps and an invaginated waist structure that is highly conserved in a wide variety of eukaryotes. Multimerization of the major vault protein (MVP) is sufficient to assemble the entire exterior shell of the barrel-shaped vault particle. Multiple copies of two additional proteins, vault poly(ADP-ribose) polymerase (VPARP) and telomerase-associated protein 1 (TEP1), as well as a small vault RNA (vRNA), are also associated with vault. Here, the crystallization of vault particles is reported. The crystals belong to space group C2, with unit-cell parameters a = 708.0, b = 385.0, c = 602.9 Å, β = 124.8°. Rotational symmetry searches based on the R factor and correlation coefficient from noncrystallographic symmetry (NCS) averaging indicated that the particle has 39-fold dihedral symmetry.
1. Introduction
Vault is a large ribonucleoprotein particle with a unique barrel-shaped structure that is widely conserved among eukaryotes (Kedersha & Rome, 1986 ▶). Vault has a molecular weight of 12.9 MDa and overall dimensions of 400 × 400 × 700 Å, both of which were determined by scanning transmission electron microscopy (Kedersha et al., 1991 ▶). In mammals, vaults contain three major proteins: the 100 kDa major vault protein (MVP; Kedersha & Rome, 1986 ▶), the 193 kDa vault poly(ADP-ribose) polymerase (VPARP; Kickhoefer, Siva et al., 1999 ▶) and the 240 kDa telomerase-associated protein 1 (TEP1; Kickhoefer, Stephen et al., 1999 ▶). Additionally, the complex contains vRNA, a small RNA molecule (Kickhoefer et al., 1993 ▶).
MVP accounts for more than 75% of the particle mass (Kedersha et al., 1991 ▶). Interestingly, expression of MVP alone in insect cells leads to the assembly of recombinant MVP-only vault, demonstrating that the multimerization of this single protein is sufficient to form the exterior shell of the vault particle (Stephen et al., 2001 ▶). VPARP is a poly(ADP-ribose) polymerase (PARP) family member that possesses a canonical PARP domain (Kickhoefer, Siva et al., 1999 ▶) and is capable of ADP-ribosylating itself and MVP. However, the physiological roles of these modifications are still unknown. Although vault particles do not show telomerase activity, experimental data suggest that TEP1 is important for vRNA binding and stabilization of the vault complex (Kickhoefer et al., 2001 ▶). Although vRNA comprises less than 5% of the vault mass, it forms important interactions with other cellular components (Kickhoefer et al., 2002 ▶) as well as with anticancer antibiotics such as mitoxantrone (Gopinath et al., 2005 ▶).
In some tumour and immune cells, vault expression correlates with resistance to a variety of structurally and chemically unrelated cytotoxins (Izquierdo et al., 1996 ▶; Kickhoefer et al., 1998 ▶; Scheffer et al., 1995 ▶; Scheper et al., 1993 ▶; Schroeijers et al., 2002 ▶). The hollow nature of vaults, as well as their subcellular localization at multiple sites, including the nucleolus (Hamill & Suprenant, 1997 ▶), nuclear pore complexes (Chugani et al., 1993 ▶) and the cytosol, in which the particles transiently interact with microtubules (Eichenmüller et al., 2003 ▶), point to a role in nucleocytoplasmic transport. In MVP-knockout mice, however, MVP−/− cells do not show increased sensitivity to various cytostatic agents (Mossink et al., 2002 ▶). Thus, the function of the vault particle has remained elusive.
