Abstract
Exposure of human and rodent cells to a wide variety of chemoprotective compounds confers resistance against a broad set of carcinogens. For a subset of the chemoprotective compounds, protection is generated by an increase in the abundance of protective enzymes like glutathione S-transferases (GST). Antioxidant responsive elements (AREs) mediate the transcriptional induction of a battery of genes which comprise much of this chemoprotective response system. Past studies identified a necessary ARE “core” sequence of RTGACnnnGC, but this sequence alone is insufficient to mediate induction. In this study, the additional sequences necessary to define a sufficient, functional ARE are identified through systematic mutational analysis of the murine GST Ya ARE. Introduction of the newly identified necessary nucleotides into the regions flanking a nonresponsive, ARE-like, GST-Mu promoter sequence produced an inducible element. A screen of the GenBank database with the newly identified ARE consensus identified 16 genes which contained the functional ARE consensus sequence in their promoters. Included within this group was an ARE sequence from the murine ferritin-L promoter that mediated induction when tested. In an electrophoretic mobility-shift assay, the ferritin-L ARE was bound by ARE–binding protein 1, a protein previously identified as the likely mediator of the chemoprotective response. A three-level ARE classification system is presented to account for the distinct induction strengths observed in our mutagenesis studies. A model of the ARE as a composite regulatory site, where multiple transcription factors interact, is presented to account for the complex characteristics of ARE-mediated chemoprotective gene expression.
Inhibition of chemical carcinogenesis by pre-exposure of rodents to protective chemical compounds was first observed over 65 years ago (1). After the discovery of carcinogen metabolism by chemoprotector-induced cellular enzymes (2), characterization of the chemoprotective induction response intensified. Further studies demonstrated that chemoprotectors can act through the induction of enzymes which metabolize carcinogens to less reactive forms (3). Numerous compounds have been identified with a range of chemoprotective effects (4), and biochemical studies have divided these compounds into two functional classes (5). These classes of chemoprotective compounds include: monofunctional inducers, which induce enzyme activities for phase II (detoxification) enzymes; and, bifunctional inducers, which induce enzyme activities for both phase II and phase I (oxidation) enzymes (5, 6). Increased expression of glutathione S-transferases (GST), one of the prominently induced phase II enzyme activities in the chemoprotective response, confers resistance against a variety of agents (7, 8). Subsequent chemoprevention studies revealed that the induction of protective enzyme activity occurs at the transcriptional level for both the phase I cytochrome P450 IAI (9) and phase II GST-Ya genes (10). Transcriptional induction of phase I genes by bifunctional inducers is mediated by the Ah-receptor (11). While an Ah-receptor-dependent induction of phase II genes by bifunctional inducers was detected, an additional Ah-receptor-independent chemoprotective response system was also discovered (11). This system is activated by both monofunctional and a subset of bifunctional inducers (11). The Ah-receptor-dependent xenobiotic response has been well characterized (reviewed in ref. 12), culminating in the cloning of the Ah-receptor and its nuclear translocator ARNT. The second mechanism, the chemoprotective response, awaits definitive characterization of its molecular components.
In early studies of the chemoprotective response system, a wide variety of chemoprotective inducers were found to be electrophiles (6, 13). The transcriptional activation of the chemoprotective response genes by these compounds has been traced to a cis-acting transcriptional enhancer called the antioxidant responsive element (ARE) (14), or, alternatively, the electrophile response element (15). AREs have been detected in the promoters of the following genes: rat and mouse GST-Ya (14, 15), rat GST-P (16), and rat and human quinone NAD(P)H:oxidoreductase/DT-diaphorase (QR; refs. 17 and 18). A region of the murine heme oxygenase-1 promoter has also been shown to have ARE-like properties, and contains three ARE-like sequences (19). All of the AREs share a common RTGACnnnGC motif (“core sequence”) originally identified by mutagenesis of the rat GST-Ya ARE (20). Subsequent study has found that the known functional AREs contain a second core-like sequence adjacent to the primary ARE core (21). The identification of the cis-acting ARE was an initial step in the elucidation of the molecular mechanism of the chemoprotective response.
Identification of the transcriptional activator mediating the chemoprotective response through the ARE has been controversial. Early suggestions that AP-1 factors mediate the response (18, 22) have been disputed by contrary findings (19, 23–28). Recent findings have indicated that the novel 160-kDa factor ARE–binding protein 1 (ARE–BP-1) is likely to mediate the chemoprotective induction of detoxification genes through the ARE (28).
ARE core sequences have been identified in the promoter regions of additional genes postulated to be members of the chemoprotective response system (29, 30); however, when tested these ARE core sequences were not sufficient to mediate a response to chemoprotective compounds. This indicates that additional sequence information is necessary to define a functional ARE. Identification of these additional sequences is essential for (i) expanding our understanding of the ARE, (ii) identifying the chemoprotective response-mediating transcription factor, and (iii) identifying functional AREs in new promoter sequences.
