Abstract
The infectious plasmid OCT, which specifies a set of dissimilatory enzymes responsible for the degradation of n-octane, has been shown to be an aggregate of a noninfectious OCT plasmid and an infectious plasmid with sex factor activity. The infectious plasmid, which can be eliminated from the cells of Pseudomonas putida by mitomycin C treatment without loss of the OCT plasmid and vice versa, has been designated as factor K. The infectious plasmid (factor K) is not only responsible for the mobilization of OCT, but also mobilizes chromosomal genes at a frequency of 10−2 to 10−3 per donor cell. Whereas OCT is incompatible with another degradative plasmid, CAM, factor K appears to be compatible with it.
Full text
PDF
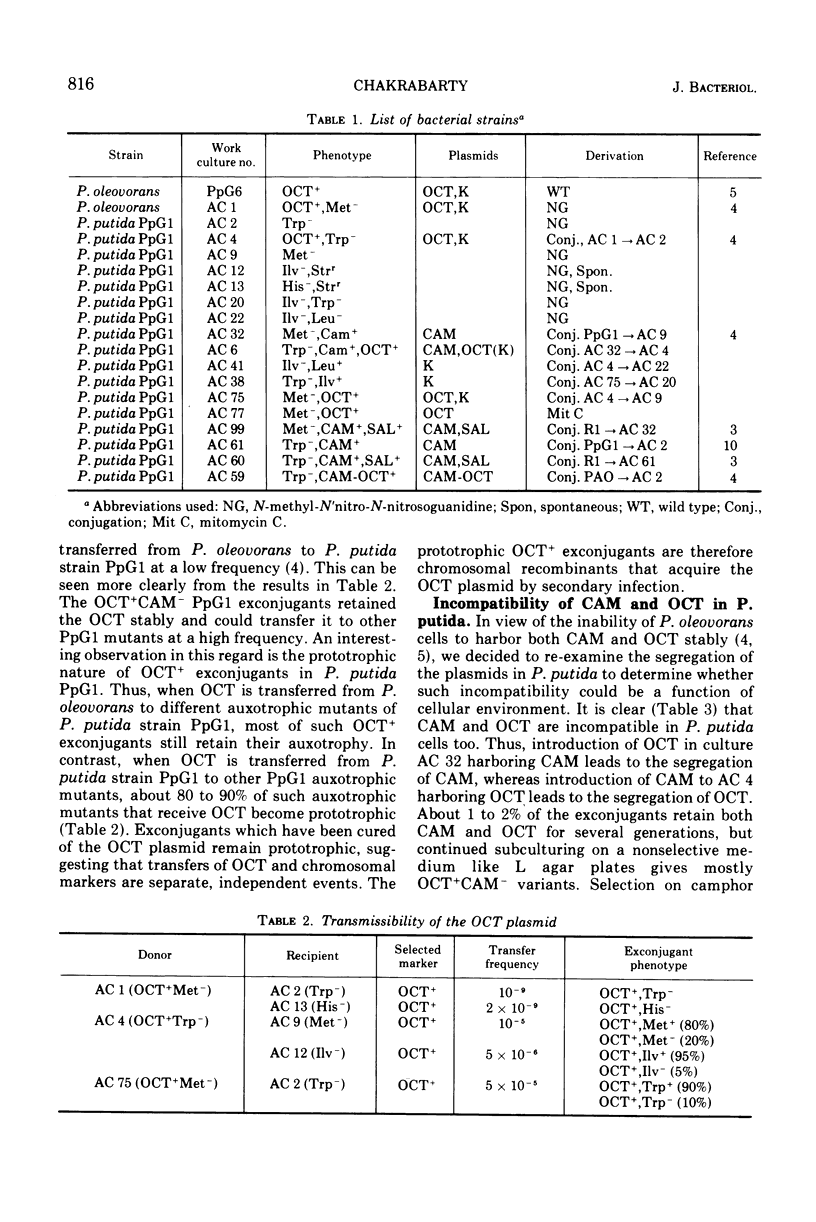
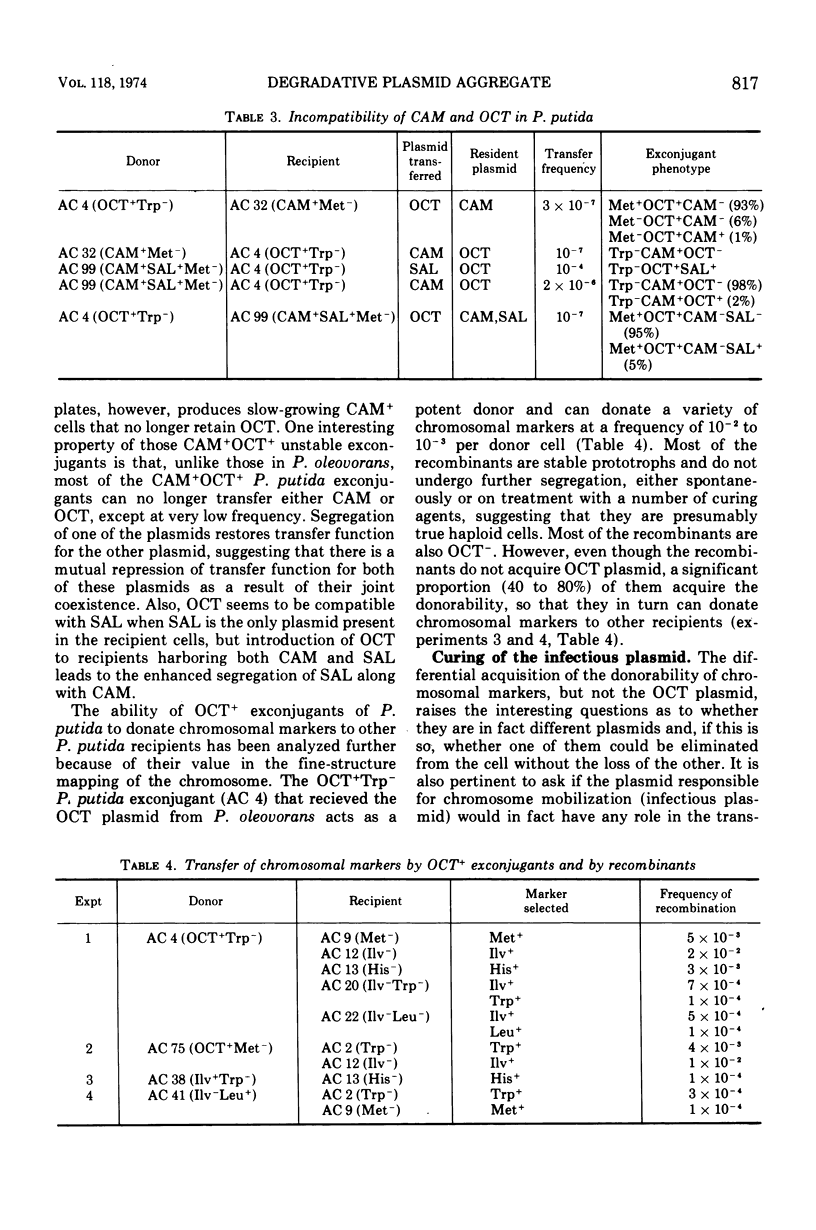



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson E. S., Lewis M. J. Characterization of a transfer factor associated with drug resistance in Salmonella typhimurium. Nature. 1965 Nov 27;208(5013):843–849. doi: 10.1038/208843a0. [DOI] [PubMed] [Google Scholar]
- Anderson E. S., Natkin E. Transduction of resistance determinants and R factors of the transfer systems by phage Plkc. Mol Gen Genet. 1972;114(3):261–265. doi: 10.1007/BF01788895. [DOI] [PubMed] [Google Scholar]
- Chakrabarty A. M., Chou G., Gunsalus I. C. Genetic regulation of octane dissimilation plasmid in Pseudomonas. Proc Natl Acad Sci U S A. 1973 Apr;70(4):1137–1140. doi: 10.1073/pnas.70.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A. M. Genetic basis of the biodegradation of salicylate in Pseudomonas. J Bacteriol. 