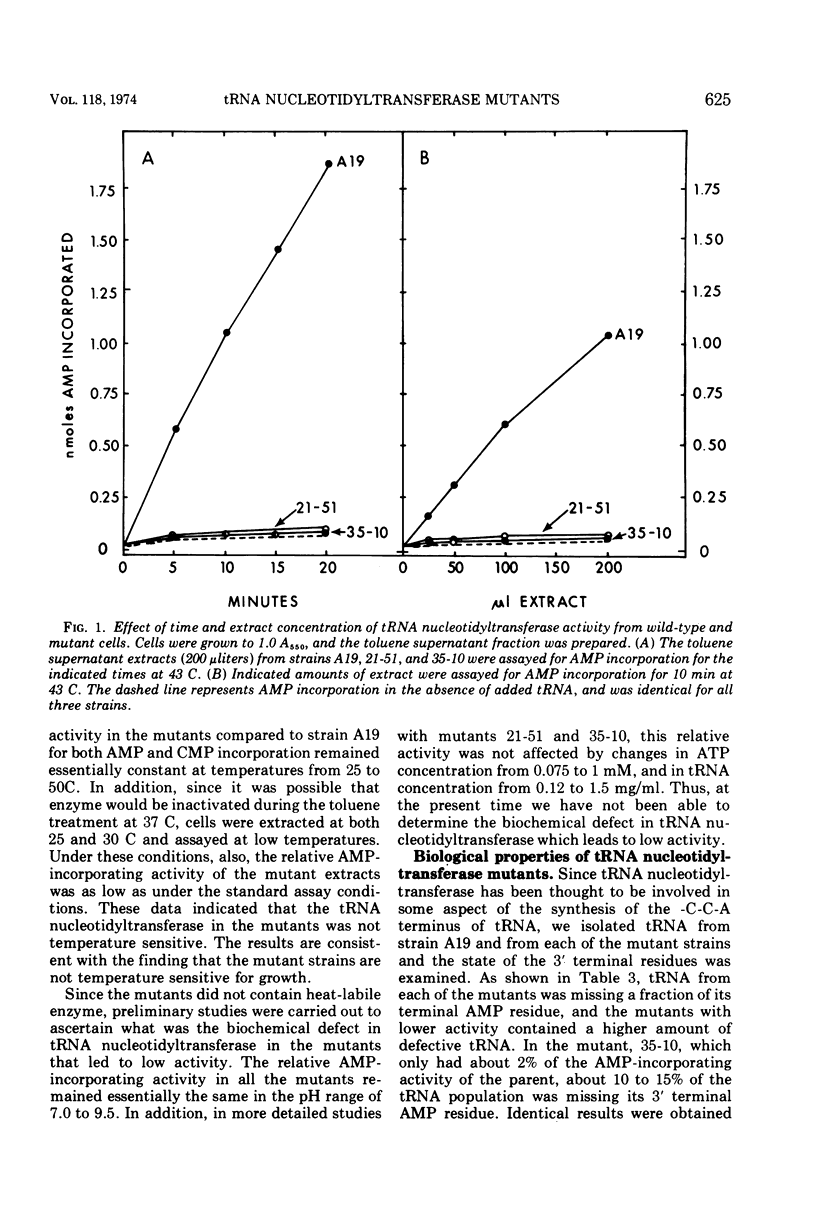
Abstract
To determine the function of the enzyme transfer ribonucleic acid (tRNA) nucleotidyltransferase in vivo, five mutants of Escherichia coli containing low levels of this enzyme were isolated. Since no selection procedure for such mutants existed, these strains were isolated by assay of large numbers of colonies from a heavily mutagenized stock. A procedure employing cells made permeable to tRNA and ATP was used to screen the large number of colonies required for the isolation. All the mutants contained less than 20% of the normal level of the AMP-incorporating activity of tRNA nucleotidyltransferase in extracts prepared by several methods, and the best mutant contained only about 2% of this activity. Three of the mutants also had equally low levels of the cytidine 5′-monophosphate-incorporating activity of the enzyme. Despite these low activities, the mutant strains displayed relatively normal growth characteristics at all temperatures examined. The enzyme in the mutant strains was not temperature sensitive, nor were any other abnormal biochemical properties detected. tRNA isolated from the mutant strains was missing significant amounts of its 3′ terminal adenosine 5′-monophosphate residue, amounting to 10 to 15% in the best mutant. However, only small amounts of the terminal cytidine 5′-monophosphate residue were missing. The results indicate that tRNA nucleotidyltransferase is involved in some aspect of synthesis or repair of the 3′ terminus of tRNA, and that the enzyme is present in large excess over its requirements for this function.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S., Smith J. D. Tyrosine tRNA precursor molecule polynucleotide sequence. Nat New Biol. 1971 Sep 8;233(36):35–39. doi: 10.1038/newbio233035a0. [DOI] [PubMed] [Google Scholar]
- Best A. N., Novelli G. D. Studies with tRNA adenylyl(cytidylyl)transferase from Escherichia coli B. II. Regulation of AMP and CMP incorporation into tRNApCpC and tRNApC. Arch Biochem Biophys. 1971 Feb;142(2):539–547. doi: 10.1016/0003-9861(71)90517-0. [DOI] [PubMed] [Google Scholar]
- Daniel V., Sarid S., Littauer U. Z. Bacteriophage induced transfer RNA in Escherichia coli. New transfer RNA molecules are synthesized on the bacteriophage genome. Science. 1970 Mar 27;167(3926):1682–1688. doi: 10.1126/science.167.3926.1682. [DOI] [PubMed] [Google Scholar]
- Daniel V., Sarid S., Littauer U. Z. Coding by T4 phage DNA of soluble RNA containing pseudouridylic acid. Proc Natl Acad Sci U S A. 1968 Aug;60(4):1403–1409. doi: 10.1073/pnas.60.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher M. P. Reactions at the 3' terminus of transfer ribonucleic acid. A single enzyme catalyzes the incorporation of adenosine monophosphate and cytidine monophosphate into transfer ribonucleic acid. J Biol Chem. 1970 Aug 25;245(16):4225–4227. [PubMed] [Google Scholar]