In this report, we define sequences flanking the RTGACnnnGC ARE core as essential parts of a functional ARE. These essential sequences are identified in comprehensive mutagenesis studies of the murine GST-Ya ARE. Modification of a nonfunctional ARE core sequence to include the newly defined flanking nucleotides created a functional ARE that conferred inducibility to a heterologous promoter. A screen of the GenBank database with our new, functional ARE consensus sequence identified 16 genes with this ARE sequence present in their promoters. One such putative ARE, from the murine ferritin-L gene, was tested and found to be inducible in a reporter–gene system. The new ferritin-L ARE sequence is bound by ARE–BP-1, the likely chemoprotective response-mediating transcription factor. Based on mutagenesis studies, three classes of AREs are defined that exhibit distinct inducibility characteristics. A model illustrating the ARE as a composite regulatory site is presented to explain how multiple transcription factors can interact at adjacent sequences, and the complexities of the ARE-mediated chemoprotective response are discussed in light of this new model.
MATERIALS AND METHODS
Cell Culture.
HepG2 human hepatoma cells were grown in DMEM with high glucose/F-12/fetal bovine serum (4.5:4.5:1); gentimycin (GIBCO/BRL) was included at 50 μg/ml. Cells were grown at 37°C with 7% atmospheric CO2.
Transfections and Assays.
HepG2 cells at 50–70% confluence were cotransfected with the indicated TATA-Inr minimal promoter: luciferase reporter constructs [pTI-luciferase (plasmid containing minimal TATA-Inr promoter) (31)], pCH-110 (simian virus 40–β-galactosidase) as an internal standard, and pUC-19 carrier DNA by calcium phosphate coprecipitation with a 2-min glycerol shock performed 5 hr after addition of the precipitate to dishes. Sixteen hours after the glycerol shock, dishes were treated with 90 μM tert-butylhydroquinone (tBHQ; Fluka) [from a 1,000× stock in dimethyl sulfoxide (DMSO)] for 20–24 hr. Cells were harvested with trypsin and lysed, and the membranes removed by centrifugation at 13,000 × g for 10 min. Supernatants were used for luciferase assays, and the results were internally standardized relative to β-galactosidase assay results. Standard deviations were based on three transfections. The procedures for cell lysate preparation, luciferase assays, and β-galactosidase activity determination were as described (32).
Plasmid Construction.
A variety of TI-luciferase (31) based plasmids were constructed with either double-stranded oligonucleotide or PCR products inserted between the MluI and BglII sites. Inserted sequences are compiled in Table 1. For technical purposes, the multiple base pair mutations were designed to introduce unique restriction endonuclease sites.
Table 1.
Sequences of the TI-luciferase inserts and double-stranded oligonucleotide gel-mobility-shift-assay competitors
| Plasmid insert/competitor | Sequence |
|---|---|
| ARE GST-Ya | TAGCTTGGAAATGACATTGCTAATGGTGACAAAGCAACTTT |
| ARE GST-Ya 3′25 | TTGCTAATGGTGACAAAGCAACTTT |
| ARE GST-Ya 3′2 | AATGGTGACAAAGCAACTTT |
| ARE GST-Ya Core | GGTGACAAAGCA |
| ARE GST-Ya flanks mt (RF) | TctggaGcgAATctttTattatATGGTGACAAAGCAAacag |
| ARE GST-Ya GC mt | TAGCTTGGAAATGACATTatTAATGGTGACAAAatAACTTT |
| ARE QR | TCAGAGATTTCAGTCTAGAGTCACAGTGACTTGGCAAAATC |
| ARE ferritin-L | GAGCTCAGCGTGACTCAGCAGAACT |
| ARE GST-P | TCGATAGTCAGTCACTATGATTCAGCAATAAA |
| MBM mutants | See Figure 2 |
| SBM mutants | See Figure 3 |
| GST-Mu ARE-like | CAGCTTCGGTGACATAGCCTCCAT |
| GST-Mu ARE mt | CAGtaaCGGTGACATAGCaaCtAT |
| γ-GCS ARE-like | AATATGTGTTGACAGAGCAATGACCTGTC |
| AP-1/TRE | TATCGATAAGCTATGACTCATCCGGGGG |
| NF-κB/RRBE | ACTTGTACTTTCCCCAGCAGGCAGC |
γ-GCS, γ-glutamylcysteine synthetase; mt, mutant; RF, randomized flanks; TRE, TPA-responsive element; RRBE, rel-related binding element.
Oligonucleotides.
Oligonucleotides were synthesized with an instrument from Applied Biosystems.
Nuclear Extracts.
Nuclear extracts were prepared as described by Dignam et al. (33). Protein concentrations were determined by the Bradford method (Pierce).
Electrophoretic Mobility-Shift Assays.
Gel-shifts were performed according to the method of Costa et al. (34). Briefly, 3× reaction buffer [40 mM Hepes, pH 7.9/80 mM KCl/4 mM MgCl2/1 mM DTT/0.2 mM EGTA/8% Ficoll (Mr = 400,000)], 2 μg nonspecific poly(dI-dC) competitor (Sigma), as indicated double-stranded oligonucleotide specific competitors, and 9 μg of nuclear extract were mixed and incubated for 10 min at room temperature. 32P-Internally labeled, gel-purified, double-stranded DNA was added, and the mixtures were incubated for an additional 10 min. The samples were electrophoresed through a pre-run 5% native acrylamide gel in 0.5× TBE. Gels were dried and the positions of retarded protein–DNA complexes visualized with a Molecular Dynamics PhosphorImager.
RESULTS
The ARE Core Sequence Is Insufficient to Mediate Induction.