Freeze-etch electron microscopy of vault from rat liver has revealed that each vault particle appears as an open pair of flower-like structures, each with eight petals (Kedersha et al., 1991 ▶). Imposing eightfold symmetry, Kong et al. (1999 ▶) analyzed the structure of rat vault using a cryoelectron-microscopic method. This revealed the reconstructed structure of the rat vault to be a hollow barrel-like structure with two protruding caps and an invaginated waist; 96 copies of MVP were predicted to form the barrel-shaped core of the particle. Difference mapping between RNase-treated and intact vaults revealed that vRNA is localized at the end of the vault caps (Kong et al., 2000 ▶). This led the authors to propose a model of the complex that has eightfold dihedral symmetry (point group D8) and that contains one copy of TEP1, four copies of VPARP and three or more copies of vRNA in each cap. One reconstituted structure of recombinant vault with a cysteine-rich tag displayed 48-fold rotational symmetry, even though only eightfold symmetry was imposed during image processing (Mikyas et al., 2004 ▶). The reconstituted structures of the recombinant vault particles have led the authors to propose a model with D 48 point-group symmetry and with the MVP N-termini located in the vault midsection.
Rat vault exhibits a crystallographic sixfold dihedral symmetry as a two-dimensional crystal, whereas monkey vault exhibits a crystallographic threefold dihedral symmetry as a three-dimensional crystal (Querol-Audí et al., 2005 ▶). These sixfold and threefold rotational symmetries agree with the 48-fold symmetry for reconstituted recombinant vault (Mikyas et al., 2004 ▶).
In this study, we crystallized vault particles from rat liver in several crystalline forms. Crystallographic studies of a monoclinic crystal, which did not have crystallographic threefold, fourfold and sixfold axes, are described in this paper. Our results demonstrate that vault has a 39-fold rotational axis.
2. Materials and methods
2.1. Purification and crystallization
Vault was purified from rat liver as described by Kong et al. (1999 ▶). The protein concentration was determined using a BCA Protein Assay Kit (Pierce). The vault purified from a CsCl density-gradient fraction was dialyzed against buffer containing 50 mM Tris pH 7.5, 75 mM NaCl and 1.5 mM MgCl2 and then concentrated to 10–80 mg ml−1 in an ultrafiltration cell (Orbital Biosciences, 150 kDa cutoff). Purified particles were verified by negative staining with 2% uranyl acetate followed by examination under an electron microscope (H-7650; Hitachi) and also by SDS–PAGE.
Vault was crystallized using the hanging-drop vapor-diffusion method at 277 or 288 K. A hanging drop was prepared by mixing equal volumes of vault solution and precipitant solution containing polyethylene glycol (PEG).
2.2. X-ray diffraction data collection
Prior to X-ray diffraction at cryogenic temperatures, crystals were soaked in a solution containing a higher concentration of PEG than that in the crystallization precipitant solution, which prevented the formation of ice at cryogenic temperatures. The PEG concentration was increased stepwise from that of the crystallization precipitant solution until it reached the final concentration. The crystals were frozen by flash-cooling after incubation in the PEG solution overnight. X-ray experiments were performed at 100 K with a DIP6040 imaging-plate detector (MAC Science/Bruker AXS) on the BL44XU beamline of the SPring-8 synchrotron facility (Harima, Hyogo, Japan). The oscillation angle of each shot was 0.3°. After five successive shots had been taken without translation of the crystal, the crystal was translated by 100 µm along its rotation axis to reduce radiation damage (the X-ray beam size was 50 × 50 µm). Diffraction data were integrated using MOSFLM (Leslie, 1992 ▶) and scaled using SCALA (Evans, 1997 ▶). Observed structure factors were evaluated with TRUNCATE (French & Wilson, 1978 ▶).
