Abstract
The purification of primitive human hematopoietic stem cells has been impaired by the absence of repopulation assays. By using a stringent two-step strategy involving depletion of lineage-positive cells followed by fluorescence-activated cell sorting, we have purified a cell population that is highly enriched for cells capable of multilineage repopulation in nonobese diabetic/severe combined immunodeficient (NOD/SCID) recipients. These SCID-repopulating cells (SRCs) were exclusively found in a cell fraction that expressed high levels of CD34 and no CD38. Through limiting dilution analysis using Poisson statistics, we calculated a frequency of 1 SRC in 617 CD34+ CD38− cells. The highly purified SRC were capable of extensive proliferation in NOD/SCID mice. Mice transplanted with 1 SRC (at limiting cell doses) were able to produce approximately 400,000 progeny 6 weeks after the transplant. Detailed flow cytometric analysis of the marrow of highly engrafted mice demonstrated both lymphoid and myeloid differentiation, as well as the retention of a significant fraction of CD34+ CD38− cells. These highly purified fractions should be useful for identification of the cellular and molecular mechanisms that regulate primitive human hematopoietic cells. Moreover, the ability to detect and purify primitive cells provides a means to develop conditions for maintaining and/or expanding these cells during in vitro culture.
The hematopoietic system in mammals is composed of a heterogeneous population of cells that range in function from mature cells with limited proliferative potential to pluripotent stem cells with extensive proliferative, differentiative, and self-renewal capacities (1–3). In addition to the key role that stem cells play in maintaining the hematopoietic system, many clinical applications are being developed to target stem cells, including purging of neoplastic cells (4), gene therapy (5), ex vivo expansion (6), and use for cord blood (CB) transplantation (7). Progress to characterize human stem cells has been impaired by their rarity in normal hematopoietic tissue, the absence of a distinctive phenotype, and the absence of a functional repopulation assay. Hence, research to purify and characterize stem cells in humans has relied primarily on surrogate in vitro assays (8). These include clonogenic assays that detect colony-forming cells (CFCs) that have limited proliferative potential and are already committed to a specific myeloid lineage and long-term cultures that detect cells capable of initiating hematopoiesis on stroma (long-term culture-initiating cells, LTC-ICs) for up to 60 days (8). Recent work suggests that a small subpopulation of LTC-ICs have more extensive proliferative capacity and can be maintained in extended LTC for up to 100 days (extended LTC-ICs, ELTC-ICs) (9). However, since growth conditions of human LTCs do not support the development of all blood lineages and these assays are unable to assess repopulating capacity, little is known about the relationship of LTC-ICs and pluripotent stem cells. Ultimately, the only conclusive assay for stem cells is their ability to reconstitute the entire hematopoietic system after transplantation.
On the basis of in vitro assays and human allogeneic transplantation, the most primitive hematopoietic cells are enriched in the CD34+ fraction (10). However, CD34+ cells, which make up approximately 1% of all normal human bone marrow (BM) cells, are heterogeneous and include committed progenitors (9). By assessing coexpression of other cell surface markers, it is possible to obtain cell fractions that are more highly enriched for primitive cells (9, 11, 12). Expression of CD38 is correlated with increased differentiation; only 1–10% of CD34+ cells do not express CD38 (9). Both the CD34+ CD38− and the CD34+ CD38+ fractions contain CFCs and LTC-ICs (9). Although LTC-ICs are more enriched in the CD34+ CD38− fraction, the absolute number of LTC-ICs in the CD34+ CD38+ fraction is higher since the proportion of CD34+ CD38+ cells in BM is larger (13). CD34+ CD38− cells do not express lineage markers, are largely quiescent prior to cytokine stimulation, and are greatly enriched for ELTC-ICs (9, 14, 15).
We have recently developed an in vivo functional assay for primitive human hematopoietic cells from BM and CB based on their ability to repopulate the BM of immune-deficient nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice after intravenous transplantation (16, 17). We have defined the engrafting cell as a SCID-repopulating cell (SRC) (18–20). Several lines of evidence indicate that the SRC is more primitive than cells detected with in vitro assays (20). Kinetic studies suggest that the rapid increase in LTC-ICs and CD34+ Thy-1lo cells in the bone marrow of NOD/SCID mice over the first 4 weeks after transplantation is due to proliferation of a more primitive cell (SRC) (26). Quantitative analysis demonstrates that the SRCs are present at lower frequency compared with LTC-ICs (27), and cell fractionation and gene marking studies have shown that SRCs are distinct from CFCs and most LTC-ICs (19). We report herein detailed fractionation of human BM and CB based on multiparameter flow sorting and evaluation of each fraction for SRC activity. The SRCs were found exclusively in the CD34+ CD38− fraction and frequency analysis demonstrated a 1,500-fold purification of SRC in this population. Moreover, the highly purified SRCs were capable of extensive proliferation and differentiation in vivo.