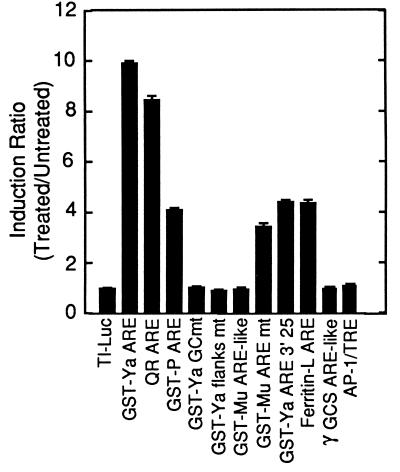
Previous studies have shown that the presence of a core sequence of RTGACnnnGC is necessary for an ARE to be functional (20, 27, 35); some recent reports, however, have indicated that this core sequence is not sufficient to mediate induction (24, 27). The insufficiency of the core sequence to mediate induction is confirmed in Fig. 1. In the context of a synthetic promoter-luciferase reporter gene plasmid (TI-luciferase), several sequences were tested. The 41-bp ARE from the murine GST-Ya promoter mediated a 10-fold induction in HepG2 cells treated with the chemoprotective response inducer tBHQ (90 μM). A similar induction level was conferred by the rat QR ARE, while the rat GST-P ARE mediated a 5-fold induction. As observed previously with the GST-Ya ARE (20, 28), mutations in the G⋅C dinucleotide portion of the core abrogated inducibility. Mutations in the regions flanking the ARE core sequence blocked induction, demonstrating that the presence of the core sequence alone is insufficient to mediate a response. An ARE-like sequence from the GST-Mu promoter (30) was not inducible, despite the presence of a perfect ARE core sequence. A core-containing ARE-like sequence from the γ-glutamylcysteine synthetase promoter (29) was also insufficient to mediate induction. Our findings demonstrate that essential cis-acting sequences are present in the sequences flanking the GST-Ya ARE core.
Figure 1.
Effect of tBHQ on luciferase production in HepG2 hepatoma cells transfected with TI-luciferase plasmids containing AREs, ARE mutants, or ARE-like sequences. The transfected plasmids carrying synthetic TI-promoter-luciferase reporter genes contained the indicated sequence in the multiple cloning site in the luciferase promoter region. The insert sequences are presented in Table 1. HepG2 cells were transfected as described with a mixture of the luciferase construct, pCH-110 (β-galactosidase reporter plasmid), and pUC19 (carrier plasmid). After 24 hr dishes of cells were treated with tBHQ (90 μM) or solvent (DMSO). Lysates were assayed for luciferase and β-galactosidase activities by standard assays. The values presented are the ratio of average luciferase activities (standardized to β-galactosidase activities) from treated versus untreated cell extracts. The averages were derived from three transfections.
Comprehensive Multiple Base Pair Mutagenesis of the GST-Ya ARE.
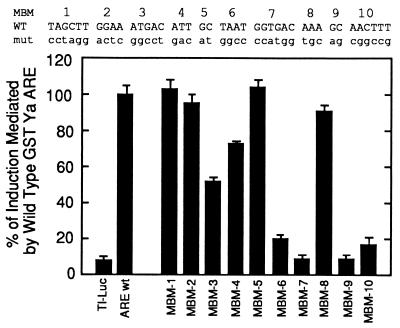
To accurately identify functional AREs in promoter sequences, it is necessary to define the ARE sequence that is sufficient to confer induction. In a first step toward identifying a sequence sufficient to mediate induction, the necessary nucleotides in the murine GST-Ya ARE were determined. To characterize the necessary regions within the murine GST-Ya ARE, 10 multiple base pair mutant AREs (MBM 1–10) were constructed which sequentially covered every base of the 41-bp element (Fig. 2). For the MBM mutants, logically grouped sets of nucleotides were substituted as shown in Fig. 2. The mutations alter the TGAC (MBM-3 and MBM-7), G⋅C dinucleotide (MBM-5 and MBM-9), or flanking sequences within the 41-bp element.
Figure 2.
MBM ARE sequences and the identification of ARE regions mediating induction by tBHQ. Ten plasmids containing ARE MBMs were constructed by insertion of the indicated sequences between the BglII and MluI sites of the TI-luciferase plasmid. Each insert contains wild-type GST-Ya ARE sequences at all positions other than those indicated for each MBM mutation. As described in Fig. 1, TI-luciferase plasmids containing the MBM inserts were transfected into HepG2 hepatoma cells. Lysates from cells treated with the chemoprotective inducer tBHQ (90 μM) or solvent (DMSO) were assayed for luciferase activities, which were standardized to β-galactosidase activity levels. The ratio of induced to uninduced expression from the GST-Ya wild-type ARE construct was defined as 100%. The experiments were performed in triplicate.
The mutations in MBM-1, MBM-2, MBM-5, and MBM-8 had no impact on induction by tBHQ (Fig. 2). The results from the MBM-2 transfection demonstrated that a putative Ets factor binding site (36) at this position is not required for induction. Similarly, the MBM-5 mutation in the 5′ G⋅C dinucleotide revealed that the G⋅C dinucleotide of the tandem core is not necessary. As expected, the MBM-9 mutation of the 3′ G⋅C dinucleotide of the primary core was not able to mediate increased expression. The 30–50% decrease in induction observed with mutants MBM-3 and MBM-4 demonstrates that the 5′ TGACnnn sequence of the tandem core performs an amplifying role. Because the MBM-3 and MBM-4 mutants still supported substantial induction, this region was not further characterized. As expected, the MBM-7 mutation that altered the primary core TGAC sequence resulted in a complete knockout of induction. Mutants MBM-6 and MBM-10, which contain alterations in the regions of the GST-Ya ARE immediately flanking the primary core sequence, were 90% reduced in induction. These data from transfection of MBMs demonstrate three distinct classes of ARE-mediated induction: (i) complete knockout of inducibility by mutations in the RTGAC and G⋅C portions of the primary core (MBM-7 and MBM-9), (ii) 90% reduction in inducibility by mutations in the sequences immediately flanking the primary core (MBM-6 and MBM-10), and (iii) 30–50% reduction in inducibility by elimination of the tandem core (MBM-3 and MBM-4).