2.3. Electron-microscopic analysis
Purified protein was embedded in vitreous ice by rapid plunging into liquid ethane using a Leica EM CPC (Leica Microsystems, Wetzlar, Germany). All images were acquired using a JEOL3000SFF electron microscope (Jeol; Tokyo, Japan) equipped with a liquid-helium stage and operated at 200 kV. Electron micrographs were recorded on Kodak SO-163 film at a magnification of ×40 000. Micrographs were scanned on a Nikon SUPER COOLSCAN9000 at 4000 dpi, which corresponded to 6.35 µm per pixel. Three-dimensional reconstruction of vault particles to produce an initiator model was performed using IMAGIC (van Heel et al., 1996 ▶). D 6 symmetry was assumed throughout the processing. The effective resolution of the final three-dimensional reconstruction was 37 Å based on a 0.5 cutoff of the Fourier shell correlation (van Heel, 1987 ▶). Low-pass filtered three-dimensional reconstruction to 37 Å resolution was performed following X-ray crystallographic analysis. Prior to use of the reconstituted structure for molecular-replacement analysis, it was modified by averaging the electron densities at equal distances from the central axis of the vault particle. Consequently, the modified model (EM model) had a circular symmetry.
2.4. Rotational symmetry search
Self-rotation functions were calculated to determine the rotational symmetry elements of the vault particles with the fast-rotation function and slow-rotation function programs of GLRF (Tong & Rossmann, 1997 ▶). The Patterson integration radius was 50 Å and data in the resolution range 50–10 Å were used.
In addition to rotation-function calculations, phase refinements from NCS averaging were used to confirm the rotational symmetry elements. NCS averaging was performed using the RAVE package from the Uppsala Software Factory (Kleywegt et al., 2001 ▶).
3. Results and discussion
3.1. Purification and crystallization of vault
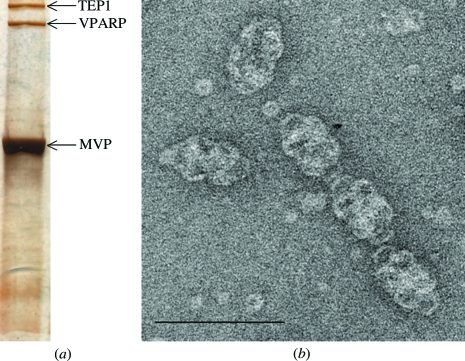
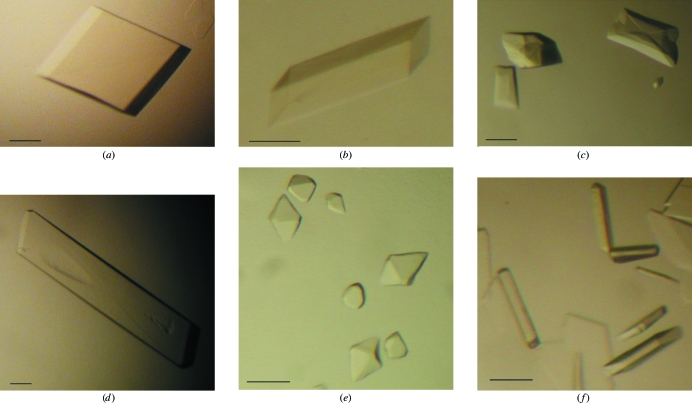
The vault samples obtained following several density-gradient centrifugations of rat liver samples were highly pure (Figs. 1 ▶ a and 1 ▶ b). Negatively stained electron-microscopic images showed barrel-like structures as previously reported (Kedersha & Rome, 1986 ▶). Purified vault particles were subjected to various crystallization conditions. Six different types of crystals were obtained with a variety of precipitant conditions. These crystals are referred to as A, B, C, D, E and F in this paper (Fig. 2 ▶). Crystallization conditions are given in Table 1 ▶. Thick crystals were grown in conditions that employed lower molecular-weight PEGs. Because we successfully identified freezing procedures for crystals B, C, D and E, X-ray experiments were performed with these crystals. Crystal B belonged to space group P1, with unit-cell parameters a = 442.0, b = 598.7, c = 638.9 Å, α = 68.1, β = 78.2, γ = 70.3°; they diffracted to 10 Å resolution. We were not able to improve the quality of crystal B despite making adjustments to the original crystallization conditions. Crystal C was a monoclinic crystal belonging to space group P21, with unit-cell parameters a = 390, b = 545, c = 672 Å, β = 94°. The crystallization conditions for crystal D were almost identical to those described by Querol-Audí et al. (2005 ▶). In 2–3 weeks, these crystals grew to as large as 1 mm in their longest dimension and diffracted to 8.0 Å resolution. The crystals belonged to space group P21, with unit-cell parameters a = 599.8, b = 385.0, c = 616.4 Å, β = 109.1°. The orientation of the vault particles in these crystals changed when they were soaked in cryoprotectant using another buffer condition, which caused the crystals to adopt space group C2, with unit-cell parameters a = 708.0, b = 385.0, c = 602.9 Å, β = 124.8°. Crystal E was a tetragonal crystal belonging to space group P4, with unit-cell parameters a = b = 625, c = 767 Å. Of these crystals, crystal D in space group C2 produced the best diffraction results; thus, the following descriptions are restricted to that crystal form.