MATERIALS AND METHODS
Human Cells.
Human BM cells were obtained from harvests of normal donors for allogeneic transplantation in accordance to procedures approved by the Human Experimentation Committee at the Ontario Cancer Institute, Toronto, Ontario. Samples of CB were obtained from discarded placental and umbilical tissues. Both BM and CB samples were diluted 1:3 in Iscove’s modified Dulbecco’s medium (IMDM, GIBCO/BRL) and enriched for mononuclear cells by centrifugation on Ficoll/Paque (Pharmacia).
Purification of Cell Populations.
As depicted in Fig. 1, mononuclear cells were stained with a mixture of lineage-specific antibodies provided by the manufacturer (Stem Cell Technologies, Paisley, Scotland), followed by addition of secondary antibody conjugated to metal colloid. Cells were then eluted through a magnetized column to enrich for cells not expressing lineage markers (Lin−); the cells remaining on the column (Lin+) were then washed off the demagnetized column with phosphate-buffered saline (PBS) containing 5% fetal calf serum (FCS). These cell fractions were then stained with anti-human CD34-fluorescein isothiocyanate (FITC) and anti-human CD38-phycoerythrin (PE), analyzed, and sorted on a FACStar Plus (Becton Dickinson) equipped with 5-W argon and 30-mW helium neon lasers. Fluorescence of FITC and PE excited at 482 nm (0.38 W) and 633 nm (30 mW), respectively, as well as known forward and side scatter properties of normal live human hematopoietic cells, were used to establish sorting gates (see Fig. 1). Data acquisition and analysis were performed with lysis ii software (Becton Dickinson).
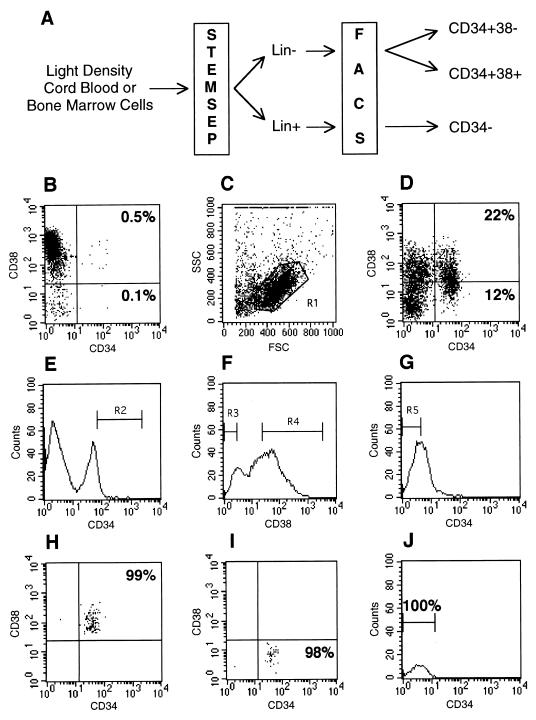
Figure 1.
Purification of CD34+ CD38−, CD34+ CD38+, and CD34− cell subsets. (A) Schematic diagram depicting the strategy used to obtain highly purified cells. Light density mononuclear cells from whole CB or BM were separated into lineage negative (Lin−) or positive (Lin+) fractions by staining with monoclonal antibodies against lineage antigens, followed by purification on an immunomagnetic column (StemSep). Lin− cells were stained with monoclonal antibodies specific for CD34 and CD38 and sorted on a FACStar Plus cell sorter into CD34+ CD38− and CD34+ CD38+ populations. Lin+ cells were stained with anti-CD34 and sorted for the CD34− population. (B) Expression of CD34 and CD38 on mononuclear cord blood cells analyzed by flow cytometry. The CD34+ cells that also express CD38 make up 0.5% of the cells in the lymphocyte/blast window (R1 in C), whereas 0.1% are CD38 negative. (C) Forward and side scatter plot of Lin− cells showing the live gate (R1) used for flow cytometry. (D) Dot plot showing that Lin− cells are highly enriched for both CD34+ and CD34+ CD38− cells. (E) Histogram of CD34 expression on Lin− cells. R2 represents the flow cytometry sorting gate used to purify CD34+ cells. (F) Histogram of CD38 expression on Lin− cells. R3 and R4 represent the gates used to sort CD38− and CD38+ cells. (G) Histogram of CD34 expression on Lin+ cells. R5 indicates the gate used to select CD34− cells (H and I). Flow cytometric reanalysis of CD34+ CD38+ (H), CD34+ CD38− (I), and CD34− (J) sorted cell fractions. Purities are given as a percentage of the cells in the particular lymphocyte/blast window.