Single Site Mutagenesis of the ARE Primary Core Flanking Regions.
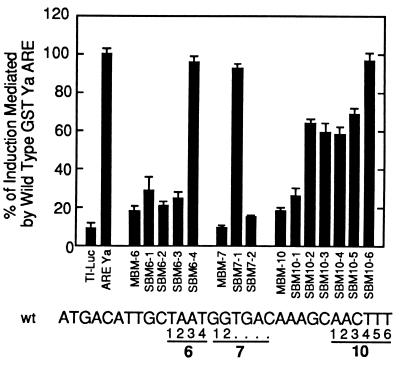
The MBM constructs demonstrated that nucleotides in the sequences flanking the primary core are necessary for induction. To identify the specific nucleotides required for inducibility, a panel of single base pair mutant ARE–TI-luciferase constructs was prepared (SBM plasmids). The individual nucleotide changes in the SBM constructs were the same as the changes in those positions in the MBM constructs (Fig. 3).
Figure 3.
Induction of reporter gene expression mediated by sequences differing by single nucleotides from the wild-type GST-Ya sequence. TI-luciferase plasmids containing SBM sequences were transfected into HepG2 cells, as described in Fig. 1. The corresponding MBM constructs were transfected for comparison. The wild-type GST-Ya ARE and parental TI-luciferase plasmids were included as controls. Lysates from cells treated with the chemoprotective inducer tBHQ (90 μM) or solvent (DMSO) were assayed for luciferase activities, which were standardized to β-galactosidase activity levels. The ratio of induced to uninduced expression from the GST-Ya wild-type ARE construct was defined as 100%. The experiments were performed in triplicate. The positions of the SBM changes are indicated on the wild-type GST-Ya ARE sequence.
Mutations within the first three SBM-6 sites dramatically reduced the chemoprotective induction response to tBHQ. The SBM6-2 (A⋅G) and SBM6-3 (A⋅C) constructs were inducible to only 10% of the wild-type induction level, and SBM6-1 (T⋅G) to only 20% of the wild-type induction. To our knowledge, this represents the first identification of these nucleotides as essential for the chemoprotective response. Also 5′ to the primary core sequence were the two SBM7 mutants. SBM7-2 (G⋅T) confirmed the purine requirement at this position (35), as substitution with a pyrimidine abrogated induction. The SBM7-1 construct supported induction equal to that mediated by the wild-type plasmid.
The set of SBM10 mutations revealed two characteristics for the region flanking the 3′ end of the primary core sequence. SBM10-1 (A⋅C) mediated only 10% of the fold-induction achieved with the wild-type ARE. This site was recently reported to be necessary in the rat QR ARE (35). Induced expression levels with the mutant plasmids SBM10-2, 10-3, 10-4, and 10-5 were 30–40% reduced relative to wild-type, indicating a preference for wild-type sequences but no single base sensitivity. The preference toward A-T base pairs in this 5-bp region (80–100% in the known AREs) will require further study.
Identification of a Consensus ARE.
In comparing the known AREs with the mutation data from Fig. 2 and Fig. 3, some positions still contained ambiguities. To develop a consensus at sites without a genetic knockout, the presence of three different nucleotides at a specific site within functional AREs was judged to indicate tolerance of any nucleotide at that position. At four positions (5-1, 6-4, 8-1, and 10-2) only two of the four possible nucleotides appeared in inducible AREs. To establish that no ambiguous base requirements exist at these positions, mutant ARE reporter plasmids SBM5-1T, SBM6-4G, SBM8-1C, and SBM10-2C were created and analyzed (data not shown). The SBM5-1, 6-4, and 8-1 mutants were all inducible, indicating no sequence requirements at these sites. The SBM10-2C mutant retained 70% of the wild-type induction, supporting a preference for A-T base pairs in the 3′ region. A recent report of purine tolerance at position 10-1 (34) was confirmed by an inducible mutant, SBM10-1G (data not shown).
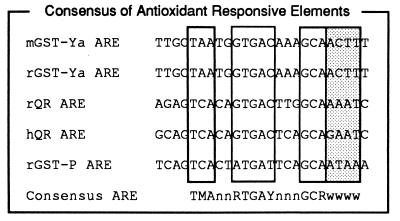
Alignment of the known AREs defines a functional ARE consensus (Fig. 4). The positions at which mutations reduced induction by at least 80% were defined as strictly as the data allowed. As shown in Fig. 4, the new consensus ARE is TMAnnRTGAYnnnGCR. A preference for A-T base pairs 3′ to the G⋅C dinucleotide in the functional AREs is observed. The inducible AREs contain 80–100% A-T content in the 5 bases 3′ to the G⋅C dinucleotide. This preference is indicated as lowercase w’s in the consensus sequence TMAnnRTGAYnnnGCRwwww.