Figure 1.
(a) SDS–PAGE of purified rat vault. Gels are silver-stained. (b) Negatively stained rat vault visualized by transmission electron microscopy. The sample was adsorbed onto a carbon-coated grid, stained with 2% uranyl acetate and air-dried. Scale bar, 100 nm.
Figure 2.
Photographs of vault crystals. (a) Crystal A. (b) Triclinic crystal B. (c) Monoclinic crystal C. (d) Monoclinic crystal D. (e) Tetragonal crystal E. (f) Crystal F. Scale bars, 0.1 mm.
Table 1. Crystallization conditions.
| Crystal | Protein concentration† (mg ml−1) | Reservoir solution |
|---|---|---|
| A | 40 | 15% PEG 400, 0.8 M NaCl, 50 mM Li2SO4 in 50 mM Tris–HCl pH 8.0 |
| B | 5 | 4% PEG 4000, 50 mM KCl, 25% ethylene glycol (EG) in 50 mM Tris–HCl pH 7.0 |
| C | 5 | 4% PEG 2000, 50 mM KCl, 25% EG in 50 mM Tris–HCl pH 7.0 |
| D | 40 | 2.4% PEG 4000, 0.8 M NaCl, 50 mM Li2SO4 in 50 mM Tris–HCl pH 8.0 |
| E | 5 | 4% PEG 3350, 50 mM ammonium acetate, 25% EG |
| F | 5 | 3% PEG 8000, 50 mM KCl, 25% EG in 50 mM Tris–HCl pH 7.0 |
Vault was dissolved in buffer containing 50 mM Tris–HCl pH 7.5, 75 mM NaCl and 1.5 mM MgCl2. The crystallization temperature for crystals A, D and E was 288 K and that for crystals B, C and F was 277 K.
3.2. Symmetry of vault
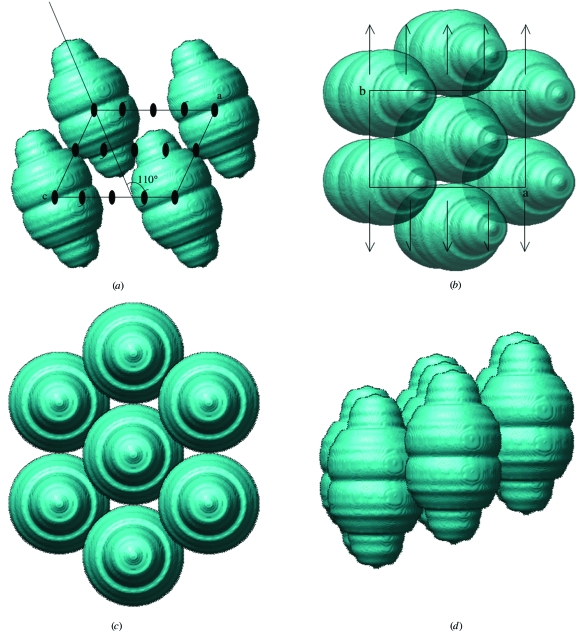
A total of 38 crystals were used to collect 1175 frames of diffraction images. The statistics of the intensity data for native crystals are given in Table 2 ▶. Intensity data to 10.0 Å resolution were acquired with 99.9% completeness and with an R merge value of 0.102. The native Patterson function calculated at 20 Å resolution showed no prominent peaks except at the origin and at (1/2, 1/2, 0), corresponding to the face-centred symmetry operation. The self-rotation function (described precisely below) located the principal rotation axis on the ac plane and a twofold axis parallel to the b axis. Particles with dimensions 400 × 400 × 700 Å were well packed in the crystalline lattice (Fig. 3 ▶). When half of a particle was packed in an asymmetric unit, the particles packed tightly and each particle did not markedly overlap with its neighbours in the crystal. Consequently, a V M value of 5.18 Å3 Da−1 was confirmed, which is outside the typical range for soluble proteins (2.5–3.5 Å3 Da−1; Matthews, 1968 ▶), probably owing to the hollow nature of the particle.