- Deutscher M. P. Reactions at the 3' terminus of transfer ribonucleic aid. IV. Extents of normal and anomalous nucleotide incorporation catalyzed by transfer ribonucleic acid nucleotidyltransferase. J Biol Chem. 1972 Jan 25;247(2):469–480. [PubMed] [Google Scholar]
- Deutscher M. P. Synthesis and functions of the -C-C-A terminus of transfer RNA. Prog Nucleic Acid Res Mol Biol. 1973;13:51–92. doi: 10.1016/s0079-6603(08)60100-2. [DOI] [PubMed] [Google Scholar]
- FRANKLIN R. M. THE INHIBITION OF RIBONUCLEIC ACID SYNTHESIS IN MAMMALIAN CELLS BY ACTINOMYCIN D. Biochim Biophys Acta. 1963 Aug 20;72:555–565. [PubMed] [Google Scholar]
- Gesteland R. F. Isolation and characterization of ribonuclease I mutants of Escherichia coli. J Mol Biol. 1966 Mar;16(1):67–84. doi: 10.1016/s0022-2836(66)80263-2. [DOI] [PubMed] [Google Scholar]
- Hilderman R. H., Deutscher M. P. Reactions at the 3' terminus of transfer ribonucleic acid. IX. tRNA nucleotidyltransferase activity in bacteriophage T4-infected Escherichia coli. Biochem Biophys Res Commun. 1973 Sep 5;54(1):205–215. doi: 10.1016/0006-291x(73)90909-1. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Merits I. Actinomycin inhibition of "soluble" ribonucleic acid synthesis in rat liver. Biochim Biophys Acta. 1965 Dec 9;108(4):578–582. doi: 10.1016/0005-2787(65)90054-7. [DOI] [PubMed] [Google Scholar]
- Milcarek C., Weiss B. Mutants of Escherichia coli with altered deoxyribonucleases. I. Isolation and characterization of mutants for exonuclease 3. J Mol Biol. 1972 Jul 21;68(2):303–318. doi: 10.1016/0022-2836(72)90215-x. [DOI] [PubMed] [Google Scholar]
- Miller J. P., Philipps G. R. Transfer ribonucleic acid nucleotidyltransferase from Escherichia coli. II. Purification, physical properties, and substrate specificity. J Biol Chem. 1971 Mar 10;246(5):1274–1279. [PubMed] [Google Scholar]
- Rosset R., Monier R. Instabilité de la séquence 3'-hydroxyle terminale du RNA de transfert chez les microorganismes. I. Renouvellement de l'AMP terminal chez Saccharomyces cerevisiae. Biochim Biophys Acta. 1965 Nov 8;108(3):376–384. [PubMed] [Google Scholar]
- Taylor A. L., Trotter C. D. Linkage map of Escherichia coli strain K-12. Bacteriol Rev. 1972 Dec;36(4):504–524. doi: 10.1128/br.36.4.504-524.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S. B., Hsu W. T., Foft J. W., Scherberg N. H. Transfer RNA coded by the T4 bacteriophage genome. Proc Natl Acad Sci U S A. 1968 Sep;61(1):114–121. doi: 10.1073/pnas.61.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. H., Kim J. S., Abelson J. N. Bacteriophage T4 transfer RNA. 3. Clustering of the genes for the T4 transfer RNA's. J Mol Biol. 1972 Nov 28;71(3):547–556. doi: 10.1016/s0022-2836(72)80022-6. [DOI] [PubMed] [Google Scholar]