Figure 4.
ARE consensus sequence. The known AREs from GST and QR genes are aligned. Nucleotides at essential positions (demonstrated in Fig. 3 and ref. 20) are outlined. Nucleotides at positions of sequence preference (demonstrated in Fig. 3) are shaded gray. The consensus for nucleotides at the essential and preference sites are indicated. The abbreviations follow standard IUPAC nomenclature (M = A or C, R = A or G, Y = C or T, W = A or T, S = G or C).
Creation of an Inducible ARE from an Inactive ARE-Like Sequence.
To determine if the nucleotides sufficient for induction have been defined, the inactive (Fig. 1) GST-Mu ARE-like sequence was altered to form a functional ARE. This mutant sequence (Table 1), including all of the essential nucleotides as well as a 3′ A-T content of 80%, was over 3-fold inducible (Fig. 4). This is comparable to the induction mediated by a GST-Ya ARE lacking the tandem core sequence (GST-Ya 3′25).
Identification of Consensus AREs in GenBank.
To begin to assess the abundance of AREs in mammalian genes, the primate and rodent portions of the GenBank database were screened for promoters containing the newly defined ARE consensus and a minimum 3′ A-T content of 60% in the 5 nucleotides adjoining the G⋅C dinucleotide. As expected, all of the previously described AREs in the database were identified (Table 2). The rat GST-Ya promoter region was not present in GenBank, explaining its absence from this list. Several genes previously unknown to contain consensus AREs in their promoters were identified. As a test of the relevancy of the database screen, the murine ferritin-L ARE consensus sequence was assayed for inducibility. When cloned into the promoter of the TI-luciferase reporter gene, the ferritin-L ARE conferred a 4.5-fold induction response to 90 μM tBHQ in HepG2 cells (Fig. 1). This is comparable to the GST-Ya ARE lacking the tandem core sequence. Whether the ferritin-L ARE sequence mediates induction within the context of the ferritin-L promoter will be determined in future studies.
Table 2.
Mammalian promoter sequences containing the ARE consensus sequence
| GenBank primate database
|
GenBank rodent database
|
||
|---|---|---|---|
| Gene | Accession no. | Gene | Accession no. |
| NAD(P)H:QR | M81596 | GST-Ya | X06723 |
| β-Globin | U01317 | GST-P | J04138 |
| Myoglobin | X00371 | NAD(P)H:QR | M58495 |
| Alzheimer gene STM2 | U50871 | Ferritin-L | L39879 |
| Collagenase | M16567 | β-Globin | Z13985 |
| P-450 aromatase | D21241 | GSH transporter | U16358 |
| Tyrosinase | X76647 | ||
| Interleukin 6 | M20572 | ||
A list of files in the GenBank database containing the ARE consensus sequence TMAnnRTGAYnnnGCR. Sequences found in cDNAs were eliminated, as were all matches outside promoter regions. Sequences with an A-T content below 60% in the 5 bp 3′ to the G·C dinucleotide were also eliminated. GSH, glutathione.
The Chemoprotective Induction Response Mediating Protein ARE–BP-1 Binds to the Ferritin-L ARE.
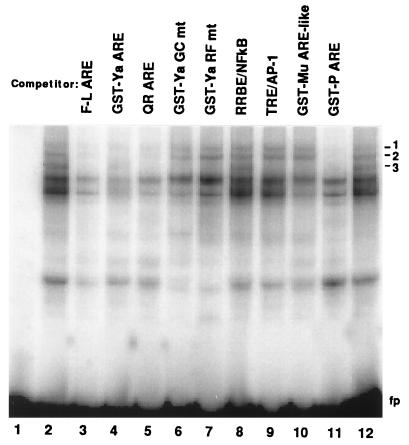
Previous studies in our laboratory identified a 160-kDa protein, ARE–BP-1, as the likely mediating factor of the chemoprotective induction response (28; unpublished data). To determine whether ARE–BP-1 or other HepG2 nuclear proteins specifically bind to the ferritin-L ARE, the newly identified ferritin-L ARE was used as a probe in a electrophoretic mobility-shift assay (Fig. 5). In the absence of specific competitors, three complexes were formed by sequence-specific binding proteins (lanes 2 and 12). Competition with the known AREs from the genes for ferritin-L (lane 3), GST-Ya (lane 4), QR (lane 5), and GST-P (lane 11) competed for all three specific bands. Oligonucleotide competitors bearing inactivating-mutations in the GST-Ya ARE failed to compete effectively for the two slowest migrating complexes (lanes 6 and 7). An NF-κB binding sequence was not competitive for any of the three specific bands (lane 8). Competitors containing consensus AP-1 binding sequences did not alter the intensities of the two slowest migrating complexes, but did compete for the third band (lanes 9 and 10), suggesting band 3 represents a basic leucine zipper (bZip) transcription factor. The competition pattern observed for the slowest migrating complexes are consistent with ARE–BP-1 specifically binding to the newly identified ferritin-L ARE.
Figure 5.