Transplantation of Purified Cells into NOD/SCID Mice.
Eight-week-old NOD/LtSz-scid/scid (NOD/SCID) mice were bred from breeding pairs originally obtained from L. Shultz (The Jackson Laboratory) and maintained in the defined flora animal facility located at the Ontario Cancer Institute. All animals were handled under sterile conditions and maintained under microisolators. Purified cell populations at the indicated dose were transplanted by tail-vein injection into sublethally irradiated mice (375 cGy from a 137Cs γ-irradiator) according to our standard protocol as described (16, 17). Mice transplanted with purified CD34+ CD38− cells were cotransplanted with nonrepopulating CD34− cells as accessory cells and/or received alternate-day intraperitoneal injections of human cytokines (10 μg of human SCF, 6 μg of human interleukin 3, and 6 μg of human granulocyte–macrophage colony-stimulating factor, from Amgen, Thousand Oaks, CA). Mice were killed 4–9 weeks after transplantation, and the BM from the femurs, tibiae, and iliac crests of each mouse were flushed into IMDM containing 10% FCS.
Analysis of Human Cell Engraftment in Transplanted Mice.
Genomic DNA was isolated from the BM of transplanted mice by standard extraction protocols (16, 17). EcoRI-digested DNA was separated by agarose gel electrophoresis, transferred onto a positively charged nylon membrane, and probed with a labeled human chromosome 17-specific α-satellite probe (p17H8). The level of human cell engraftment was determined by comparing the characteristic 2.7-kb band with those of human/mouse DNA mixtures as controls (limit of detection 0.05% human DNA). The presence of human progenitors in the BM of transplanted mice was determined by plating BM cells in methylcellulose cultures under conditions that are selective for the growth of human cells (17). The number of colonies scored at 12–15 days correlated with the percentage of human cells obtained from DNA analysis.
Statistical Analysis.
For limiting dilution analysis, a transplanted mouse was scored as positive (engrafted) if any human cells were detected in the murine BM by Southern blot analysis. The data from several limiting dilution experiments were pooled and analyzed by applying Poisson statistics to the single-hit model. The major assumptions of this model are that transplantation of only one cell of only one cell type (i.e., SRC) is required for a positive response (an engrafted mouse) and that every transplanted SRC will generate a positive response (21). The frequency of SRCs in each cell source was calculated with the maximum likelihood estimator (21).
Flow Cytometric Analysis of Murine BM.
To prepare cells for flow cytometry, contaminating red cells were lysed with a 6% ammonium chloride solution and the remaining cells were washed in PBS containing 5% FCS. Approximately 106 cells were resuspended in 1 ml of PBS/5% FCS containing 5% human serum (to block Fc receptors) for 30 min at 4°C, washed, and then incubated with monoclonal antibodies at a concentration of 5 μg/ml for 30 min at 4°C. The antibody combinations used are indicated in Fig. 3 (CD45 was conjugated to PerCP; CD34, CD14, CD20, sIgM, and CD15 were conjugated to FITC; and CD38, CD33, CD19, and CD13 were conjugated to PE). CD45, CD34, and CD38 antibodies were purchased from Becton Dickinson; all other antibodies were obtained from Coulter. Cells were then washed three times in PBS/5% FCS and analyzed on a FACScan. For each mouse analyzed, an aliquot of cells was also stained with mouse IgG conjugated to FITC, PE, and PerCP as an isotype control. Bone marrow cells from an untransplanted NOD/SCID were stained in parallel as an additional negative control. Fluorescence levels excluding greater than 99% of the cells in these negative controls were considered to be positive and specific for human staining.