1972 Nov;112(2):815–823. doi: 10.1128/jb.112.2.815-823.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A. M. Genetic fusion of incompatible plasmids in Pseudomonas. Proc Natl Acad Sci U S A. 1973 Jun;70(6):1641–1644. doi: 10.1073/pnas.70.6.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowes R. C. Molecular structure of bacterial plasmids. Bacteriol Rev. 1972 Sep;36(3):361–405. doi: 10.1128/br.36.3.361-405.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtiss R., 3rd Bacterial conjugation. Annu Rev Microbiol. 1969;23:69–136. doi: 10.1146/annurev.mi.23.100169.000441. [DOI] [PubMed] [Google Scholar]
- Holloway B. W., Krishnapillai V., Stanisich V. Pseudomonas genetics. Annu Rev Genet. 1971;5:425–446. doi: 10.1146/annurev.ge.05.120171.002233. [DOI] [PubMed] [Google Scholar]
- Milliken C. E., Clowes R. C. Molecular structure of an R factor, its component drug-resistance determinants and transfer factor. J Bacteriol. 1973 Feb;113(2):1026–1033. doi: 10.1128/jb.113.2.1026-1033.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinwald J. G., Chakrabarty A. M., Gunsalus I. C. A transmissible plasmid controlling camphor oxidation in Pseudomonas putida. Proc Natl Acad Sci U S A. 1973 Mar;70(3):885–889. doi: 10.1073/pnas.70.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. W., Gyles C. L. The relationship between different transmissible plasmids introduced by F into the same strain of Escherichia coli K12. J Gen Microbiol. 1970 Aug;62(3):227–285. doi: 10.1099/00221287-62-3-277. [DOI] [PubMed] [Google Scholar]