Analysis of the proteins that interact with the ARE consensus sequence from the murine ferritin-L promoter. Proteins that specifically interact with the ferritin-L ARE were determined by electrophoretic mobility-shift assay as outlined in Materials and Methods. A 32P-labeled ferritin-L ARE was mixed with nuclear extracts prepared from untreated human HepG2 hepatoma cells. Lanes: 1, no protein; 2 and 12, gel mobility shift with no specific competitors; 3–11, the indicated specific competitors were included at concentrations 200-fold greater than that of the probe. The specific binding protein–DNA complexes are numbered from the slowest to fastest mobility. Competitor sequences are listed in Table 1.
DISCUSSION
The goal of our work presented here was to identify those nucleotides that compose a functional ARE and to use this information to identify genes that compose the chemoprotective response. Our results demonstrate that (i) nucleotides outside of the previously described RTGACnnnGC primary core are essential for an inducible ARE, (ii) a nonfunctional GST-Mu ARE-like sequence is converted into an inducible ARE by applying our criteria for a functional ARE consensus, (iii) genes that contain inducible ARE sequences are identified by screening the GenBank database with our consensus sequence, and (iv) the nuclear protein ARE–BP-1 preferentially interacts with ARE sequences that support the chemoprotective induction response.
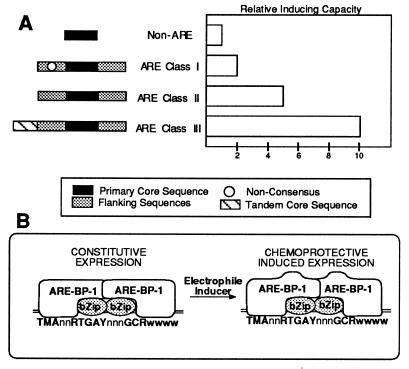
Three functional classes of ARE mutants were observed in our studies: (i) mutations in the primary core sequence that abrogated induction completely, (ii) mutations in primary core-flanking sequences that reduced induction by 90%, and (iii) mutations in the TGACnnn motif of the tandem core that reduced induction by 50%. A classification scheme for the known AREs is depicted (Fig. 6A) in which the level of ARE inducibility is predictable based upon the presence or absence of specific ARE domains. Class III AREs include both primary and tandem core sequences, match the newly defined ARE consensus, and are 5- to 10-fold inducible by 90 μM tBHQ in HepG2 cells. Class II AREs lack the tandem core, but include the primary core plus the newly defined flanking sequence consensus and are 2.5- to 5-fold inducible. While class I AREs contain the primary core and may have a tandem core, they are only weakly inducible due to nonconsensus flanking sequences. The known AREs would be classified as follows: the GST-Ya, GST-P, and QR AREs would fit into class III; the ferritin-L ARE into class II; and the SBM6-1 mutant would fit into class I. Variability within the range of induction strength is also likely to result from the A-T content of the 5 nucleotides 3′ to the G⋅C dinucleotide. Differences in induction strengths for gene products (e.g., GST isozymes) would be expected to be associated with genetic variability in ARE sequences. These differences would be expected to carry associated risks for cancer or other diseases related to damage by electrophiles or oxygen radicals.
Figure 6.
Models for ARE-mediated expression in response to chemoprotective compounds. (A) A classification system for AREs. Three distinct classes of tBHQ-inducible sequences have been established (Figs. 2 and 3). Class I sequences contain the original ARE “core” sequence (20), but differ slightly from the ARE consensus (Fig. 4). Class II AREs contain the entire consensus, but do not contain a tandem core region. Class III AREs contain the consensus and the tandem core region. (B) A composite site model consistent with the new ARE consensus and ARE binding proteins observed. A transcription factor from the leucine-zipper family is proposed to bind to the RTGAYnnn portion of the core. The chemoprotective induction response-mediating protein ARE–BP-1 interacts with the bZip proteins and binds to flanking sequences to confer the unique induction properties of the ARE.
Our studies have identified essential ARE sequences not previously determined in studies of the rat GST-Ya ARE (20), rat QR ARE (35), and human QR ARE (27). In 1991 Rushmore et al. (20) identified the TGACnnnGC primary core within the rat GST-Ya ARE by single base mutagenesis. In this landmark study, no significant changes in constitutive or induced expression levels were observed at any of the tested nucleotides outside of the primary core site. Mutation of either base in the G⋅C dinucleotide of the primary core abrogated induction but did not alter constitutive expression levels. In Favreau and Pickett’s (35) more recent mutagenesis study of the rat QR ARE, different characteristics were presented. The primary core was now extended to RTGACnnnGCA and mutations in the G⋅C dinucleotide reduced both constitutive and induced expression. Xie et al. (27) constructed multiple base mutations in the human QR ARE. Mutations within the tandem core decreased induction by 80% compared with the wild-type ARE. While the rat and human QR mutagenesis studies were originally analyzed within a “tandem core” paradigm, the data are consistent with a “flanking sequence” view. In the GST-Ya ARE, the tandem core is separated from the flanking sequences which allowed us to more clearly delineate the contributions from each region.
The new ARE consensus allows accurate identification of AREs in promoter sequences. When used as a probe against the GenBank sequence database, the new consensus identified the known AREs as well as potential AREs in 12 additional promoter sequences. Some of the potential AREs were found in genes that might plausibly be components in a detoxification response system, while others were associated with oxygen content or neurodegenerative disease. The ARE consensus sequence from the ferritin-L promoter was tested and found to be functional in a reporter system.