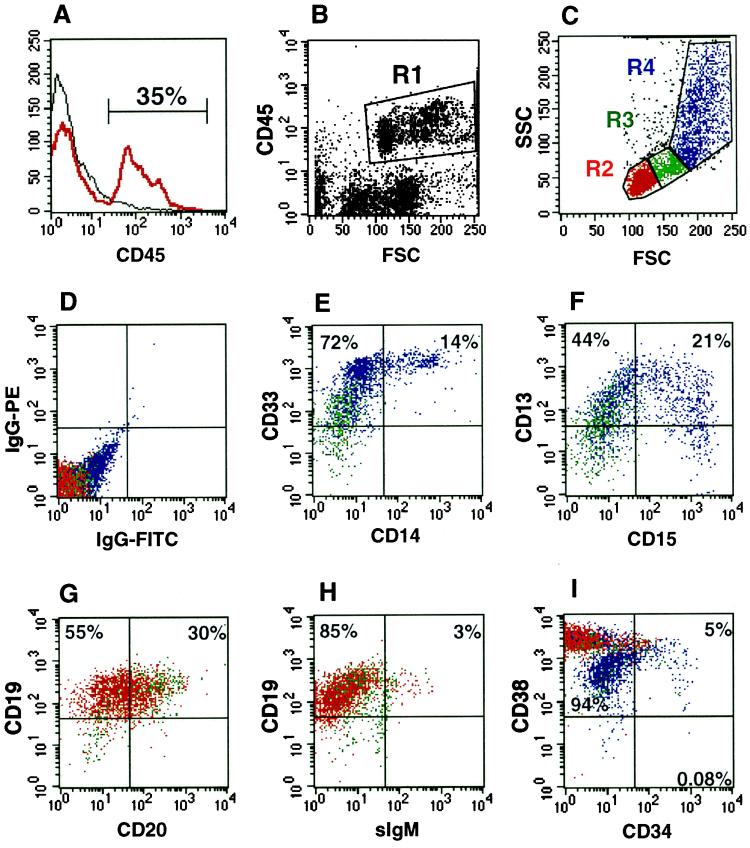
Figure 3.
Multilineage differentiation of human CD34+ CD38− cells in NOD/SCID mice. Bone marrow from a highly engrafted mouse transplanted with 5,000 CD34+ CD38− CB cells was stained with various human-specific monoclonal antibodies and analyzed by flow cytometry. (A) Histogram of CD45 (pan-leukocyte marker) expression indicating that 35% of the cells present in the murine bone marrow are human. (B) Analysis of lineage markers was done on cells within gate R1 (CD45+). (C) Cells in R1 were further gated based on forward and side scatter properties into lymphoid (R2), blast (R3) and myeloid (R4) windows. (D) Isotype control for nonspecific IgG staining of PE and FITC fluorescence. (E–I) Expression of the following markers is shown: E, myeloid marker CD33 and monocytic marker CD14 (R3 and R4); F, myeloid marker CD13 and mature granulocyte marker CD15 (R3 and R4); G, pan-B cell markers CD19 and CD20 (R2 and R3); H, CD19 and mature B cell marker sIgM (R2 and R3); I, CD38 and the immature hematopoietic marker CD34 on cells from R2, R3, and R4.
RESULTS
Purification of Cell Populations from Human CB and BM.
Fig. 1A shows the strategy used to obtain purified populations of CD34+ CD38−, CD34+ CD38+, and CD34− cells. The first step in the purification procedure was to enrich for cells that do not express lineage-specific antigens by magnetic depletion of lineage committed cells stained with a mixture of 15 monoclonal antibodies directed against a variety of lymphoid, myeloid, and erythroid antigens. The Lin− and Lin+ fractions were further purified by flow cytometry. Fig. 1 B–J shows the results of an analysis of the purified populations from a representative CB sample. Fig. 1B is the analysis of the unseparated CB cells. Consistent with other studies, the CD34+ fraction represents only 0.6% (0.5% CD34+ CD38+ plus 0.1% CD34+ CD38−) of the population in R1. Analysis of CD34 and CD38 expression on cells in the lymphocyte/blast window R1 (Fig. 1C) demonstrates a 45- and 120-fold enrichment of the CD34+ CD38+ and CD34+ CD38− populations in the Lin− fraction, respectively (Fig. 1D). The sorting gates were set to obtain highly purified populations of CD34+ CD38−, CD34+ CD38+, and CD34− cells with minimal contamination (Fig. 1 E–G). Reanalysis of sorted cell fractions demonstrated the extremely high level of purity (>98%) of the cell populations obtained in this experiment. Purity ranges for the 16 blood samples used in this study were 94–100% for CD34+ CD38+ (Fig. 1H), 84–100% for CD34+ CD38− (Fig. 1I), and 99–100% for CD34− (Fig. 1J) populations.