Table 2. Intensity data.
Values in parentheses are for the highest shell.
| Resolution (Å) | 210–10.0 (10.5–10.0) |
| Total No. of frames | 1175 |
| Total No. of reflections | 613655 |
| Unique reflections | 71967 |
| Completeness (%) | 99.9 (100.0) |
| Multiplicity | 8.5 (8.8) |
| I/σ(I) | 18.4 (17.7) |
| Rmerge† | 0.102 (0.113) |
R
merge = 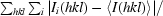
 , where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the averaged intensity over equivalent reflections from different measurements.
, where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the averaged intensity over equivalent reflections from different measurements.
Figure 3.
Packing of vault particles with dimensions 400 × 400 × 700 Å in crystal D with unit-cell parameters a = 708.0, b = 385.0, c = 602.9 Å, β = 124.8°. Packing is shown (a) in the ac plane, (b) in the ab plane, (c) parallel to the N-fold rotation axis and (d) perpendicular to the N-fold rotation axis. A surface-rendered three-dimensional image of vault was obtained from cryoelectron microscopy and single-particle reconstruction.
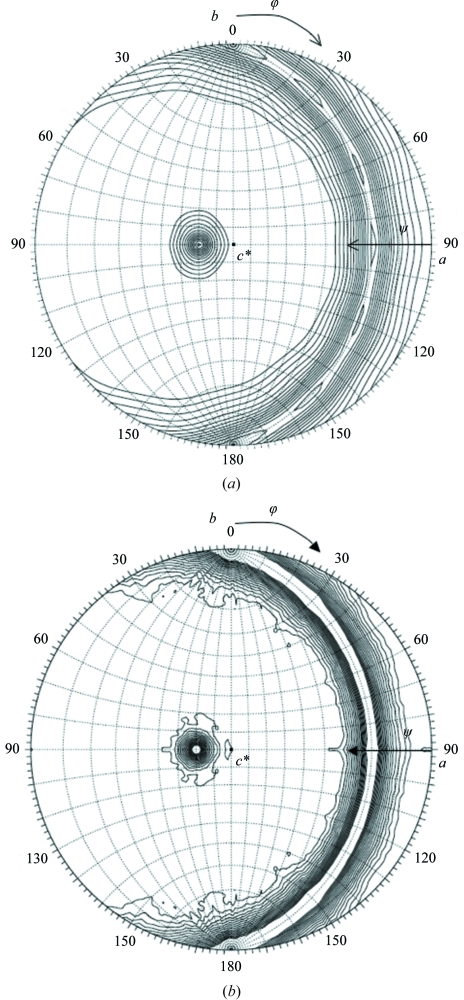
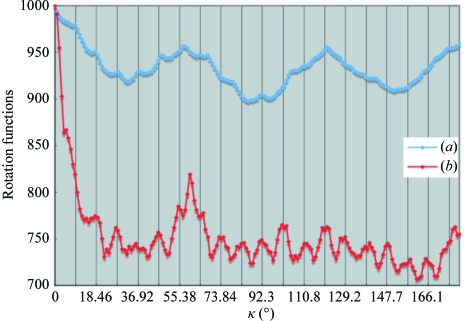
The self-rotation functions were calculated at 10 Å resolution using the fast-rotation function and the slow-rotation function software GLRF (Tong & Rossmann, 1997 ▶). Using reflections in the resolution range 50–10 Å, each rotation function was calculated at an equal step of 1.0° with an integration radius of 50 Å. Twofold rotational symmetries are shown in κ = 180° sections of stereo diagrams in Fig. 4 ▶. Fig. 5 ▶ shows plots of the rotation functions against κ at (ϕ, ψ) = (90°, 110°). There were multiple twofold symmetry axes with a strong peak in the stereographic drawings of the κ = 180° section; the axes were perpendicular to the rotation axis at (ϕ, ψ) = (90°, 110°). The fast-rotation function is plotted against κ at (ϕ, ψ) = (90°, 110°) with broad peaks at κ = 60°, 120° and 180°, indicating that the particle had a sixfold rotation symmetry axis at (ϕ, ψ) = (90°, 110°) (Fig. 5 ▶). Multiple twofold symmetry axes perpendicular to the sixfold rotation axis were detected every 30° (Fig. 4 ▶ a). The fast-rotation function calculation at 10 Å resolution indicated that the particle exhibited point group D 6. The