The range of ARE induction potencies, which result from variations in ARE nucleotide composition, allows for precise control of induced gene expression. The presence of an inducible ARE sequence in the promoter of the murine ferritin-L gene is consistent with a recent report of increased ferritin-L mRNA levels in chemoprotector-treated cells (37). The inducibility of this ferritin-L ARE sequence must be confirmed in the context of the ferritin-L promoter, but ferritin-L is a reasonable candidate gene for the chemoprotective response battery. An increased ferritin-L/ferritin-H molar ratio shifts the properties of the ferritin complex to favor the storage of iron in its center (38). This iron storage reduces the available iron in the cell and thus inhibits formation of reactive oxygen species via the Fenton reaction (39). There is a report that ferritin-H mRNA is also induced, albeit at a level below that observed for ferritin-L (37). This lower level of induction would allow for the formation of more ferritin complexes, but with the iron storage favoring high L/H ratio. A recently reported regulatory sequence in the mouse ferritin-H promoter could be a class I ARE as it matches our new consensus at all but 1 nucleotide (40). The inducibility differences between AREs of differing classes may allow for precise control of cellular iron levels.
Previous studies have generated controversy over the transcription factor(s) mediating the chemoprotective induction response. Initial observations of factors interacting with the ARE focused upon the AP-1 family of leucine-zipper transcription factors as potential mediating proteins (18, 22). A dominant role for AP-1 regulation has been refuted in recent years (19, 23, 26–28). It has been suggested that Maf/NF-E2 factors, also in the bZip family, may mediate the response (23, 41). Of interest, the β-globin promoter sites identified in the GenBank search for ARE consensus sequences (Table 2) have previously been identified as sites regulated by NF-E2 family members in a tissue specific manner (42).
Our demonstration of the necessity of core flanking sequences strongly suggests a transcription factor distinct from bZip family members is essential for a functional response. In our previous studies, the in vitro binding of ARE–BP-1 was perfectly correlated with in vivo inducibility of ARE sequences (28). The identification of the ferritin-L ARE and the binding of ARE–BP-1 in electrophoretic mobility-shift assays further implicates this 160-kDa transcription factor as a mediating protein.
The requirement for a primary core sequence and our additional 5′ and 3′ flanking sequence requirements, the extended length of these combined motifs, and the numerous proteins observed in gel shifts with an ARE probe all support the concept of the ARE as a composite regulatory element (Fig. 6B). In our model, members of the bZip transcription factor family (AP-1, NF-E2, or others) interact with the TGACnnn portion of the primary core sequence, while ARE–BP-1 binds to the G⋅C dinucleotide and flanking sequences. The binding of the ferritin-L ARE gel shift complex 3 is consistent with a bZip factor (Fig. 6B). The schematic model also allows for three classes of AREs and accounts for the observed ARE–BPs. Class III AREs would feature bZip factors binding to both the primary and tandem core sequences, while ARE–BP-1 would interact with primary core flanking sites. Class II AREs lack the tandem core and only support binding of a single bZip dimer. Class I AREs have decreased ARE–BP-1 binding strength due to lower affinity binding sites. This model is testable in future studies.
Purification and cloning of ARE–BP-1 will enable mechanistic studies to determine how chemoprotector-induced stress activates transcription through the ARE.
Acknowledgments
We thank Andrea Mast and Susan Johnston for expert assistance, and the other members of the Fahl laboratory for helpful suggestions. W.W.W. was a Cremer Scholar. This work was supported by National Institutes of Health Grant CA-22484 (W.E.F.).
ABBREVIATIONS
- ARE
antioxidant responsive element
- ARE–BP
ARE-binding protein
- bZip
basic leucine zipper
- GST
glutathione S-transferase
- tBHQ
tert-butylhydroquinone
- QR
quinone NAD(P)H:oxidoreductase/DT-diaphorase
- DMSO
dimethyl sulfoxide
- MBM
multiple base pair mutant
- SBM
single base pair mutant
- pTI-luciferase
plasmid containing minimal TATA-Inr promoter
References
- 1.Berenblum I. J Pathol Bacteriol. 1929;32:425–434. [Google Scholar]
- 2.Conney A H, Miller E C, Miller J A. Cancer Res. 1956;16:450–459. [PubMed] [Google Scholar]
- 3.Miller E C, Miller J A, Brown R R, MacDonald J C. Cancer Res. 1958;18:469–477. [PubMed] [Google Scholar]
- 4.Wattenberg L W. Cancer Res. 1985;45:1–8. [PubMed] [Google Scholar]