SRC Are Exclusively CD34+ CD38−.
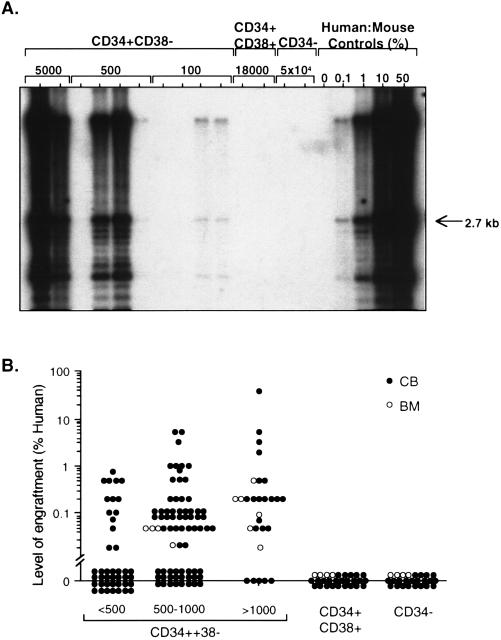
To determine which fractions contained SRC activity, sorted cells were transplanted into NOD/SCID mice and the level of human cell engraftment was determined by Southern blot analysis 4–8 weeks later. Fig. 2A shows the results of a representative experiment using one CB donor. Engraftment was detected only after transplantation with CD34+ CD38− cells. Mice transplanted with CD34+ CD38+ cells were not engrafted, even though up to 180-fold more cells than the minimum CD34+ CD38− cell dose were transplanted. Mice that received 500-fold more CD34− cells compared with CD34+ CD38− cells also were not engrafted. As few as 100 CD34+ CD38− cells were sufficient to engraft a portion of the transplanted mice to low levels (0.1%). There appeared to be a dose-response relationship as mice that received 500 cells contained 1% human cells and those transplanted with 5,000 cells had high levels of engraftment (>10%). Fig. 2B summarizes the results of transplanting purified populations from 14 CB and 2 BM samples. The repopulating cells were exclusively in the CD34+ CD38− fraction; cells from the CD34+ CD38+ or the CD34− fraction never contained repopulating activity even at doses of 60,000 or 400,000 cells, respectively. We have also demonstrated that up to 2.6 million CD34− cells directly sorted from CB cells had no repopulating capacity (data not shown).
Figure 2.
Human SRCs are exclusively CD34+CD38−. (A) Representative Southern blot analysis of individual NOD/SCID mice transplanted with CD34+ CD38−, CD34+ CD38+, and CD34− cells from CB. DNA was extracted from the murine bone marrow 8 weeks after the transplant and hybridized with a human chromosome 17-specific α-satellite probe. (B) Summary of the level of human cell engraftment in the BM of mice transplanted with purified cell fractions from 14 CB (solid circles) and two BM (open circles) samples. Numbers indicate the dose of CD34+ CD38− cells transplanted. Up to 60,000 CD34+ CD38+ cells and 400,000 CD34− cells were transplanted.
Through limiting dilution analysis, we have established that the NOD/SCID model provides a quantitative assay for SRCs (27). To determine the frequency of SRCs in the purified CD34+ CD38− fraction, Poisson statistics were used to analyze the data from mice transplanted over a dose range that resulted in no engraftment in a fraction of the mice. The pooled data from 11 experiments are given in Table 1. The frequency of SRCs as determined by the maximum likelihood estimator, is 1 SRC in 617 CD34+ CD38− cells (95% confidence interval; 1 in 341 to 1 in 1,115). This represents a 1,500-fold enrichment compared with unseparated CB, which has a frequency of 1 SRC in 9.3 × 105 cells (27).
Table 1.