section at κ = 180° (Fig. 4 ▶ b) and the plot against κ at (ϕ, ψ) = (90°, 110°) (Fig. 5 ▶) were calculated using the slow-rotation function. Although Fig. 4 ▶(b) shows multiple twofold axes perpendicular to the rotational axis at (ϕ, ψ) = (90°, 110°) as observed in Fig. 4 ▶(a), six peaks were not detected, which contrasted with the results obtained with the fast-rotation function. The slow-rotation function plot shows a number of peaks at intervals of 360°/39, indicating 39-fold symmetry. However, there is an additional peak at 60° that would be in conflict with 39-fold symmetry. The plot did not show a clear peak at κ = 180°, which was one of the essential peaks for D 6 symmetry. Consequently, the rotational symmetry of vault was not consistently determined using the two rotation functions.
Figure 4.
Stereo diagrams of (a) the fast-rotation function and (b) the slow-rotation function for twofold rotational symmetry (κ = 180° section) in the 50–10 Å resolution range. Contours were drawn at equal intervals of 0.5σ.
Figure 5.
(a) The fast-rotation function and (b) the slow-rotation function plotted against κ at (ϕ, ψ) = (90°, 110°). The integration radii of the calculations were five times the highest resolution limits. Each rotation function was calculated with 71 269 reflections in the resolution range 50–10 Å. The height of the crystallographic symmetry was normalized to 1000.
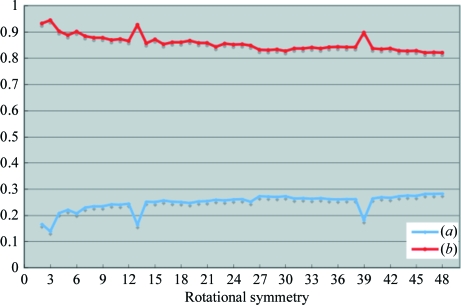
To determine the rotational symmetry at (ϕ, ϕ) = (90°, 110°), phase refinements were performed using NCS averaging by assuming twofold to 48-fold rotational symmetry. Initial phases were calculated at 30 Å resolution using the EM model and phase extension was performed to 10 Å resolution. R factors and correlation coefficients between observed and calculated structure factors from each NCS averaging are plotted in Fig. 6 ▶. The R factors at the threefold, 13-fold and 39-fold symmetries were significantly smaller than their neighbouring symmetries and the correlation coefficients exhibited significantly higher peaks at the threefold, 13-fold and 39-fold symmetries. Although an R factor and correlation coefficient for sixfold symmetry were observed in local peaks, these criteria were less significant than those for the threefold, 13-fold and 39-fold symmetries.