- 5.Prochaska H J, De Long M J, Talalay P. Proc Natl Acad Sci USA. 1985;82:8232–8236. doi: 10.1073/pnas.82.23.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talalay P, De Long M J, Prochaska H J. Proc Natl Acad Sci USA. 1988;85:8261–8265. doi: 10.1073/pnas.85.21.8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puchalski R B, Fahl W E. Proc Natl Acad Sci USA. 1990;87:2443–2447. doi: 10.1073/pnas.87.7.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gulick A M, Fahl W E. Proc Natl Acad Sci USA. 1995;92:8140–8144. doi: 10.1073/pnas.92.18.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitlock J P, Jr, Denison M S, Fisher J M, Shen E S. Mol Biol Med. 1989;6:169–178. [PubMed] [Google Scholar]
- 10.Pearson W R, Reinhart J, Sisk S C, Anderson K S, Adler P N. J Biol Chem. 1988;263:13324–13332. [PubMed] [Google Scholar]
- 11.Prochaska H J, Talalay P. Cancer Res. 1988;48:4776–4782. [PubMed] [Google Scholar]
- 12.Swanson H I, Bradfield C A. Pharmacogenetics. 1993;3:213–230. doi: 10.1097/00008571-199310000-00001. [DOI] [PubMed] [Google Scholar]
- 13.Prestera T, Holtzclaw W D, Zhang Y, Talalay P. Proc Natl Acad Sci USA. 1993;90:2965–2969. doi: 10.1073/pnas.90.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rushmore T H, King R G, Paulson K E, Pickett C B. Proc Natl Acad Sci USA. 1990;87:3826–3830. doi: 10.1073/pnas.87.10.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friling R S, Bensimon A, Tichauer T, Daniel V. Proc Natl Acad Sci USA. 1990;87:6258–6262. doi: 10.1073/pnas.87.16.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okuda A, Imagawa M, Maeda Y, Sakai M, Muramatsu M. J Biol Chem. 1989;264:16919–16926. [PubMed] [Google Scholar]
- 17.Favreau L V, Pickett C B. J Biol Chem. 1991;266:4556–4561. [PubMed] [Google Scholar]
- 18.Li Y, Jaiswal A K. J Biol Chem. 1992;267:15097–15104. [PubMed] [Google Scholar]
- 19.Prestera T, Talalay P, Alam J, Ahn Y I, Lee P J, Choi A M K. Mol Med. 1995;1:827–837. [PMC free article] [PubMed] [Google Scholar]
- 20.Rushmore T H, Morton M R, Pickett C B. J Biol Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 21.Daniel V. CRC Crit Rev Biochem Mol Biol. 1993;28:173–207. doi: 10.3109/10409239309086794. [DOI] [PubMed] [Google Scholar]
- 22.Friling R S, Bergelson S, Daniel V. Proc Natl Acad Sci USA. 1992;89:668–672. doi: 10.1073/pnas.89.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen T, Rushmore T H, Pickett C B. J Biol Chem. 1994;269:13656–13662. [PubMed] [Google Scholar]
- 24.Favreau L V, Pickett C B. J Biol Chem. 1993;268:19875–19881. [PubMed] [Google Scholar]
- 25.Yoshioka K, Deng T, Cavigelli M, Karin M. Proc Natl Acad Sci USA. 1995;92:4972–4976. doi: 10.1073/pnas.92.11.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B, Williamson G. Biochim Biophys Acta. 1994;1219:645–652. doi: 10.1016/0167-4781(94)90223-2. [DOI] [PubMed] [Google Scholar]
- 27.Xie T, Belinsky M, Xu Y, Jaiswal A K. J Biol Chem. 1995;270:6894–6900. doi: 10.1074/jbc.270.12.6894. [DOI] [PubMed] [Google Scholar]
- 28.Wasserman W W, Fahl W E. In: The Oxygen Paradox. Davies K J A, Ursini F, editors. Padova, Italy: CLEUP Press; 1995. pp. 413–424. [Google Scholar]
- 29.Mulcahy R T, Gipp J J. Biochem Biophys Res Commun. 1995;209:227–233. doi: 10.1006/bbrc.1995.1493. [DOI] [PubMed] [Google Scholar]
- 30.Reinhart J, Pearson W R. Arch Biochem Biophys. 1993;303:383–393. doi: 10.1006/abbi.1993.1299. [DOI] [PubMed] [Google Scholar]
- 31.Miltenberger R J, Cortner J, Farnham P J. J Biol Chem. 1993;268:15674–15680. [PubMed] [Google Scholar]
- 32.Jin H M, Brady M L, Fahl W E. Proc Natl Acad Sci USA. 1993;90:7563–7567. doi: 10.1073/pnas.90.16.7563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1480. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costa R H, Lai E, Grayson D R, Darnell J E., Jr Mol Cell Biol. 1988;8:81–90. doi: 10.1128/mcb.8.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Favreau L V, Pickett C B. J Biol Chem. 1995;270:24468–24474. doi: 10.1074/jbc.270.41.24468. [DOI] [PubMed] [Google Scholar]
- 36.Bergelson S, Daniel V. Biochem Biophys Res Commun. 1994;200:290–297. doi: 10.1006/bbrc.1994.1447. [DOI] [PubMed] [Google Scholar]
- 37.Primiano T, Kensler T W, Kuppusamy P, Zweier J L, Sutter T R. Carcinogensis. 1996;17:2291–2296. doi: 10.1093/carcin/17.11.2291. [DOI] [PubMed] [Google Scholar]
- 38.Levi S, Santambrogio P, Cozzi A, Rovida E, Corsi B, Tamborini E, Spada S, Albertini A, Arosio P. J Mol Biol. 1994;20:649–654. doi: 10.1006/jmbi.1994.1325. [DOI] [PubMed] [Google Scholar]
- 39.Aust S D. Toxicol Lett. 1995;82:941–944. doi: 10.1016/0378-4274(95)03605-9. [DOI] [PubMed] [Google Scholar]
- 40.Beaumont C, Seyhan A, Yachou A-K, Grandchamp B, Jones R. J Biol Chem. 1994;269:20281–20288. [PubMed] [Google Scholar]
- 41.Hayes J D, Pulford D J. CRC Crit Rev Biochem Mol Biol. 1995;30:445–500. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 42.Shivdasani R A, Orkin S H. Blood. 1996;87:4025–4039. [PubMed] [Google Scholar]