Determination of the frequency of SRC in the CD34+ CD38− population by limiting dilution analysis
| Cell dose | n positive mice | n negative mice | n total | % negative |
|---|---|---|---|---|
| 1,000 | 18 | 8 | 26 | 31 |
| 575 | 21 | 13 | 34 | 38 |
| 100 | 7 | 21 | 28 | 75 |
| 50 | 3 | 8 | 11 | 73 |
NOD/SCID mice (n = 99) were transplanted with various doses of CD34+ CD38− CB cells (range, 50–1,000 cells) with cotransplantation of CD34− cells as accessory cells and/or intraperitoneal administration of growth factors. Murine BM was analyzed after 6–9 weeks. Mice were considered to be engrafted if a band was detectable by Southern blot analysis of the extracted DNA followed by hybridization with a human-specific probe (limit of detection, approximately 0.05% human cells). Poisson statistics were applied to the single-hit model, and the frequency of SRCs, calculated with the maximum likelihood estimator, was 1 in 617 CD34+ CD38− cells (95% confidence limits; 1 in 341 to 1 in 1,115).
Phenotypic Analysis of the BM of Mice Transplanted with Highly Purified CD34+ CD38− cells.
The differentiative and proliferative capacity of the highly purified SRCs were assessed by flow cytometric analysis of engrafted NOD/SCID mice 8 weeks after the transplant. The representative analysis of a NOD/SCID mouse transplanted with 5,000 CD34+ CD38− cells is shown in Fig. 3. The BM of this mouse contained 35% CD45+ human cells (Fig. 3A). CD45 is a human specific pan-leukocyte marker. Human CD45+ cells with medium to high forward scatter (region R1 in Fig. 3B) were further gated based on characteristic forward and side scatter properties into lymphoid (R2), blast (R3), and myeloid (R4) windows as shown in Fig. 3C. The isotype control is shown in Fig. 3D. Fig. 3E demonstrates the presence of mature monocytes (CD14+) among myeloid cells (CD33+), and staining for CD13 and CD15 (Fig. 3F) demonstrates granulocytic differentiation. B lymphoid cells were also present in the murine BM as shown by staining for CD19 and CD20 (Fig. 3G). The presence of a small number of cells positive for sIgM illustrates the capacity of these cells to differentiate into mature B cells (Fig. 3H). In addition to differentiated human cells, a large proportion of CD34+ cells (5%) were detected in the R1 fraction, along with CD34+ CD38− cells (0.08%) (Fig. 3I), providing evidence that immature cells are produced and maintained in the murine BM. This engraftment pattern of mice transplanted with purified CD34+ CD38− cells is similar to what is observed after transplantation of unsorted CB cells (16).
DISCUSSION
Many clinical applications have been developed to target human hematopoietic stem cells (e.g., gene transfer, purging, purification, ex vivo expansion, and CB transplantation). However, progress to develop these applications has been impeded by the absence of an assay that detects the repopulating activity of primitive human cells. The SRC assay we have developed provides a foundation for gaining a better understanding of human repopulating stem cells, enabling analysis of the proliferative, differentiative, and self-renewal capacity of these cells in vivo. We have extensively purified and characterized SRCs based on the expression of cell surface markers. The SRCs were found exclusively in the CD34+ CD38− cell fraction; in contrast, many studies have shown that CFCs and LTC-ICs are also found in the CD34+ CD38+ fraction (9, 13). Thus, these data support the conclusions of our recent gene marking study (19) and provide further evidence that the majority of SRCs are unique and more primitive than cells detected with in vitro assays. Recent studies have suggested that some murine stem cells do not express the CD34 antigen (22). The fact that we were unable to repopulate mice with even 2.6 × 106 CD34− cells suggests that the phenotype of primitive human cells may be different from murine stem cells.
Through limiting dilution analysis, we were able to quantitate SRCs and have determined that the SRCs were enriched by 1,500-fold in the CD34+ CD38− fraction from CB resulting in a frequency of 1 SRC in 617 cells. Although the number of BM samples tested was too small to precisely determine the frequency of SRCs, the number of transplanted CD34+ CD38− cells purified from BM that were capable of engrafting NOD/SCID mice was similar to that for CB, suggesting that the frequency of the SRCs in purified CB and BM is comparable. The calculated frequency of SRCs in purified CB cells was not corrected for seeding efficiency, so the actual frequency could be even higher in the CD34+ CD38− fraction. Since this fraction is not homogeneous for SRCs, additional techniques need to be employed to purify the cells to homogeneity. These studies provide the basis to incorporate into purification strategies additional techniques that assess other cellular properties such as cell cycle status, cell size, and metabolic status.