Figure 6.
R factors (a) and correlation coefficients (b) plotted against various NCS averaging values at 10 Å resolution.
Fig. 3 ▶ shows the packing of vaults in the crystal. Because the large vault particles (400 × 40 × 700 Å) were very thin, at about 30 Å, the ratio of the cross-vector and the self-vector of the Patterson function within a radius of 50 Å is higher than that usually observed for crystals consisting of spherical proteins. The large peak at κ = 60° detected with both the fast-rotation and slow-rotation function may have been caused by the packing of the vault particles, which exhibited a hexagonal arrangement in the projection along the principal rotational axis (Fig. 3 ▶ c).
Rat liver vault in a crystalline state showed D 39 symmetry. To our knowledge, such 39-fold symmetry has not been previously detected in any supramolecular complex. The eightfold symmetry observed in the half vault particle from rat (Kedersha et al., 1991 ▶), the 48-fold rotation symmetry of the rat recombinant vault particle elucidated by cryoelectron microscopy (Mikyas et al., 2004 ▶) and the D 6 symmetry of rat vault (Querol-Audí et al., 2005 ▶) were not detected in the present study of rat liver vault. Of note, three-dimensional crystals of monkey vault exhibit crystallographic D 3 symmetry (Querol-Audí et al., 2005 ▶) consistent with the 39-fold symmetry observed in the present study.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research in a Priority Area (10188101 and 10179101 to TT) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by a Ground-based Research Announcement for Space Utilization provided by the Japan Space Forum (to TT). This work was also supported in part by a Grant-in-Aid for Young Scientists (B) 19770082 (to HT) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Chugani, D. C., Rome, L. H. & Kedersha, N. L. (1993). J. Cell Sci.106, 23–29. [DOI] [PubMed]
- Eichenmüller, B., Kedersha, N., Solovyeva, E., Everly, P., Lang, J., Himes, R. H. & Suprenant, K. A. (2003). Cell Motil. Cytoskeleton, 56, 225–236. [DOI] [PubMed]
- Evans, P. R. (1997). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.33, 22–24.
- French, S. & Wilson, K. (1978). Acta Cryst. A34, 517–525.
- Gopinath, S. C., Matsugami, A., Katahira, M. & Kumar, P. K. (2005). Nucleic Acids Res.33, 4874–4881. [DOI] [PMC free article] [PubMed]
- Hamill, D. R. & Suprenant, K. A. (1997). Dev. Biol.190, 117–128. [DOI] [PubMed]
- Heel, M. van (1987). Ultramicroscopy, 21, 111–123. [DOI] [PubMed]
- Heel, M. van, Harauz, G., Orlova, E. V., Schmidt, R. & Schatz, M. (1996). J. Struct. Biol.116, 17–24. [DOI] [PubMed]
- Izquierdo, M. A., Scheffer, G. L., Flens, M. J., Schoemaker, R. H., Rome, L. H. & Scheper, R. J. (1996). Cytotechnology, 19, 191–197. [DOI] [PubMed]
- Kedersha, N. L., Heuser, J. E., Chugani, D. C. & Rome, L. H. (1991). J. Cell Biol.112, 225–235. [DOI] [PMC free article] [PubMed]