Two key criteria that define stem cells are extensive proliferation and multilineage differentiation. A crude calculation can be used to determine the proliferative potential of the engrafting cells when transplanted into NOD/SCID mice at limiting dose (1 SRC). Approximately 80 million mononuclear cells are present in murine BM. The BM from engrafted mice transplanted at limiting doses contained between 0.05 and 0.5% human cells, indicating there must be at least 4 to 40 × 104 human cells present in the BM of engrafted mice that originated from the single transplanted SRC. This result implies that a single SRC has an enormous proliferative capacity. Progenitor and flow cytometric analysis of human cells in the BM of NOD/SCID mice transplanted with these highly purified cells revealed multilineage differentiation, similar to what is observed in mice transplanted with unseparated CB (16). Mice transplanted with purified cells also contained a large proportion of CD34+ cells, indicating that the murine microenvironment supports the production and maintenance of primitive cells. Thus, these data provide evidence of the extensive proliferative capacity of the engrafting cells and of their ability to give rise to both immature cells and cells committed to the erythroid, myeloid, and B cell lineages.
Mice transplanted with low numbers of highly purified CD34+ CD38− cells were cotransplanted with CD34− accessory cells, which themselves are devoid of repopulating capacity (Fig. 2). In an independent study, we have found that accessory cells, treatment with growth factors, or both are required to achieve engraftment with this highly purified cell population (unpublished results). Further studies will determine the exact phenotype of these accessory cells.
Due to potentially important differences in the sorting criteria used, it is somewhat difficult to compare cell purification studies from different laboratories that are based on analysis of cell surface marker expression. The sorting gates used in this study are similar to those reported by Hao et al. (9, 23) who demonstrated that the CD34+ CD38− cells were almost exclusively quiescent and that the CD34+ CD38+ cells were virtually all cycling. In addition, they found that ELTC-ICs, like SRCs, were highly enriched in the CD34+ CD38− fraction; a more recent study demonstrates that this population is refractory to retroviral infection (23). These data suggest that ELTC-ICs may be closely related to SRCs.
Definitive proof of the self-renewal capacity and differentiation potential of a single SRC requires the use of techniques such as retrovirus-mediated gene transfer to mark the cell in vitro and then to track the fate of that cell in both primary and secondary transplantation recipients. However, recent gene marking experiments have indicated that SRCs and pluripotent BNX repopulating cells are inefficiently transduced (19, 24). This poses a serious barrier to clinical gene therapy. The highly purified SRCs obtained by our sorting strategy can be used to determine the basis of this barrier to retrovirus infection and to develop methods to transduce them more efficiently during ex vivo culture. Moreover, the availability of highly purified normal SRCs provides a means to undertake molecular analysis of genes involved in self-renewal and differentiation processes (25). Comparisons can also be made between normal SRCs and leukemic stem cells to identify the genes involved in leukemic progression.
Acknowledgments
We thank I. McNiece at Amgen for cytokines, L. McWhirter for providing CB specimens, N. Jamal and H. Messner for providing BM samples, and members of the laboratory for critically reviewing the manuscript. This work was supported by grants to J.E.D. from the Medical Research Council of Canada (MRC), the National Cancer Institute of Canada (NCIC) with funds from the Canadian Cancer Society, the Canadian Genetic Diseases Network of the National Centers of Excellence; a Research Scientist award from the NCIC (J.E.D.); an MRC Scientist award (J.E.D.); postdoctoral fellowships from the NCIC (M.B.); the Leukemia Research Fund of Canada and the MRC (J.C.Y.W); the Deutsche Krebshilfe (U.K.); and the Human Frontier Science Organization Program and the French Cancer Research Association (D.B.).