- Kedersha, N. L. & Rome, L. H. (1986). J. Cell Biol.110, 895–901. [DOI] [PMC free article] [PubMed]
- Kickhoefer, V. A., Liu, Y., Kong, L. B., Snow, B. E., Stewart, P. L., Harrington, L. & Rome, L. H. (2001). J. Cell Biol.152, 157–164. [DOI] [PMC free article] [PubMed]
- Kickhoefer, V. A., Poderychi, M. J., Chan, E. K. & Rome, L. H. (2002). J. Biol. Chem.277, 41282–41286. [DOI] [PubMed]
- Kickhoefer, V. A., Rajavel, K. S., Scheffer, G. L., Dalton, W. S., Scheper, R. J. & Rome, L. H. (1998). J. Biol. Chem.273, 8971–8974. [DOI] [PubMed]
- Kickhoefer, V. A., Searles, R. P., Kedersha, N. L., Garber, M. E., Johnson, D. L. & Rome, L. H. (1993). J. Biol. Chem.268, 7868–7873. [PubMed]
- Kickhoefer, V. A., Siva, A. C., Kedersha, N. L., Inman, E. M., Ruland, C., Streuli, M. & Rome, L. H. (1999). J. Cell Biol.146, 917–928. [DOI] [PMC free article] [PubMed]
- Kickhoefer, V. A., Stephen, A. G., Harrington, L., Robinson, M. O. & Rome, L. H. (1999). J. Biol. Chem.274, 32712–32717. [DOI] [PubMed]
- Kleywegt, G. J., Zou, J.-Y., Kjeldgaard, M. & Jones, T. A. (2001). International Tables for Crystallography, Vol. F, edited by M. G. Rossmann & E. Arnold, pp. 354–355. Dordrecht: Kluwer Academic Publishers.
- Kong, L. B., Siva, A. C., Kickhoefer, V. A., Rome, L. H. & Stewart, P. L. (2000). RNA, 6, 890–900. [DOI] [PMC free article] [PubMed]
- Kong, L. B., Siva, A. C., Rome, L. H. & Stewart, P. L. (1999). Structure, 7, 371–379. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Mikyas, Y., Makabi, M., Raval-Fernandes, S., Harrington, L., Kickhoefer, V. A., Rome, L. H. & Stewart, P. L. (2004). J. Mol. Biol.344, 91–105. [DOI] [PubMed]
- Mossink, M. H., van Zon, A., Franzel-Luiten, E., Schoester, M., Kickhoefer, V. A., Scheffer, G. L., Scheper, R. J., Sonneveld, P. & Wiemer, E. A. (2002). Cancer Res.62, 7298–7304. [PubMed]
- Querol-Audí, J., Perez-Luque, R., Fita, I., Lopéz-Iglesias, C., Castón, J. R., Carrascosa, J. L. & Verdaguer, N. (2005). J. Struct. Biol.151, 111–115. [DOI] [PubMed]
- Scheffer, G. L., Wijingaard, P. L., Flens, M. J., Izquierdo, M. A., Slovak, M. L., Pinedo, H. M., Meijer, C. J., Clevers, H. C. & Scheper, R. J. (1995). Nature Med.1, 578–582. [DOI] [PubMed]
- Scheper, R. J., Broxterman, H. J., Scheffer, G. L., Kaaijk, P., Dalton, W. S., van Heijningen, T. H., van Kalken, C. K., Slovak, M. L., de Vries, E. G., van der Valk, P., Meijer, C. J. & Pinedo, H. M. (1993). Cancer Res.53, 1475–1479. [PubMed]
- Schroeijers, A. B., Reurs, A. W., Scheffer, G. L., Stam, A. G., de Jong, M. C., Rustemeyer, T., Wiemer, E. A., de Gruijl, T. D. & Scheper, R. J. (2002). J. Immunol.168, 1572–1578. [DOI] [PubMed]
- Stephen, A. G., Raval-Fernandes, S., Huynh, T., Torres, M., Kickhoefer, V. A. & Rome, L. H. (2001). J. Biol. Chem.276, 23217–23220. [DOI] [PubMed]
- Tong, L. & Rossmann, M. G. (1997). Methods Enzymol.276, 594–611. [PubMed]