ABBREVIATIONS
- NOD/SCID
nonobese diabetic/severe combined immunodeficient
- SRC
SCID-repopulating cell
- LTC-IC
long-term culture-initiating cell
- ELTC-IC
extended LTC-IC
- CFC
colony-forming cell
- BM
bone marrow
- CB
cord blood
- FCS
fetal calf serum
- FITC
fluorescein isothiocyanate
- PE
phycoerythrin
References
- 1.Morrison S J, Uchida N, Weissman I L. Annu Rev Cell Dev Biol. 1995;11:35–71. doi: 10.1146/annurev.cb.11.110195.000343. [DOI] [PubMed] [Google Scholar]
- 2.Ogawa M. Blood. 1993;81:2844–2853. [PubMed] [Google Scholar]
- 3.Orlic D, Bodine D M. Blood. 1994;84:3991–3994. [PubMed] [Google Scholar]
- 4.Verfaillie C M, Miller W J, Boylan K, McGlave P B. Blood. 1992;79:1003–1010. [PubMed] [Google Scholar]
- 5.Kohn D B. Curr Opin Pediatr. 1995;7:56–63. doi: 10.1097/00008480-199502000-00011. [DOI] [PubMed] [Google Scholar]
- 6.Williams D A. Blood. 1993;81:3169–3172. [PubMed] [Google Scholar]
- 7.Broxmeyer H E. Curr Opin Pediatr. 1995;7:47–55. doi: 10.1097/00008480-199502000-00010. [DOI] [PubMed] [Google Scholar]
- 8.Eaves C J, Sutherland H J, Udomsakdi C, Lansdorp P M, Szilvassy S J, Fraser C C, Humphries R K, Barnett M J, Phillips G L, Eaves A C. Blood Cells. 1992;18:301–307. [PubMed] [Google Scholar]
- 9.Hao Q L, Shah A J, Thiemann F T, Smogorzewska E M, Crooks G M. Blood. 1995;86:3745–3753. [PubMed] [Google Scholar]
- 10.Link H, Arseniev L, Bahre O, Kadar J G, Diedrich H, Poliwoda H. Blood. 1996;87:4903–4909. [PubMed] [Google Scholar]
- 11.Craig W, Kay R, Cutler R B, Lansdorp P M. J Exp Med. 1993;177:1331–1342. doi: 10.1084/jem.177.5.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang S, Terstappen W M M. Blood. 1994;83:1515–1526. [PubMed] [Google Scholar]
- 13.Terstappen L W W M, Huang S, Safford M, Lansdorp P M, Loken M R. Blood. 1991;77:1218–1227. [PubMed] [Google Scholar]
- 14.Shah A J, Smogorzewska E M, Hannum C, Crooks G M. Blood. 1996;87:3563–3570. [PubMed] [Google Scholar]
- 15.Leemhuis T, Yoder M C, Grigsby S, Aguero B, Eder P, Srour E F. Exp Hematol. 1996;24:1215–1224. [PubMed] [Google Scholar]
- 16.Vormoor J, Lapidot T, Pflumio F, Risdon G, Patterson B, Broxmeyer H E, Dick J E. Blood. 1994;83:2489–2497. [PubMed] [Google Scholar]
- 17.Lapidot T, Pflumio F, Doedens M, Murdoch B, Williams D E, Dick J E. Science. 1992;255:1137–1141. doi: 10.1126/science.1372131. [DOI] [PubMed] [Google Scholar]
- 18.Larochelle A, Vormoor J, Lapidot T, Sher G, Furukawa T, Li Q, Shultz L, Oliveri N F, Stamatoyannopoulos G, Dick J E. Hum Mol Genet. 1995;4:163–172. doi: 10.1093/hmg/4.2.163. [DOI] [PubMed] [Google Scholar]
- 19.Larochelle A, Vormoor J, Hanenberg H, Wang J C Y, Bhatia M, Lapidot T, Moritz T, Murdoch B, Xiao L X, Kato I, Williams D A, Dick J. Nat Med. 1996;2:1329–1337. doi: 10.1038/nm1296-1329. [DOI] [PubMed] [Google Scholar]
- 20.Dick J E. Sem Immunol. 1996;8:197–206. doi: 10.1006/smim.1996.0025. [DOI] [PubMed] [Google Scholar]
- 21.Taswell C. J Immunol. 1981;126:1614–1619. [PubMed] [Google Scholar]
- 22.Osawa M, Hanada K, Hamada H, Nakauchi H. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 23.Hao Q, Thiemann F T, Peterson D, Smogorzewska E M, Crooks G M. Blood. 1996;88:3306–3313. [PubMed] [Google Scholar]
- 24.Nolta J A, Hanley M B, Kohn D B. Blood. 1994;83:3041–3051. [PubMed] [Google Scholar]
- 25.Brady G, Billia F, Knox J, Hoang T, Kirsch I R, Voura E B, Hawley R G, Cumming R, Buchwald M, Siminovitch K, Miyamoto N, Boehmelt G, Iscove N N. Curr Biol. 1995;5:909–922. doi: 10.1016/S0960-9822(95)00181-3. [DOI] [PubMed] [Google Scholar]
- 26.Cashman, J. D., Lipidot, T., Wang, J. C. Y., Doedens, M., Shultz, L. D., Lansdorp, P., Dick, J. E. & Eaves, C. J. (1997) Blood, in press. [PubMed]
- 27.Wang, J. C. Y., Doedens, M. & Dick, J. E., (1997) Blood, in press. [PubMed]